Abstract
Purpose
Genetic instability is a hallmark of glioblastoma multiforme (GBM). Microsatellite instability (MSI) is a significant event in the tumorigenesis of many sporadic malignancies. The aim of our investigation was to study microsatellite instability in newly diagnosed glioblastomas.
Methods
MSI was investigated in 109 GBMs with 15 microsatellite markers. Immunohistochemistry was performed for the mismatch repair (MMR) proteins hMLH1, hMSH2, hPMS2, and hMSH6 in cases showing MSI. Sequence and promoter methylation status of hMLH1 were analyzed in the case of a decreased hMLH1 protein expression as well. To further investigate MSI(+) GBMs we carried out studies of LOH at selected chromosome regions, EGFR amplification, and sequence of p53 and PTEN.
Results
MSI was observed in six GBMs (5.5%) and it was more frequent in GBMs with a previous lower grade astrocytoma (18.8% vs. 3.2%). MMR protein staining was positive in all MSI(+) GBMs except in one case, which showed an aberrant expression of hMLH1 and hPMS2 without hMLH1 inactivation. Among MSI(+) GBMs, one tumor corresponded to the GBM molecular type 1 (p53 mutation, no EGFR amplification), another tumor to type 2 (wild-type p53, EGFR amplification), and four tumors to neither type (wild-type p53, no EGFR amplification). None of the six tumors carried a PTEN mutation.
Conclusions
MSI in GBM might be caused by inactivation of minor MMR genes rather than by a deficiency of hMLH1 or hMSH2 and it appears not to play a decisive role in the pathogenesis of these tumors. MSI(+) GBMs predominantly showed a profile which included wild-type of p53 and PTEN and absence of EGFR amplification but MSI occurred in all GBM molecular subtypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma multiforme (GBM) is the most frequent intracranial tumor in adults and accounts for approximately 15% of all brain neoplasms (Kleihues et al. 2002). The prognosis is dismal with a median survival time of 12 months after diagnosis despite aggressive therapy (Lacroix et al. 2001). In the pathogenesis of GBM two molecular pathways are characterized, type 1 and type 2 glioblastomas (von Deimling et al. 1993). Type 1 GBMs typically harbor p53 inactivation and most of type 2 GBMs carry EGFR amplification without p53 mutations (von Deimling et al. 1995; Reifenberger et al. 1993). However, in about one-third of all GBMs neither of these molecular alterations can be found (von Deimling et al. 1995; Louis 1997).
Genetic instability in GBMs is characterized by chromosomal instability with multiple imbalances (Hui et al. 2001), an event observed in the tumorigenesis of numerous solid tumors (Kinzler and Vogelstein 1997). A second form of instability, denominated microsatellite instability (MSI), was first described in patients with hereditary non-polyposis colorectal cancer (HNPCC) syndrome, which is caused by a deficiency of the mismatch repair (MMR) system and shows length mutations of microsatellite sequences (Thibodeau et al. 1993; Ionov et al. 1993). MSI was observed in HNPCC-associated malignancies in 80–90% of the cases (Ionov et al. 1993) as well as in many sporadic neoplasms such as colorectal, gastric, pancreatic, endometrial, and breast carcinomas (Ionov et al. 1993; Peltomäki et al. 1997; Wooster et al. 1997) in up to 40% of these tumors. HNPCC-related colorectal cancers show distinct pathological features as compared to their MSS homologues (Dietmaier et al. 1997; Jass et al. 1999), and MSI in those malignancies was observed to be predominantly associated with an inactivation of hMLH1 or hMSH2 (Boland et al. 1998). To our knowledge, an investigation of the MMR system in a large series of more than 100 glioblastomas has not been systematically performed. Furthermore, since 30% of GBMs show molecular alterations which do not conform to one of the known pathways, the analysis of the genetic events associated with MSI is of high interest. Therefore, we studied MSI in 109 sporadic newly diagnosed GBMs, evaluated the expression of the MMR proteins in MSI(+) cases, and performed an analysis of hMLH1 including sequencing and promoter methylation status in the case of hMLH1 reduced expression. Additionally, we characterized the molecular features of MSI(+) GBMs according to type 1 and 2 pathways.
