Abstract
Purpose
We investigated whether miRNA expression profiles can distinguish and predict outcome of non-small-cell lung carcinoma (NSCLC) patients with different histological subtypes.
Methods
High-throughput microarray was used to measure miRNA expression levels in six NSCLC samples. Subsequently, quantitative reverse transcription polymerase chain reaction (qRT-PCR) was used to verify findings in an independent set of 54 squamous-cell lung carcinomas (SCC), 51 lung adenocarcinomas (AD), and paired adjacent non-neoplastic lung tissue.
Results
We showed that, compared to adjacent non-neoplastic lung tissues, the expressions of miR-125a-5p and let-7e were decreased in AD and SCC samples, while increased expressions of miR-93, miR-205, and miR-221 were observed in SCC samples. In addition, miR-205 expression was significantly higher in SCC patients with lymph node metastasis. Lower let-7e expression was associated with lymph node metastasis, >3 cm tumor size, and differentiation of the NSCLC AD subtype. High levels of miR-100 expression also correlated with the AD subtype in current smokers. Moreover, induction of miR-93 and miR-205 expressions and reduction of let-7e were strongly associated with shorter overall survival in SCC patients, whereas AD patient survival was only associated with reduced let-7e.
Conclusions
We identified differential expression profiles of miRNAs in AD and SCC. More importantly, in addition to morphology and immunocytochemistry approaches, we report that miR-93, miR-205, miR-221, and let-7e may represent novel biomarkers for differential diagnosis and prognosis of certain NSCLC subtypes or be new targets of histology-specific treatments. Furthermore, our results suggest a strong correlation between high expression of miR-100 and AD patients with history of heavy smoking.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung carcinoma is the most common cause of cancer incidence and cancer-related deaths worldwide. Among the non-small-cell lung cancer (NSCLC) patients, squamous-cell lung carcinoma (SCC) and lung adenocarcinoma (AD) represent the two major subtypes. Despite improvements in early diagnosis and development of new chemo-or/and targeted therapies, the overall 5-year survival rate remains low (Miller 2005). Patients with AD and SCC are normally treated with surgical resection, followed by radiation and/or chemotherapy. However, the outcomes for the two NSCLC subtypes are different, possibly due to the unique histopathological characteristics between them.
MicroRNAs (miRNAs) are a group of small non-protein-coding RNAs that demonstrate distinctive temporal and/or spatial expression patterns. These expression profiles have been successfully used to distinguish tumors from normal tissues and from different developmental origin, and have great potential to diagnose the cytological and histological origins of tissues with unclear differentiated phenotypes (Lu et al. 2005). Recently, study of miRNA profiles in NSCLC patients has unveiled a potential new tool in predicting diagnosis and prognosis (Lu et al. 2005). However, the actual role miRNAs played in distinguishing patients with certain NSCLC subtypes has not been established; this approach is particularly promising for helping to identify novel therapeutic targets that may enhance our ability to understand and treat the two types of patients.
Studies have shown that miRNAs can be used to define some NSCLC subtypes (Bishop et al. 2010; Landi et al. 2010; Raponi et al. 2009). Yanaihara and colleagues (Yanaihara et al. 2006) identified six miRNAs (miR-205, miR-99b, miR-203, miR-202, miR-102, and miR-204) that were expressed at distinct levels in lung AD and SCC subtypes. miR-99b and miR-102 expressions, for example, were higher in AD than in SCC. Landi and colleagues (Landi et al. 2010) also reported different miRNA expression profiles in patients with AD and SCC and demonstrated their role in prognosis. For example, high levels of miR-155 and miR-146b were strongly associated with poor overall survival of SCC patients. Thus, an aberrant expression profile of miRNAs might be a significant indicator for the different subtypes of lung cancer.
In this study, we investigated the miRNA expression profile in patients with AD and SCC subtypes and examined the potential correlation of different miRNA expression to treatment outcomes and prognosis.
Materials and methods
Patients and samples
NSCLC and matched adjacent non-cancerous tissues were excised from patients undergoing surgery at the Zhoushan Hospital from January 2008 to December 2010. The tissue specimen was immediately transported to the clinical pathologic laboratory. Each sample was placed in a cryovial, flash-frozen in liquid nitrogen for 30 min, and then stored at −80 °C until use in analysis. The cytology specimens, which contained the majority of tumor cells and were used to study the expression of miRNAs, were formalin-fixed and stained with hematoxylin and eosin (H&E). All cases were reviewed by two pathologists, and the diagnosis was confirmed according to the recently published guidelines from the recent national comprehensive cancer network (NCCN). Moreover, there was no major evidence of inflammation or other underlying lung diseases in the tumor species. Figure 1a shows the standard H&E staining of SCC, while Fig. 1b shows the pathological changes in the AD subtype. The clinicopathological parameters are presented in Table 1.
