Abstract
It is known that tetraspanin proteins are involved in many physiological somatic cell mechanisms. Additionally, research has indicated they also have a role in various infectious diseases and cancers. This review focuses on the molecular interactions underlying the tetraspanin web formation in gametes. Primarily, tetraspanins act in the reproductive tract as organizers of membrane complexes, which include the proteins involved in the contact and association of sperm and oocyte membranes. In addition, recent data shows that tetraspanins are likely to be involved in these processes in a complex way. In mammalian fertilization, an important role is attributed to CD molecules belonging to the tetraspanin superfamily, particularly CD9, CD81, CD151, and also CD63; mostly as part of extracellular vesicles, the significance of which and their potential in reproduction is being intensively investigated. In this article, we reviewed the existing knowledge regarding the expression of tetraspanins CD9, CD81, CD151, and CD63 in mammalian spermatozoa, oocytes, and embryos and their involvement in reproductive processes, including pathological events.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Key molecular processes and mechanisms of the immune system are also engaged in mammalian reproduction. Based on current knowledge, an important role in fertilization is attributed to the surface molecules of gametes and reproductive cell tissue. Cluster of differentiation (CD) molecules such as CD9, CD81, and CD151 belonging to the tetraspanin superfamily take a prominent place in gamete interaction [1, 2]. CD63, is another member of the tetraspanins superfamily, and is known to be a part of extracellular vesicles (EVs), but its significance and potential in reproduction has not been fully addressed [3].
Tetraspanin proteins are involved in many cellular mechanisms. They can interact with a range of other proteins such as integrins, members of the immunoglobulin superfamily and proteases, and with each other creating a large network called the tetraspanin web [4]. The association of tetraspanins with proteins and lipids leads to an organization of specific microdomains located in membranes, these are the so-called tetraspanin-enriched microdomains (TEMs), and these are different from lipid rafts. Although the tetraspanin family includes distinct proteins, all of them are defined by a common structure consisting of four transmembrane domains containing conserved polar residues, small (SEL) and large (LEL) extracellular loops with four, six, seven or eight conserved cysteine residues located in the variable region, and short cytoplasmic tails. Most tetraspanins are glycosylated and also palmitoylated [5,6,7,8,9]. In addition, research points to the fundamental role of tetraspanins in the pathogenesis of viral, bacterial, parasitic, and fungal infections (reviewed in [10,11,12]), and cancer (reviewed in [13, 14]).
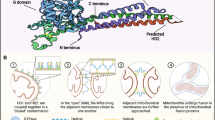
In this study, the participation of tetraspanins CD9, CD81, CD151, and CD63 in mammalian reproduction (summarized in Fig. 1), including certain pathological processes were addressed. It was long believed that the first recognition and primary binding of spermatozoa to the glycoprotein coat of oocytes, the zona pellucida (ZP), induces acrosomal exocytosis in sperm that enables them to interact with oocytes [15]. According to the real time observations made by Jin et al. [16], most fertilizing mouse spermatozoa begin their acrosome reaction before contact with the ZP. Spermatozoa undergo the acrosome reaction at the end of sperm maturation process called capacitation, occurring within the female reproductive tract [17,18,19], which represents a sequence of many biochemical and biophysical changes (reviewed in [20]). During the acrosome reaction, the sperm plasma membrane (PM) fuses with the underlying outer acrosomal membrane (OAM) leading to the release of acrosomal content (hydrolytic enzymes) and the exposure of the inner acrosomal membrane (IAM) proteins on the sperm head surface. Only completely acrosome-reacted spermatozoa are able to penetrate the ZP and perivitelline space (PVS, the space between the oolema and ZP) and fuse with the oocyte plasma membrane (oolema) [15]. After the membrane fusion, the sperm nucleus, mitochondria, centriole, and flagellum enter the oocyte and male and female gamete nuclei merge together to form a zygote. A monospermic fertilization where an egg is fertilized by only one sperm, is ensured by the blocking of other sperm passing through the ZP and fusing with oolema by a polyspermy block [15].