Material and methods
GBM samples
One hundred and nine paired samples of blood and tumor tissue were obtained from GBM patients treated at the Department of Neurosurgery of the University of Dresden. Portions of the fresh tumor samples were frozen in liquid nitrogen and stored at −80 °C. Tumor tissue was evaluated at the Institute of Pathology, University of Dresden, Germany, according to the World Health Organization (WHO) criteria (Kleihues et al. 2002). Formalin-fixed, paraffin-embedded specimens were used for immunohistochemistry. This study was conducted with the consent of the institutional ethics committee and informed consent from each patient or patient caretaker was obtained. Sixteen patients had a previous astrocytoma, 11 patients an astrocytoma grade II, and further five patients had an astrocytoma grade III. Seven of the 109 GBM had a survival time of more than 36 months after surgery (so-called long-term survivors). Four patients had a personal history of cancer (three colorectal carcinoma and one breast cancer). There was no evidence for an association between MSI and family history of cancer (Table 1).
MSI analysis
Matched genomic tumor and blood DNA samples (n=109) were amplified by PCR using 15 primer pairs (Dietmaier et al. 1997): mononucleotides (BAT25, BAT26, BAT40, TGFß-RIIpA, GTBPIn5, GTBPIn9), dinucleotides (D5S346, D17S250, D10S197, D2S123, D18S58, D13S153, D3S1300, D3S1619), and tetranucleotide (MYCL1). PCR was performed containing 50–100 ng DNA. Amplified PCR products were electrophoresed on a 6.5% Long Ranger polyacrylamide gel on an Automated Laser Fluorescence (A.L.F. express) sequencing device (Amersham Pharmacia Biotech, Freiburg, Germany) and analyzed using the ALLELELINKS 1.00 software (Amersham Pharmacia Biotech). MSI was defined by the presence of novel alleles in tumor DNA, which were absent in corresponding leukocyte DNA and confirmed in two independent analyses. Informative dinucleotide repeats without MSI were assessed for LOH as described (Cawkwell et al. 1994). Three categories were defined according to the MSI findings: high-level MSI (MSI-H, ≥40% of studied loci altered), low-level MSI (MSI-L, <40%), and MSS (no analyzed marker unstable) (Dietmeier et al. 1997; Boland et al. 1998).
Immunohistochemistry of MMR proteins
Immunostaining for hMLH1 (clone G168–15, PharMingen Int., San Diego, Calif., USA), hMSH2 (clone FE11, Oncogene Res. Products, Cambridge Mass., USA), hMSH6 (clone 44, Transduction Lab., Lexington, UK), and hPMS2 (A16–4, PharMingen Int.) was performed in all six MSI(+) tumors as described (Plaschke et al. 2002). The relative densitometry of MMR protein expression (compared to respective negative controls) was performed using the Meta-Vue software (Universal Imaging Systems, Penn., USA).
Mutational and epigenetic analyses of hMLH1
We sequenced all coding exons and exon-intron boundaries of hMLH1 from leukocyte and tumor DNA in the case of a decreased hMLH1 protein expression. After PCR amplification, the products were electrophoresed in 0.8% agarose gel and subjected to cycle sequencing reactions using the Thermo Sequenase Fluorescent Cycle Sequencing kit (Amersham Pharmacia Biotech). After denaturation, the cycle sequencing products were resolved using a denaturing 6.5% Long Ranger polyacrylamide gel on a sequencing device. Primer sequences and amplification conditions were as described elsewhere (Kolodner et al. 1995). To evaluate the hMLH1 promoter methylation in the case mentioned above, DNA were digested with methylation-sensitive or insensitive restriction endonucleases, HpaII (Life Technologies, Karlsruhe, Germany) or MspI (New England Biolabs, Frankfurt am Main, Germany), respectively. After digestion, 25 ng from each digest was used as the template for PCR amplification of an 829-bp fragment which contains the promoter region, the exon 1 and a part of the intron 1 of the hMLH1, and harbors five HpaII sites. The amplification products were visualized using a 1% agarose gel. Primer sequences and PCR conditions were as described previously (Kane et al. 1997). The breast cancer cell line CAL51 was used as positive control.