Immunohistochemical morphology (H&E staining) of lung adenocarcinoma and squamous-cell lung carcinoma subtypes. Lung tumors from a SCC and b AD patients were inflated, fixed in formalin, paraffin-embedded, and sectioned for histological analysis. Slides were washed and counterstained with H&E to observe the morphological changes. Original magnification ×400
All samples were obtained with patient informed consent. The study was approved by the Hospital’s Ethical Research Committee.
RNA and miRNA isolation
Total RNA was isolated from six lung carcinomas and matched adjacent non-neoplastic tissues using the TRIzol reagent (Invitrogen, USA), according to the manufacturer’s instructions. The purified RNA was subsequently used in microarray analysis. miRNAs were extracted from 100 mg of cancer samples and adjacent non-cancerous tissues using a commercially available miRNA isolation kit (Applied Biosystems Inc. (ABI), USA) and following the manufacturer’s protocol. RNA concentration was determined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, USA).
miRNA microarray and data analysis
Three–five microgram of total RNA was labeled by ligation of an RNA linker, pCp-Cy3, to the miRNA 3′-end. Agilent microarray slides were then incubated with the labeled RNA and washed, according to the manufacturer’s instructions. Arrays were scanned using the Agilent Microarray Scanner at the fixed PMT setting of 600 and a scan resolution of 61 × 21.6 mm. A total of 509 miRNAs passed the initial screening criteria of median normalized fluorescence signal. The microarray hybridization images were analyzed using SpotReader software (Niles Scientific, USA).
Quantitation of miRNAs by quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Expression of miRNA was quantified by qRT-PCR using ABI’s human TaqMan MicroRNA Assay Kit. The reverse transcription reaction was carried out with ABI’s TaqMan MicroRNA Reverse Transcription Kit at 16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min. PCR amplification was performed using miRNAs-specific primers. The reaction mixtures were incubated in an ABI 7500 Fast Real-Time PCR system at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The cycle threshold (Ct) values were calculated by the SDS 2.0.1 software from ABI.
Sample score and statistical analysis
The differences in the expression levels detected by miRNA microarray were assessed by a two-sided paired t test within significance analysis of microarrays (SAM). The nine differentially expressed miRNAs and two small nuclear (sn) RNAs, U6 snRNA and U48 snRNA, were measured in triplicate by qRT-PCR. The average Ct for each triplicate (AvgCt miR205, AvgCt mir93, AvgCt mir221, AvgCt mir30e, AvgCt mir29b, AvgCt mirlet7e, AvgCt mir100, AvgCt mir125a-5p, AvgCt U6, and AvgCt U48) was calculated. Fold changes in gene expression were calculated by the equation for \( 2^{ - \Updelta \Updelta Ct} , \) normalized with ΔCt = [AvgCt miRNA − (AvgCt U6 + AvgCt U48)/2] and ΔΔCt = [ΔCt NSCLC − ΔCt non-neoplastic lung tissue] (Livak and Schmittgen 2001). Statistical analysis was performed with SPSS 11.5 and Graphpad Prism 5.0 statistical software packages. Homogeneity variance, one-way ANOVA, or Mann–Whitney test was used to analyze the correlation between expression of miRNAs and clinicopathological features of the patients. The Pearson correlation test was used to analyze the correlation between expression of miR-100 and smoking status of AD patients. Survival was estimated by the Kaplan–Meier method, and the log-rank test was used to compare the survival distribution between groups. Cox’s hazard regression model was used to analyze the risk factors for patients with lung cancer. A P value ≤0.05 was considered significant.
Results
Differential miRNA profiles of adenocarcinoma and squamous-cell lung cancer assessed by microarray
We initially performed miRNA profiling on six NSCLC resected tumors and matched adjacent non-neoplastic lung tissues, including three squamous-cell lung carcinomas and three adenocarcinomas, using an Agilent miRNA microarray containing 723 human miRNA probes (from version 10.0 of the miRBase sequence database). We compared the miRNA expression levels detected from SCC and AD and identified nine differentially expressed miRNAs (Table 2). Among them, hsa-miR-205, hsa-miR-93, hsa-miR-221, and hsa-miR-30e were found to be overexpressed in SCC (P values: 0.024, 0.042, 0.028, and 0.027, respectively). In contrast, hsa-miR-29b, hsa-miR-29c, hsa-let-7e, hsa-miR-100 and hsa-miR-125a-5p were found to be more highly expressed in lung AD (P values: 0.017, 0.012, 0.023, 0.040, and 0.012, respectively).