The engagement of tetraspanins during the sperm and egg interaction. 1. Recognition and primary binding of capacitated sperm to the cumulus cells (CC) and zona pellucida (ZP) of the egg is assisted by CD9, CD81, and CD63 and followed by 2. the sperm acrosome reaction and CD9 and CD81 relocation (mouse) that facilitates 3. a secondary sperm binding in presence of CD9, CD81 (mouse, human), and CD151 (mouse, human, cattle). This results in 4. the sperm penetration through ZP and 5. adhesion to the oocyte plasma membrane (PM) both in assistance of CD9, CD81 and CD151. Finally, 6. in presence of CD9, CD81, and CD151, gamete membrane fusion occurs followed by pronuclei fusion and a zygote formation. 7. shortly after a sperm fuses with the oocyte, the impermeability of ZP to other sperm is ensured by the polyspermy block. (PB) polar body, (PVS) perivitelline space
CD9 in oocytes and embryos
CD9 is the most studied tetraspanin in terms of its involvement in fertilization. The significance of CD9 was shown by experiments using female mice with the knockout (KO) Cd9 gene. KO females were born and grew normally, but they were mostly infertile [21,22,23]. During in vitro fertilization, spermatozoa that penetrated the ZP stayed imprisoned in the perivitelline space [21,22,23], while oocytes directly injected with spermatozoa were fertilized [21]. Similarly, Kaji et al. [24] and Zhu et al. [25] documented the “recovery” of sperm–egg fusion after the application of CD9 mRNA into Cd9-deficient oocytes. The CD9 molecule in mice was located in the plasma membrane of oocytes [21,22,23, 26]. It was detected on the oocyte surface covered by microvilli, the place is thought to be an initial contact point for spermatozoa with the PM but was not observed in the area overlying the second meiotic spindle, thus its role in the adhesion was proposed [23]. This assumption was later supported by Runge et al. [27]. They documented that CD9 may affect the rearrangement of microvilli capturing the sperm; the microvilli of the Cd9-deficient oocytes were short, thick, and denser in comparison to the long and thin microvilli of natural oocytes.
CD9 has also been documented in humans, where CD9 was found in oocytes in the germinal vesicle stage (GV), in metaphase I (MI), metaphase II (MII) [28, 29], and embryos [30, 31]. In pig, CD9 was present on the whole PM of oocytes, including the area over the second meiotic spindle. The CD9 level significantly increased during oocyte maturation [32]. Similarly, an increasing fluorescent signal of CD9 was detected on the PM of sheep oocytes and embryos [33] and in the mature cumulus-oocyte complex of yak [34].
In bovine oocytes, CD9 was observed for the first time in the plasma membrane of ZP-free oocytes by Zhou et al. [35]. The following study of Jankovicova et al. [36] described localization of CD9 in the PM of ZP-intact oocytes in different maturation stages as well as in the vesicles released into the perivitelline space of oocytes and embryos, that were most apparent after fertilization and division of the zygote. CD9 detection was dependent on the type of antibody used; the polyclonal antibody recognized the unevenly distributed clusters on the PM surface, while staining with monoclonal antibody appeared more homogenous over the PM of oocytes except for the region over the metaphase plate. Moreover, the fluorescent signal was also observed in ZP. It should be noted that the epitopes of applied antibodies probably differ. While polyclonal anti-CD9 antibody (ABIN741015) was directed to a part of the large extracellular loop (AA120-165 peptide), the monoclonal antibody IVA-50 (used also in the study of Zhou et al. [35]) was obtained after immunization with bovine thrombocytes [37] and thereby could probably recognize more extensive conformational epitope. Moreover, it was documented that tetraspanins are organized in small nanoclusters, detectable only by STED microscopy [38]. The reaction pattern of the polyclonal antibody detected by confocal microscopy in all probability represents a portion of the CD9 molecule within the larger domain, while due to more accessible epitope, IVA-50 stained CD9 more extensively. Similarly, a different reaction pattern of two anti-CD53 antibodies was documented in B cells [38]. As we mentioned previously, filament-like structures resembling transzonal projections (TZPs) were detected by monoclonal antibody in bovine oocytes passing through the ZP in a similar way to actin filaments. CD9 could be a part of a transzonal projection via interaction with actin binding proteins [39] or it could be a part of extracellular vesicles, the presence of