Characterization of type 1 and type 2 pathways in MSI(+) GBMs
LOH at 10q23–24 was studied with the markers D10S215 and D10S541 (flanking PTEN), PTENCA (intragenic marker), D10S583 and D10S579 (telomeric and centromeric to PTEN, respectively). Analysis of LOH at 17p13 (p53) was done with the TP53 and a p53 intragenic marker. Allelic losses at 9p21 (p16/CDKN2A, p14ARF and p15/CDKN2B) and 13q14 (RB1) were studied with the primers D9S1748, D9S171, D9S1749, D13S153, and D13S267, respectively. Loss of heterozygosity at 1p35–36 and 19q13 were assessed with the markers D1S468, D1S482, D19S112, and D19S412 (Smith et al. 2001). Primer sequences and PCR amplification conditions are based on Genome Database entries (www.gdb.org). To detect EGFR amplification a differential PCR with the CFTR- (cystic fibrosis transmembrane regulator) gene was carried out, as previously described (Hunter et al. 1995). Sequencing of all coding exons and intron-exon boundaries of p53 and PTEN was performed using primers and conditions previously described, which are available from the authors upon request (Liaw et al. 1997; Steck et al. 1997).
Statistical analysis
Contingency analyses between variables were performed using a two-tailed Fisher’s exact test and t-test. The relationship between MSI and other variables was evaluated by multiple regression analysis. Odds ratios (OR) and 95% confidence intervals (CI) were obtained through logistic regression. A value of P <0.05 was considered to be statistically significant.
Results
MSI analysis
MSI-L was observed in six glioblastomas (5.5%) (Fig. 1). Nine of the 872 evaluable dinucleotides (1.03%) and one of 109 tetranucleotides (0.92%) were unstable. Instability of mononucleotide repeats was not evidenced. The most sensitive markers were D3S1619, D13S153 and D17S250, comprising 60% of the observed length mutations.
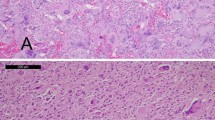
Left: microsatellite instability at marker D10S197 (case G1). N and T mean constitutional and tumor DNA, respectively. The arrow indicates a novel peak observed in tumor DNA. Right: analysis of hMLH1 promoter methylation of the giant-cell GBM (case G5). The HpaII- and MspI digested DNA (H and M, respectively) showed no amplification of the promoter fragment (829 bp) of hMLH1, indicating no methylation of the five cytosine residues within the CpG islands of the five restriction sites within the fragment. Leukocyte DNA (B5) from the patient was also analyzed. As negative control (NC) we used leukocyte DNA from a healthy Caucasian adult. The breast cancer cell line CAL51 was used as positive control. U and W stand for undigested DNA and water, respectively
We plotted the relative frequency of microsatellite mutation in respect to patient age at tumor resection. The MSI-L rate in patients older than 55 years was 9.5% (this cut-off level showed the lowest P-value), whereas in patients younger than 55 years 4.4% (P=0.26). No difference was observed between MSI-L and MSS groups in regards to age (P=0.32) or gender (P=0.23). The incidence of MSI-L in newly diagnosed GBMs was 3.2% but 18.8% in GBM patients who previously showed an astrocytoma grade II or III (P=0.15).