Validation of differentially expressed miRNAs by qRT-PCR
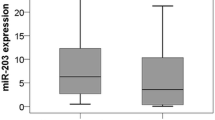
The levels of the nine differentially expressed miRNAs identified by microarray were confirmed by qRT-PCR using a subset of 105 samples that included 51 AD and 54 SCC. Since high expression of U48 and U6 snRNA has been previously reported in both AD and SCC patients, these molecules were used as positive controls (Thomson et al. 2006). Results showed that, compared to adjacent non-neoplastic lung tissues, the expression levels of miR-125a-5p and let-7e were significantly decreased in AD samples (P values: 0.0008 and 0.0166, respectively). In the SCC subtype, the expression levels of miR-93, miR-205, and miR-221 were increased (P values: 0.0013, 0.0347, and 0.0471, respectively; Fig. 2), but miR-125a-5p and let-7e were decreased (P values: <0.0001 and 0.0034, respectively; Fig. 2). In addition, partly consistent with the findings from microarray, the results of qRT-PCR showed that the expression level of miR-205, miR-221, and miR-30e were increased (P values: 0.0003, <0.0001, and 0.0253, respectively; Fig. 3), whereas the levels of miR-29b, miR-125a-5p, and let-7e were decreased (P values: 0.0082, 0.0187, and 0.0002, respectively; Fig. 3) in SCC patients compared to AD patients. Note that the expression levels of miR-29c, miR-93, and miR-100 had no significant differences between the two lung carcinoma subtypes (P values: 0.1168, 0.1024, and 0.8949, respectively; Fig. 3).
qRT-PCR analysis of miRNAs in lung carcinomas and matched non-cancerous tissues. Total miRNA was isolated from a lung adenocarcinoma (n = 51), b lung SCC (n = 54), and matched noncancerous tissues. qRT-PCR was performed to detect expression levels of miR-93, miR-100, miR-205, miR-221, miR-125a-5p, and let-7e. Paired t test was used to compare the differential expressions. In a, the P values for miR-125a-5p and let-7e were 0.0008 and 0.0166, respectively. In b, the P values for miR-93, miR-205, miR-221, miR-125a-5p, and let-7e were 0.0013, 0.0347, 0.0471, <0.0001, and 0.0034, respectively. *P < 0.05, as compared to the control
qRT-PCR analysis of miRNAs in two subtypes of lung carcinoma. Total miRNA was isolated from lung adenocarcinoma (n = 51), lung SCC (n = 54), and matched non-neoplastic tissues. qRT-PCR was performed to detect expression levels of miR-29b, miR-29c, miR-93, miR-100, miR-205, miR-221, miR-125a-5p, miR-30e, and let-7e. Unpaired t test was used to compare the differential expressions between the two subtypes of lung carcinoma. The expression levels of miR-205, miR-221, and miR-30e were increased (P values: 0.0003, <0.0001, and 0.0253, respectively), whereas miR-29b, miR-125a-5p, and let-7e levels were decreased (P values: 0.0082, 0.0187, and 0.0002, respectively) in SCC patients compared to AD patients. However, the expression level of miR-29c, miR-93, and miR-100 was not significantly different between the two lung carcinoma subtypes (P values: 0.1168, 0.1024, and 0.8949, respectively)
Differentially expressed miRNAs correlated to clinicopathological parameters of NSCLC
To identify the clinical relevance of miRNA expression in AD and SCC, the clinicopathological parameters, including tumor size, lymph node metastasis, differentiation, and clinical TNM stages, were subjected to correlation analysis (Table 3). Our results showed that the low expression level of let-7e was uniquely associated with >3 cm tumor size and differentiation of lung cancer in AD subtype, but not with those parameters in SCC. However, SCC patients with lymph node metastasis and advanced stage were associated with high-level expression of miR-205 and miR-221, respectively (Table 3).