which was detected in the TZPs by Macaulay et al. [40]. CD9 as a component of extracellular vesicles was already documented by several authors. Barraud-Lange et al. [41] reported that the transfer of CD9-containing oocyte fragments to fertilizing spermatozoa resembled trogocytosis, the process by which lymphocytes capture the membrane fragments of antigen presenting cells. In the studies of Miyado et al. [42] and Barraud-Lange et al. [43], release of vesicles (including CD9-positive vesicles) from PM into the PVS of oocytes was related to the ability of facilitating the sperm–egg fusion. Similar findings were described in hamster [42]. Recently, in human oocytes, the CD9-positive vesicles were observed in the PVS but not within the ZP of matured (MII) oocytes [31]. This observation corresponds with the findings of Miller et al. [44] in mice. Furthermore, Vyas et al. [31] documented an abundance of CD9-positive vesicles in the ZP after fertilization of human zygotes, continuing throughout cleavage to the blastocyst stage. Interestingly, similar extensive staining of CD9-positive clusters (probably vesicles within the PVS) has been already documented in bovine and porcine embryos, but CD9-fluorescent signal within the ZP was detected only in bovine but not in porcine zygotes and embryos [36]. It could be hypothesized that the different pattern detected in the ZP of mouse, porcine, bovine, and human oocytes is caused by antibody epitope availability or non-availability (as mentioned above), or could reflect the species-specific traits of CD9 in the processes associated with gamete adhesion and intercellular communication during fertilization.
Differing results were also observed, when the role of CD9 in the fertilization process (particularly in the sperm-egg binding and fusion) was analyzed using specific anti-CD9 antibodies in in vitro fertilization assays, whereas Chen et al. [26] and Takahashi et al. [45] observed significant inhibition of sperm–egg binding and fusion in mice [26], Miyado et al. [21] and Miller et al. [44] recorded the inhibitory effect of the same antibody on fusion without a decrease in sperm binding. In human, the same anti-CD9 antibody that inhibited the fusion in mice did not affect the fusion of sperm with ZP-free oocytes [29]. However, when human intact oocytes were maintained in the presence of the antibody during ZP removal and gamete fusion, a strong inhibitory effect on gamete fusion was recorded [29].
In non-rodent species, the antibody treatment significantly reduced the sperm binding and fusion with ZP-free oocytes in pig [32], sheep [33], and cattle [35]. In contrast, the antibody treatment of ZP-intact bovine oocytes did not cause any decrease in the number of fertilized oocytes [36]. The diversity of the findings probably reflects (a) the differing experimental design (at least the use of ZP-free vs. ZP-intact oocytes), (b) interspecies variability, or (c) the fact, that the antibodies did not recognize the crucial epitopes [36]. The application of the same antibody to oocytes with or without ZP (in human, mice [29], and cattle [35, 36]) with different effects on fertilization pointed to the limitation of experiments with ZP-free oocytes. The chemical or enzymatic removal of ZP may result in impairment or loss of function of the egg proteins critical to sperm–egg interaction [46, 47]. Moreover, in experiments using ZP-free oocytes, it is not clear whether sperm binds to the PM, or ZP residues [48]. Furthermore, the results could depend on the method of removing loosely bound spermatozoa and also the fact that the acrosome reaction is not induced by ZP [49]. So far, it seems impossible to reliably distinguish between binding/adhesion and fusion defect after oocyte antibody treatment of ZP-free oocytes [49].
The importance of the CD9 molecule in the pre-fusional adhesion steps was later proved by Jegou et al. [50]. The authors documented that CD9 generates a strong adhesion site in contact with gamete membranes that was necessary for successful fusion. This suggestion was later supported by Chalbi et al. [51], who showed that this accumulation of CD9 on the oocyte membrane was controlled by adhesion of Juno, an essential egg molecule interacting with sperm essential protein Izumo [52]. Furthermore, Ravaux et al. [53] documented that recruitment of egg CD9 to the egg–sperm interface is initiated by sperm oscillations (flagellum beating) after the contact of acrosome-reacted sperm with the egg membrane. Immediately after gamete fusion, CD9 together with associated proteins leave the PM and thus, it probably participates in the prevention of polyspermy.