The relationship between MSI status and LOH pattern was evaluated as well. Overall, 108 instances of LOH were observed among 63 tumors. No significant differences between the MSI-L and MSS GBMs could be detected regarding allelic losses at chromosome regions mapping hMLH1 (3p21, P=0.51), hPMS2 (7p22, P=0.54), hMSH6 (2p16, P=0.42), and hMSH2 (2p22, P=0.43).
Immunohistochemical evaluation of MMR proteins
Five MSI-L GBMs had a positive nuclear staining for all MMR proteins. A loss of hPMS2 and a reduced staining of hMLH1 was found in a giant cell GBM (Fig. 2). Interestingly, only the giant cell phenotype showed this altered staining pattern, whereas the non-giant tumor cells had a positive pattern. There was a significant difference of the relative densitometry of hMLH1 expression (compared to respective negative controls) between giant (n=50) and non-giant (n=50 ) cells (P=0.0005, t-test). No significant differences in the relative densitometry of MMR proteins was found in twenty MSI(-) GBMs as compared to MSI(+) GBMs (P>0.05).
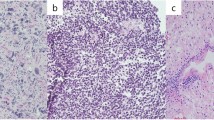
Immunohistochemical studies of hMLH1 (×200), hPMS2 (×200), hMSH2 (×400), and hMSH6 (×400) of the giant-cell GBM (case G5). They show a reduced expression of hMLH1 and a loss of expression of hPMS2 in the giant tumor cells. The non-giant tumor cells show a positive expression of both gene products. Positive nuclear patterns of hMSH2 and hMSH6 are observed in both giant and non-giant glioblastoma cells. Arrows indicate giant cells.
Sequence and promoter methylation analysis of hMLH1
Neither germline nor somatic genetic alterations were detected by sequencing hMLH1 in the giant cell GBM with reduced hMLH1- and loss of hPMS2 expression. By the methylation analysis of the promoter of hMLH1, no fragment were obtained from tumor DNA after digestion with HpaII. These findings indicated that the 5-tested cytosine residues in the promoter region of hMLH1 were not hypermethylated (Fig. 1).
Characterization of the molecular profile of MSI(+) GBMs
Mutations of p53 were found only in one giant cell GBM consisting in two transitions, on exon 5 (codon 175, CGC→CAC) and exon 6 (codon 199, GGA→AGA) predicting the amino acid substitution of arginine and glycine by histidine and arginine, respectively. In this specimen, an LOH at 17p13 was observed as well. An EGFR amplification was detected in one GBM. PTEN mutations were not found in any case. However, LOH at 10q23 was evidenced in three cases. According to the results of p53 and EGFR analyses, four MSI(+) GBM had a profile which did not correspond either to type 1 or to type 2 pathways, another GBM was classified as type 1, and a further tumor as type 2 GBM. Additional molecular features of the MSI(+) GBMs are shown in Table 2.
Discussion
In the pathogenesis of neoplasms, mechanisms responsible for the maintenance of the fidelity of DNA replication play a crucial role (Kinzler and Vogelstein 1997). A deficient MMR was observed to be the underlying mechanism in HNPCC-associated colon cancers and up to 40% of sporadic colon cancers. Such a deficiency leads to MSI and accumulation of somatic and/or germline alterations in target genes (Thibodeau et al. 1998; Boland et al. 1998). However, in most non-colonic sporadic cancers MSI is not caused by inactivation of MMR genes (Boland et al. 1998; Parc et al. 2000). Some reports described MSI in GBMs by either investigating small series (Dams et al. 1995; Zhu et al. 1996; Sobrido et al. 2000) or using few microsatellite markers (Alonso et al. 2001). Furthermore, different instability criteria and methods were used. We investigated MSI with 15 repeats as recommended by the National Cancer Institute (Boland et al. 1998) and observed low-level MSI in six of 109 (5.5%) GBMs. However, out of the six MSI(+) tumors, only one GBM showed a deficient MMR protein expression. This is in concordance with the observations in other MSI(+) sporadic non-colonic cancers such as gastric or endometrial neoplasms (Wu et al. 1997; Katabuchi et al. 1995). Thus, our findings suggest that MSI in the analyzed GBMs might rather be explained by inactivation of minor MMR genes (such as hMSH3 or hPMS1). This was supported by the results of our LOH studies in MSI and MSS GBMs at chromosome regions mapping the MMR genes. Alternatively, MSI might be caused by MMR genes mutations leading to a dominant negative phenotype as reported for hPMS2 (Nicolaides et al. 1998) or an imbalance in the relative amount of MMR proteins as shown for hMSH3 and hMSH6 (Marra et al. 1998).