Effect of cigarette smoking on miRNA expression in NSCLC subtypes
We also tested whether tobacco smoking affected miRNA expression by comparing smokers versus non-smokers with the AD and SCC subtypes. Mann–Whitney test found that miR-100 was significantly higher in current smokers than that in non-smokers with the AD subtype (P = 0.0395), while no significant difference was observed in the SCC subtype patients (P > 0.05). Moreover, expression of miR-100 showed a significant increase in AD patients who were smoking more than 20 cigarettes/day, or for 20 years or more (Hecht 1999) (P values: 0.0017 and 0.048, respectively; Pearson: r = 0.4355, P = 0.0014, and r = 0.298, P = 0.047, respectively; Fig. 4). Note that none of the other miRNAs showed any significant correlations with tobacco smoking in AD or SCC subtypes.
Correlation between the expressions of miR-100 and smoking status of lung adenocarcinoma patients. The correlation between the expressions of miR-100 and smoking status of lung adenocarcinoma patients was carried out by Pearson correlation test. Pearson’s correlation coefficient between the expression level of miR-100 and a cigarette smoking (per day) (r = 0.435, P = 0.0014) and b smoking duration (years) (r = 0.298, P = 0.047) is shown
Correlation between expression of miRNAs and survival of NSCLC patients
Due to the observed differences in the miRNA expression between the different subtypes, survival analysis was conducted separately for AD and SCC patients. In patients with AD, only let-7e (median: 0.19) was identified in the analysis restricted to the overall survival time for the associations (log-rank test: P = 0.0015; Fig. 5a). A risk prediction analysis by Cox regression for miRNA expression also showed that decreased let-7e was significantly associated with overall survival in AD patients [P = 0.004, hazard ratio (HR) = 1.04, 95 % confidence interval (CI) 1.01–1.07; Table 4]. In the SCC patients, we found that expressions of miR-93 (median: 4.29), miR-205 (median: 19.36), and let-7e (median: 0.56) were predictive of overall survival. By Kaplan–Meier curves, Cox univariate, or multivariate analysis, these miRNAs showed a significantly higher HR for poor prognosis (Fig. 5b–e). Patients with high miR-205 and low let-7e levels had an increased probability of mortality, which was potentially related to dose dependence (P = 0.000, HR = 10.11, 95 % CI 3.01–33.96; Table 4).
Kaplan–Meier survival curves for AD and SCC patients according to let-7e, miR-93 and miR-205 expression. Kaplan–Meier survival curves for NSCLC patients were obtained based on the median level of fold change detected for each miRNA. The P value was calculated using the log-rank analysis showing the difference between patients with high and low expression levels. Overall survival time of patients with high versus low expression levels of let-7e are shown for the a AD subtype and d SCC subtype, and of b miR-93, c miR-205, and e miR-205 for the SCC subtype. P < 0.05 indicates significant difference between the two groups
Discussion
Lung cancer is the leading cause of cancer-related deaths worldwide, and NSCLC accounts for 80 % of all cases of lung cancer. Early diagnosis for NSCLC is difficult. Biopsy and fine-needle aspirations are capable of confirming some advanced lung malignancies; however, recent clinical trials have indicated that distinct histological subtypes of NSCLC respond differently to chemotherapy and numerous target-specific treatment agents (Hirsch et al. 2008; Scagliotti et al. 2008). Even when tissues are obtained by resection, tumor classification can be difficult by the conventional diagnostic techniques, including traditional light microscopy and immunohistochemical staining (Jorda et al. 2009). However, efficacy of targeted therapies is dependent not only on the presence of particular genetic alterations but also on the precise NSCLC subtype; otherwise, treatment outcome is at best suboptimal and at worst ineffective.
Several studies have shown that miRNA signatures can distinguish squamous-cell lung carcinomas from adenocarcinomas and are useful prognostic predictors (Bishop et al. 2010; Landi et al. 2010; Raponi et al. 2009). In the study presented herein, we found that, as compared to adjacent non-neoplastic lung tissues, levels of miR-125a-5p and let-7e were decreased in AD and SCC tumor samples, while miR-93, miR-205 and miR-221 were uniquely increased in the SCC samples. This was further confirmed by the “gold standard” test of TaqMan real-time RT-PCR (Wang et al. 2011) using samples from NSCLC patients. Our results suggested that, in addition to well-known morphology and immunocytochemistry techniques, miR-93, miR-205, miR-221 and let-7e could be used as an additional tool to distinguish SCC from AD NSCLC subtypes, as suggested by other studies (Bishop et al. 2010; Lebanony et al. 2009).