The association of oocyte CD9 with other proteins was already proposed by Chen et al. [26]. According to their model, which did not take into account CD9 in sperm, CD9 through direct or non-direct association with integrin α6β1 on the mouse oocyte surface could assist the integrin α6β1 in binding to fertilin β located on the sperm head. However, the role of integrin α6β1 is controversial, because another study in mice did not confirm the participation of the integrin α6β1 in this binding process. The disagreement could likewise be caused by modification of plasma membrane proteins after ZP removal [44]. On the other hand, the study of Ziyyat et al. [29] confirmed that redistribution of α6β1 within the PM of human oocyte is controlled by CD9. Based on the findings that anti-CD9 antibody added prior to the ZP removal inhibited the formation of α6β1 patches formed in the absence of antibody, authors hypothesized that CD9 controls the lateral mobility of α6β1 and maintains the tetraspanin web, to which it linked. As described above, CD9 was detected on the surface of mouse oocyte, which was covered by microvilli. The microvilli rearrangement may be related to the functional connection of CD9 with EWI-2 and EWI-F [54], immunoglobulins that are able to bind ezrin, radixin, moesin (ERM proteins), connecting to the actin filaments of cytoskeleton [55]. The CD9/EWI-2/ERM complex on the oocyte plasma membrane is able to regulate the structure and dynamics of microvilli optimal for the sperm–egg contact [27]. In very recent study based on the crystal structure of CD9 and the cryo-electron microscopic structure of human CD9, Umeda et al. [56] showed the interaction between CD9 and EWI-2, mediated by small residues in the transmembrane region and protein/lipid interaction. Authors suggested the possible role of other (not only flexible) LEL regions of CD9 in fertilization and moreover the role of EWI-2 as a bridge between tetraspanins and other proteins, important for remodeling of membrane resulting in the formation of complex protein network and vesicles. Tetraspanins clustering can directly induce the membrane curvature and thus facilitate the exosome budding or control the vesicular cargo sorting through the association with other partner proteins [56].
Taken all together, CD9 on the oocyte plasma membrane could probably participate in the reorganization and curvature of the membrane, which presumably facilitates the mutual protein–protein communication leading to successful fertilization. This ability to control the lateral mobility of protein within the tetraspanin web, connects to actin filaments and thus, reorganize the membrane applied in particular processes like microvilli rearrangement, adhesion site generation or releasing of vesicles that could prevent polyspermy. Moreover, it could enable the intercellular communication during oocyte and embryonic development, and embryo-endometrial cross-talk [30, 57, 58].
CD9 in spermatozoa
In contrast with the negative results in mice [26] and pig sperm [32], CD9 was detected in rat and mouse spermatogonia [59, 60], and later also in mouse spermatozoa [61]. CD9 was found in the cytoplasm of mouse testicular germ cells, in spermatogonia, spermatocytes, and round spermatids. In mature spermatozoa with permeabilized membrane and spermatozoa in the initial stage of acrosome reaction, CD9 was present in the inner acrosomal membrane, mainly in the marginal region of the anterior acrosome, extended to the equatorial region with advancing acrosomal reaction [61]. In contrast, Barraud-Lange et al. [43] observed only a low portion (10%) of mature or capacitated spermatozoa positive for CD9, which appear mainly as a thin line in the acrosomal region. Conversely, 60–75% of acrosome-reacted sperm showed bright CD9 fluorescent dots. Frolikova et al. [62] described the presence of tetraspanin CD9 in both the inner and outer acrosomal membrane. During the acrosome reaction, CD9 relocated to the equatorial segment and partially over the post-acrosomal region [62]. The different pattern of CD9 localization on mouse spermatozoa might have been caused by dissimilar methods of sample fixation. Sperm possess both the outer and inner acrosomal membranes and these are unavailable for antibody staining without permeabilization treatment. While Ito et al. [61] detected CD9 on frozen-thawed spermatozoa, Barraud-Lange et al. [43] found this molecule on fresh sperm fixed with paraformaldehyde (non-permeabilizing agent). Obviously, freezing can cause drastic changes in the sperm membranes such as deterioration in integrity but it may also expose the inner acrosomal membrane, which is normally revealed after the acrosomal exocytosis. The CD9-staining in acrosome-reacted sperm was revealed by both research teams using epifluorescence microscopy. An additional signal of CD9 on the outer acrosomal membrane was detected by Frolikova et al. [62] probably due to the use of super-resolution microscopy.