The correlation between the clinicopathological data and MSI status revealed that the incidence of MSI was higher in patients who previously had an astrocytoma WHO grade II or III than in those clinically de novo glioblastomas (18.8% vs 3.2%, P=0.15). This might indicate a trend between MSI and progression of lower grade astrocytoma to glioblastoma multiforme, although this finding should be confirmed by investigating a larger number of GBMs. A similar observation was reported in gastric cancers since MSI was observed to be more frequent in advanced than in early cancers (Strickler et al. 1994).
According to our results of p53, EGFR, and PTEN, MSI(+) GBMs showed wild-type p53 and PTEN more frequently as well as an absence of EGFR amplification. Mutations of p53 were detected only in one MSI(+) giant cell GBM, a subgroup which typically shows a higher rate of p53 mutations than common GBMs (Meyer-Puttlitz et al. 1997). Additionally, only missense p53 mutations were detected but not insertions/deletions at iterated bases, thus making unlikely the possibility that MSI caused p53 alterations. A similar observation was found in MSI(+) gastric carcinomas (Renault et al. 1996). In the studied GBMs, PTEN mutations and MSI were mutually exclusive events.
We have shown a reduced hMLH1 expression and a loss of hPMS2 expression in a giant cell GBM. This GBM subtype is characterized by specific clinicopathological and molecular features (Kleihues et al. 2002). Typically, they show numerous multinucleated giant cells. They usually arise de novo (like type 2 GBMs) but carry genetic alterations which are different from those corresponding to type 1 and type 2 GBMs (Meyer-Puttlitz et al. 1997). In the studied specimen, the reduced hMLH1 expression was not associated with mutational or epigenetic inactivation of hMLH1. Furthermore, LOH at 3p21 (hMLH1) and 7p22 (hPMS2) was not evidenced. Taking into account the histological characteristics of giant cell GBMs, the possibility that hMLH1 mutations and/or hypermethylation of the hMLH1 promoter only affected the giant cell phenotype cannot be excluded. Alternatively, a transcriptional silencing affecting only the giant cells might lead to this condition as well.
In summary, our data show that MSI-L is infrequent in sporadic newly diagnosed GBMs. This instability phenotype was not associated with a deficient expression of MMR proteins in most of the cases. Although MSI(+) GBM predominantly showed a molecular profile with wild-type p53 and PTEN and absence of EGFR amplification, MSI was present in all different glioblastoma molecular subtypes.