miR-205 is a well-characterized miRNA that is highly expressed in SCC, as compared to AD (Bishop et al. 2010; Lebanony et al. 2009; Xing et al. 2010) and similar to our findings. However, a recent study showed that miR-205 levels in NSCLC were only slightly capable of distinguishing AD from SCC (Vescovo et al. 2011). These discordant results may be explained by the fact that each study used relatively few NSCLC samples (25 AD, 24 SCC, and 1 adenosquamous case). Thus, the biologic role played by miR-205 in SCC remains to be determined. As a member of the miR-200 family, miR-205 is capable of suppressing epithelial to mesenchymal transition by targeting the transcriptional factors ZEB1 and SIP1 (Gregory et al. 2008). Majid and colleagues reported that induced expression of miR-205 stimulated apoptosis and cell cycle arrest, but inhibited cell growth, migration, and invasiveness of prostate cancer cells. In addition, miR-205 was demonstrated to specifically activate the tumor suppressor genes IL24 and IL32 by targeting specific sites in their promoters (Majid et al. 2010). Our results demonstrated a strong correlation between overexpression of miR-205 and lymph node metastasis, suggesting that miR-205 might activate cancer-associated dysregulated genes, thereby contributing to tumor migration and eventual mortality. Further studies should explore the detailed biological functions of miR-205 in NSCLC, especially in the SCC subtype.
Yu and colleagues evaluated the expression of 157 miRNAs and found that five miRNA signatures could be used as an independent predictor of overall survival in patients with NSCLC (Yu et al. 2008). Raponi and colleagues reported that miR-146b and miR-155 were associated with overall survival in SCC (Raponi et al. 2009; Yanaihara et al. 2006). Yanahaira et al. (2006) presented data that suggested high levels of miR-155, along with low let-7a expression, were related to poor prognosis in lung AD (Yanaihara et al. 2006). Herein, we explored the association of miRNAs expression with survival from different subtypes of lung cancer. Our results demonstrated a distinct expression pattern of let-7e, miR-93, and miR-205 that was correlated with overall survival in different NSCLC subtypes, suggesting a potential unique role for this profile in predicting survival of patients with different NSCLC subtypes. Reduced expression of the let-7 family was correlated with increased tumorigenesis and poor survival in lung cancer (Landi et al. 2010; Ragusa et al. 2010; Takamizawa et al. 2004). Consistent with this, we found that low let-7e expression was associated with lymph node metastasis and >3 cm tumor size in the NSCLC AD subtype. Taken together with the survival data, this finding suggested that let-7e might play an important prognostic role in this type of NSCLC. Let-7e has been shown to target RAS and other oncogenes involved in regulation of cell cycles, such as the high mobility group AT-hook 2 (HMGA2), myelocytomatosis (MYC), cyclin-dependent kinase 6 (CDK6), and cell division cycle 25 (CDC25) (Medina and Slack 2008; Johnson et al. 2007; Lee and Dutta 2007). Patients with high miR-205 and low let-7e had significantly increased risk of cancer death, suggesting the existence of dynamic miRNA expression that may play a role in tumor progression (Li et al. 2009). Further studies of the regulatory mechanism involving miR-205 and let-7e would likely increase our understanding of the molecular pathogenesis of certain NSCLC subtypes.
Tobacco smoking is a well-known risk factor for lung cancer mortality (Boffetta et al. 2011; Frost et al. 2011); however, the effect of tobacco smoking on miRNA expression has not been completely elucidated. Izzotti and colleagues reported that miRNAs were decreased in rats exposed to cigarette smoke (Izzotti et al. 2009). Likewise, a screening cohort study by Van Pattelberge and colleagues showed that 34 miRNAs were differentially expressed between human non-smokers and current smokers without airflow limitation (Van Pottelberge et al. 2010). In our study, we demonstrated a strong correlation between high expression of miR-100 and tobacco smoking status in AD patients, suggesting that miR-100 might play an important role in contributing to cigarette smoke-induced lung cancer. More studies are needed to confirm this. Since the majority of SCC patients enrolled in our study were tobacco smokers, the link between miR-100 expression and tobacco smoking in SCC patients needs to be more precisely evaluated in future studies.