In non-rodent species, Li et al. [32] did not detect CD9 in frozen-thawed pig spermatozoa, probably due to the fact that boar semen is more sensitive to cryo-damage than other species. In contrast, CD9 was detected in frozen-thawed as well as freshly ejaculated bull spermatozoa, where CD9 was localized in the apical part or through the entire anterior region of the plasma membrane of ejaculated and capacitated spermatozoa. The molecule was lost from the sperm after acrosome reaction [63]. A similar pattern was observed in human, when CD9 was localized in the apical region within the acrosomal cap of ejaculated and capacitated spermatozoa [62]. After the acrosome reaction, CD9 was located only over the equatorial segment. It seems that the localization of CD9 in bull spermatozoa is more similar to human than mouse sperm. This may be due to the different shape of mouse sperm head in comparison with bull and human spermatozoa, which probably effects spatial protein arrangement and distribution.
The authors [62] also suggested the involvement of CD9 in the tetraspanin web formation in the sperm membrane. The results of immunoprecipitation, western blot analysis, and molecular modeling identified a possible presence of CD9 dimers in human spermatozoa. The large extracellular domain of CD9 would be involved in the dimer formation and could potentially mediate the trans-interaction [62]. The post-translational modification by palmitoylation of CD9 [6] could participate in the CD9 web stabilization during the sperm maturation and acrosome reaction [62]. Moreover, the knowledge that CD9 is able to associate with EWI-2 and EWI-F [64] and these proteins are major partners of CD9 and CD81 acting as linkers connecting the tetraspanin microdomains to the actin cytoskeleton through their direct interaction with ERM proteins (ezrin/radixin/moesin) [65], supports the possible role of the tetraspanin web in the stabilization of the sperm acrosome. It has been previously suggested that ezrin is involved in the activation of the Rho GTPase family members followed by actin polymerization during human sperm capacitation [66].
The significance of CD9 (and also ezrin, F-actin, cdc42, and β-tubulin) in the maintenance of sperm function can be illustrated by a lower expression of these proteins in asthenozoospermic semen (sperm with reduced motility), compared to normospermic semen. It was hypothesized to be probably related to cytoskeletal reorganization and disturbance of sperm plasma membrane [67].
CD81 in oocytes and embryos
Another tetraspanin studied in relation to reproduction, human and mouse CD81 shares about 43% amino acid sequence identity with CD9 [68]. Deletion of the Cd81 gene in mice showed that the fertility of Cd81-deficient females decreased by 40% [69]. The reduction of fertility was also apparent when the ZP-free oocytes were incubated with anti-CD81 antibody [45], while fertilization of ZP-intact mouse oocytes in the presence of antibody did not affect sperm penetration or the rate of two-cell embryos [70]. Similarly, pre-treatment of ZP-intact bovine oocytes with the anti-CD81 antibody did not decrease the fertilization rate [36]. Its detection and localization (similar to CD9) varies in dependence on application of the antibody, using ZP-free/ZP-intact oocytes; additionally, species specificity could not be excluded. CD81 was first observed on the surface of mouse ZP-free oocytes [45], but later the molecule was found only in the inner area of ZP [70]. The fact that CD81 production is predicted to be predominantly by wild-type cumulus cells and then re-localized to ZP is in contrast with the later detection of CD81 clusters on the surface of ZP-free Cd81-deficient oocytes after injection of mRNA encoding CD81 [70]. This discrepancy can be explained by the detection of tetraspanin within larger or smaller domains depending on the antibody epitope [38]. In human, CD81 has been localized over the whole PM of unfertilized ZP-intact oocytes with the formation of patches after ZP removal [29]. Recently, the presence of CD81 in the intact bovine oocytes (GV, MI, and MII) was detected on the PM and moreover after fertilization in PVS of zygotes and embryos [71]. In porcine, besides the oocyte PM, CD81 was also observed in the inner part of the ZP, and additionally released within vesicles concentrated into several clusters in the PVS of zygotes and embryos [36].