References
Alonso M, Hamelin R, Kim M, Porwancher K, Sung T, Parhar P, Miler DC, Newcomb EW (2001) Microsatellite instability occurs in distinct subtypes of pediatric but not adult central nervous system tumors. Cancer Res 61:2124–2128
Boland R, Thibodeau S, Hamilton S, Sidransky D, Eshleman J, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S A (1998) National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination for microsatellite instability in colorectal cancer. Cancer Res 58:5248–5257
Cawkwell L, Lewis FA, Quirke P (1994) Frequency of allele loss of DCC, p53, RB1, WT1, NF1, NM23 and APC/MCC in colorectal cancer assayed by fluorescent multiplex polymerase chain reaction. Br J Cancer 70:813–818
Dams E, van der Kelft EJ, Martin JJ, Verlooy J, Willems PJ (1995) Instability of microsatellites in human gliomas. Cancer Res 55:1547–1549
Dietmaier W, Wallinger S, Bocker T, Kullman F, Fishel R, Ruschoff J (1997) Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res 57:4749–4756
Hui AB, Lo KW, Yin XL, Poon WS, Ng HK (2001) Detection of multiple gene amplifications in glioblastoma multiforme using array-based comparative genomic hybridization. Lab Invest 81:717–723
Hunter SB, Abbott K, Varma VA, Olson JJ, Barnett DW, James D (1995) Reliability of differential PCR for the detection of EGFR and MDM 2 gene amplification in DNA extracted from FFPE glioma tissue. J Neuropathol Exp Neurol 54:57–64
Ionov Y, Peinado A, Malkhosyan D, Shibata D, Perucho M (1993) Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363:558–561
Jass JR (1999) Towards a molecular classification of colorectal cancer. Int J Colorectal Dis 14:194–200
Kane MF, Loda M, Gaida G, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R (1997) Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 57:808–811
Katabuchi H, van Rees B, Lambers AR, Ronnett BM, Blazes MS, Leach FS, Cho KR, Hedrick L (1995) Mutations in DNA mismatch repair genes are not responsible for microsatellite instability in most sporadic endometrial carcinomas. Cancer Res 55:5556–5560
Kinzler KW, Vogelstein B (1997) Cancer susceptibility genes. Gatekeepers and caretakers. Nature 386:761–763
Kleihues P, Louis DN, Scheithauer BW, Rorke L, Reifenberger G, Burger PC Cavenee WK (2002) The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol 61:215–225
Kolodner RD, Hall NR, Lipford J, Kane MF, Morrison PT, Finan PJ, Burn J, Chapman P, Earabino C, Merchant E (1995) Structure of the human MLH1 locus and analysis of a large hereditary nonpolyposis colorectal carcinoma kindred for mlh1 mutations. Cancer Res 55:242–248
Lacroix M, Abi-Said D, Fourney DR, Gokaslan ZL, Shi W, De Monte F, Lang FF, McCutcheon IE, Hassenbusch SJ, Holland E, Hess K, Michael C, Miller D, Sawaya R (2001) A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection and survival. J Neurosurg 95:190–198
Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacock M, Eng C, Parsons R (1997) Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet 16:64–67
Louis DN (1997) A molecular genetic model of astrocytoma histopathology. Brain Pathol 7:755–764
Marra G, Iaccarino I, Lettieri T, Roscilli G, Delmastro P, Jiricny J (1998) Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc Natl Acad Sci USA 95:8568–8573
Meyer-Puttlitz B, Hayashi Y, Waha A, Rollbrocker B, Boström J, Wiestler O, Louis DN, Reifenberger G, von Deimling A (1997) Molecular genetic analysis of giant cell glioblastomas. Am J Pathol, 151:853–857
Nicolaides NC, Littman SJ, Modrich P, Kinzler KW, Vogelstein B (1998) A naturally occurring hPMS2 mutation can confer a dominant negative mutator phenotype. Mol Cell Biol 18:1635–1641
Parc YR, Halling HC, Wang L, Christensen ER, Cunningham JM, French AJ, Burgert LJ, Price-Troska TL, Roche PC, Thibodeau SN (2000) hMSH6 alterations in patients with microsatellite instability-low colorectal cancer. Cancer Res 60:2225–2231