In summary, we identify differential expression profiles of miRNAs in AD and SCC. More importantly, we show that, in addition to the established morphology and immunocytochemistry approaches, miR-93, miR-205, miR-221 and let-7e may represent novel biomarkers for differential diagnosis and prognosis of certain NSCLC subtypes. These miRNAs may also represent new targets of histology-specific treatment approaches. Finally, miR-100 appears to play a role in cigarette smoke-induced lung cancer, although larger sample population and functional tests need to be performed to confirm this observation.
References
Bishop JA, Benjamin H, Cholakh H, Chajut A, Clark DP, Westra WH (2010) Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res 16(2):610–619
Boffetta P, Jayaprakash V, Yang P et al (2011) Tobacco smoking as a risk factor of bronchioloalveolar carcinoma of the lung: pooled analysis of seven case–control studies in the international lung cancer consortium (ILCCO). Cancer Causes Control 22:1–7
Frost G, Darnton A, Harding AH (2011) The effect of smoking on the risk of lung cancer mortality for asbestos workers in Great Britain. Ann Occup Hyg 55(3):239–241
Gregory PA, Bert AG, Paterson EL et al (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10(5):593–601
Hecht SS (1999) Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst 91(14):1194–1210
Hirsch FR, Spreafico A, Novello S, Wood MD, Simms L, Papotti M (2008) The prognostic and predictive role of histology in advanced non-small cell lung cancer: a literature review. J Thorac Oncol 3(12):1468–1481
Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S (2009) Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J 23(3):806–812
Johnson CD, Esquela-Kerscher A, Stefani G et al (2007) The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67(16):7713–7722
Jorda M, Gomez-Fernandez C, Garcia M et al (2009) P63 differentiates subtypes of non-small cell carcinomas of lung in cytologic samples. Cancer Cytopathol 117(1):46–50
Landi MT, Zhao Y, Rotunno M et al (2010) MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res 16(2):430–441
Lebanony D, Benjamin H, Gilad S et al (2009) Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol 27(12):2030–2037
Lee YS, Dutta A (2007) The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21(9):1025–1030
Li Y, VandenBoom TG, Kong D et al (2009) Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res 69(16):6704–6712
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-[delta][delta] CT method. Methods 25(4):402–408
Lu J, Getz G, Miska EA et al (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043):834–838
Majid S, Dar AA, Saini S et al (2010) MicroRNA-205 directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer 116(24):5637–5649
Medina PP, Slack FJ (2008) MicroRNAs and cancer: an overview. Cell Cycle 7:2485–2492
Miller YE (2005) Pathogenesis of lung cancer: 100 year report. Am J Respir Cell Mol Biol 33(3):216–223
Ragusa M, Majorana A, Statello L et al (2010) Specific alterations of microRNA transcriptome and global network structure in colorectal carcinoma after cetuximab treatment. Mol Cancer Ther 9(12):3396–3403
Raponi M, Dossey L, Jatkoe T et al (2009) MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res 69(14):5583–5776
Scagliotti GV, Parikh P, von Pawel J et al (2008) Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol 26(21):3543–3551
Takamizawa J, Konishi H, Yanagisawa K et al (2004) Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64(11):3753–3756
Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM (2006) Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20(16):2202–2207
Van Pottelberge GR, Mestdagh P, Bracke KR et al (2010) MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 183:898–906
Vescovo VD, Cantaloni C, Cucino A et al (2011) MiR-205 expression levels in non-small cell lung cancer do not always distinguish adenocarcinomas from squamous cell carcinomas. Am J Surg Pathol 35(2):268–275
Wang B, Howel P, Bruheim S et al (2011) Systematic evaluation of three microRNA profiling platforms: microarray, beads array, and quantitative real-time PCR array. PLoS One 6(2):e17167
Xing L, Todd NW, Yu L, Fang HB, Jiang F (2010) Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol 23(8):1157–1164
Yanaihara N, Caplen N, Bowman E et al (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9(3):189–198
Yu SL, Chen HY, Chang GC et al (2008) MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 13(1):48–57
Acknowledgments
This research was supported by grants from the Medical Science Foundation of Zhejiang Province (No. 2009A210), the Science and Technology Program of Zhoushan (Nos. 20081059 and 091042), and the Medical Science Foundation of Zhoushan (No. 2009B03).
Conflict of interest
The authors declare no conflict of interests exist in relation to the publication of this study.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhang, YK., Zhu, WY., He, JY. et al. miRNAs expression profiling to distinguish lung squamous-cell carcinoma from adenocarcinoma subtypes. J Cancer Res Clin Oncol 138, 1641–1650 (2012). https://doi.org/10.1007/s00432-012-1240-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-012-1240-0