Initially, it was proposed that CD81 plays a complementary role with CD9, because Cd81 and Cd9 double knockout female mice were completely infertile and microinjection of CD81 mRNA partially compensated CD9 in Cd9-deficient oocytes [1]. Ohnami et al. [70] later suggested that in mice both CD81 and CD9 work independently as extracellular components in gamete fusion, whereas microinjection of CD9 mRNA reversed a fusion defect in Cd81- and Cd9-deficient oocytes, however injection of CD81 mRNA failed. It seems that the expression and co-localization of both tetraspanins might be species-specific or their detection is dependent on the antibody used. Rubinstein et al. [69] supposed that on the surface of mouse oocytes, CD81 is associated with tetraspanin enriched microdomains. According to Ohnami et al. [70], the association of CD81 and CD9 in mouse oocytes may be limited, because the localization of these two proteins has been detected in different areas of the oocytes. CD81 localized at the inner part of ZP may help to form a complex with CD9 released within the vesicles to the PVS of mouse oocytes [42] and thus help transfer the CD9 to the sperm [70]. The above mentioned localization of these two tetraspanins on plasma membrane of human, bovine, and porcine oocytes seems to be a prerequisite for the complex formation and possible cooperation of these tetraspanins not only within the vesicles, but also within the oocyte plasma membrane.
CD81 in spermatozoa
CD81 was detected in rat spermatogonia [60] and by western blot analysis of protein extracts from mouse capacitated spermatozoa [71]. The precise localization in spermatozoa has been newly described in mouse [71], cattle [71], and human [62]. In the mouse, CD81 was localized on the PM covering the apical acrosome of the epididymal spermatozoa and relocated during the acrosome reaction across the equatorial segment over the whole sperm head. In the bull, CD81 was apparent on the apical part and partially in the equatorial region of the epididymal and ejaculated spermatozoa, disappearing from the sperm head after the acrosome reaction [71]. In human, the localization of CD81 has been observed in the apical acrosomal area and partially in the post-acrosomal area of ejaculated and capacitated spermatozoa. After acrosome reaction, it disappeared from the apical part of the acrosome, but stayed detectable in the post-acrosomal region.
The high rate of CD81 and CD9 co-localization without the co-localization occurring after the acrosome reaction suggested that CD81 may interact and form complexes with CD9 on the membrane of ejaculated human spermatozoa [62]. The probable cooperation of these two tetraspanins was predicted by molecular modeling based on the open/closed (cholesterol-free/bound) conformation of CD81 that was previously proven by molecular dynamics simulation using the crystal structure model of full-length human CD81 [72] and from the data obtained on human spermatozoa [62]. The dynamics of the CD9 and CD81 interaction network is directly related to the presence or absence of cholesterol in the CD81 cavity. The CD81 cholesterol-free molecule has a more compact transmembrane region as well as a change from conical to cylindrical conformation. This change in the membrane curvature may possibly play a role in sperm–egg fusion because the convex part of the equatorial segment has been considered as a site of the sperm–egg membrane interaction. The cholesterol efflux during sperm capacitation possibly influences CD81-cholesterol binding and might lead to a more compact transmembrane region and greater positive curvature to adapt at the moment of fusion [62]. It appears that cholesterol binding modulates the activity of CD81 in cells that suggest a potential mechanism for the regulation of tetraspanin function. The assumption that CD9 and CD81 participate in sperm–egg interaction seems to be in conflict with the fact that Cd81−/− and Cd9−/− and even double knockout Cd9−/− Cd81−/− mouse males were normally fertile [69]. The cooperation of these two tetraspanins is estimated on the basis of data obtained from the human model, where the mutual co-localization detected by immunofluorescent staining was supported by co-immunoprecipitation experiments. These findings, however, were in contrast to the mouse model where both methods failed to reveal any mutual interaction of CD9 and CD81 [62]. According to Miyado et al. [42], the fertility of deficient mice could be rescued by transfer of CD9-containing vesicles from wild-type oocytes; however, these suggestions were later challenged by Gupta et al. [73] and Barraud-Lange et al. [43]. To ensure reproduction, it appears therefore that some species have developed unique mechanisms of fertilization that may also include several “backup” mechanisms, for example; the replacement of tetraspanin function by another protein.