Peltomäki P, de la Chapelle A (1997) Mutations predisposing to hereditary nonpolyposis colorectal cancer. Adv Cancer Res 71:93–119
Plaschke J, Kruger S, Pistorius S, Theissig F, Saeger HD, Schackert HK (2002) Involvement of hMSH6 in the development of hereditary and sporadic colorectal cancer revealed by immunostaining is based on germline mutations, but rarely on somatic inactivation. Int J Cancer 97:643–648
Reifenberger G, Liu L, Ichimur K, Schmidt KK, Collins VP (1993) Amplification and overexpression of the MDM 2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res 53:2736–2739
Renault B, Calistri D, Buonsanti G, Nanni O, Amadosi D, Ranzani GN (1996) Microsatellite instability and mutations of p53 and TGF-ßRII genes in gastric cancers: Hum Genet 98:601–607
Smith JS, Tachibana I, Lee HK, Qian J, Pohl U, Mohrenweiser HW, Borell TJ, Hosek SM, Soderberg CL, von Deimling A, Perry A, Scheithauer BW, Louis DN, Jenkins RB (2001) Mapping of the chromosome 19 q-arm glioma tumor suppressor gene using fluorescence in situ hybridization and novell microsatellite markers. Genes Chromosomes Cancer 29:16–25
Sobrido MJ, Rodriguez P, Barros F, Forteza J, Carracedo A, Lema M (2000) Low frequency of replication errors in primary nervous system tumors. J Neurol Neurosurg Psychiatry 69:369–375
Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV (1997) Identification of a candidate tumor suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advances cancers. Nat Genet 15:356–362
Strickler JG, Zheng J, Shu Q, Burgart LJ, Alberts SR, Shibata D (1994) p53 mutations and microsatellite instability in sporadic gastric cancer: when guardians fail. Cancer Res 54:4750–4755
Thibodeau SN, Bren G, Schaid D (1993) Microsatellite instability in cancer of the proximal colon. Science 260:816–819
Thibodeau SN, French AJ, Cunningham JM, Teter D, Burgart LJ, Roche PC, McDonnell SK, Schaid DJ, Vockley CW, Michels VV, Farr GH Jr, Connell MJ (1998) Microsatellite instability in colorectal cancers: different mutator phenotypes and the principal involvement of hMLH1. Cancer Res 58:1713–1718
Von Deimling A, von Ammon K, Schoenfeld D, Wiestler OD, Seizinger BR, Louis DN (1993) Subsets of glioblastoma multiforme defined by molecular genetic analysis. Brain Pathol 3:19–26
Von Deimling A, Louis DN, Wiestler OD (1995) Molecular pathways in the formation of gliomas. Glia 15:328–338
Wooster R, Cleton-Jansen AM, Collins VP, Mangion J, Cornelis RS, Cooper CS, von Deimling A, Wiestler OD, Stratton MR (1994) Instability of short tandem repeats (microsatellites) in human cancer. Nat Genet 6:152–156
Wu MS, Sheu JC, Shun CT, Lee WJ, Wang JT, Wang TH, Cheng AL, Lin JT (1997) Infrequent hMSH2 mutations in sporadic gastric adenocarcinoma with microsatellite instability. Cancer Lett 112:161–166
Zhu JJ, Guo SZ, Beggs AH, Muruyama, T, Santarius T, Dashner K, Olsen N, Wu J, Black PM (1996) Microsatellite instability analysis of primary human brain tumors. Oncogene 12:1417–1423
Acknowledgements
We thank C. Voelter at ENT Department, University of Würzburg, Germany for helpful discussion, and L. Moser at ENT Department, University of Würzburg, Germany for assessment of statistical analyses. We also thank A. Serra for advice with the densitometry studies and M. Reichmann for technical assistance. The authors are indebted S. Scherneck at the Department of Surgery and Surgical Oncology, Robert-Roessle Clinic, Berlin, Germany for kindly providing the breast cancer cell line CAL51
Author information
Authors and Affiliations
Corresponding author
Additional information
Grant sponsor: supported by a grant from the Deutsche Forschungsgemeinschaft, DFG (MA 2448/1)
Rights and permissions
About this article
Cite this article
Martinez, R., Schackert, H.K., Appelt, H. et al. Low-level microsatellite instability phenotype in sporadic glioblastoma multiforme. J Cancer Res Clin Oncol 131, 87–93 (2005). https://doi.org/10.1007/s00432-004-0592-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-004-0592-5