CD151 in oocytes
Research has shown that Cd151 knockout mice were capable of reproduction [74]; however, the inhibition of sperm–egg fusion (up to 50%), by the anti-CD151 antibody suggested a significant but not essential role of CD151 in the gamete interaction in human [29]. The CD151 molecule was detected on ZP-intact human oocytes, where it was evenly distributed in PM. However, on ZP-free oocytes, CD151 formed patches together with α6β1 integrin. Interestingly, the addition of anti-CD9 antibody during ZP removal inhibited the formation of these patches involved in gamete fusion in human [29]. In mice, the CD9 antibody affects the formation of α6β1 but not CD151 patches. The authors [29] hypothesized that a well-developed hyaluronan-containing matrix in the PVS of human but not mouse oocytes [75] could attach the plasma membrane of oocyte via hyaluronan receptor CD44 [76]. Association of CD44 with tetraspanins [77] may have a stabilizing effect which is disturbed by ZP removal resulting in the elimination of proteins and vesicles from the PVS [29]. Tetraspanin CD151 is associated with various tetraspanins, integrins, or other molecules [78] within plasma membrane domains, and tetraspanin-enriched microdomains, the composition of which differ in particular cell types. Based on chemical crosslinking and stability in strong detergents, it seems that within TEMs CD151 directly associates with laminin-binding integrins [79,80,81,82]. On the cell surface, CD151 links the laminin-binding integrins to the other tetraspanins, including CD9 and CD81 which interact with their partners. CD9 and CD81 can interact with immunoglobulin superfamily proteins EWI-2 and EWI-F and link them via CD151 to α3β1 integrin [82, 83].
The participation of the CD151 in the fertilization process has also been demonstrated using a protein interaction network approach. This network described a set of possible candidates involved in human sperm–egg interaction. In addition, the network also proposed the interaction of CD9 and CD151 in the PM of the egg with integrin α3 (CD49C) in the sperm membrane and interaction of integrin α4 (CD49D) of sperm with CD81 in the oocyte [84]. These data have relevance in relation to the recently documented localization of integrins in mouse spermatozoa [85]. This could suggest that CD151 (and also CD9) could interact with integrins not only within the oocyte (cis interaction) but also in trans interaction with integrins in sperm.
CD151 in spermatozoa
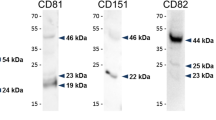
In our very recent study, we report for the first time, the presence of CD151 in the sperm of three species. This protein is expressed in germ cells during spermatogenesis and remains during epididymal transport and ejaculation of mouse and bull spermatozoa, and in human sperm a similar feature is assumed. CD151 is located in the equatorial segment, the initial fusion region of sperm that is exposed after the acrosomal exocytosis [86]. Based on the recently obtained data confirming the presence of the α6β1 and α6β4 heterodimers on mouse sperm [85] and the knowledge that CD151 share the same location in the equatorial segment with α6β4 but not with α3β1 integrin, it is possible that CD151 could stabilize the equatorial domain via α6β4 and plectin [86] surrounding the sperm nucleus [87] at least in mouse spermatozoa. Although, the pattern of CD9 and CD81 differs between mouse, bull, and human spermatozoa [62, 63, 71], CD151’s location was consistent among all the three species (Fig. 2).
Localization of tetraspanins in mouse, bull, and human spermatozoa. The diagram presents the localization of CD9 (green), CD81 (red), and CD151 (blue) in mouse, bull, and human spermatozoa before and after the acrosome reaction. Plasma membrane (PM), outer acrosomal membrane (OAM), inner acrosomal membrane (IAM), and equatorial region (ER).
CD63 in extracellular vesicles
CD63 is one of the most prominent tetraspanins present on the surface of late endosomes, multivesicular bodies (MVBs), late lysosomes, and extracellular vesicles. It was also documented in the cell membrane in smaller quantities [88].
CD63 has also been found incorporated in tetraspanin-enriched microdomains, typically consisting of tetraspanins and other molecules, such as cholesterol, integrins (β2, α4β1, α3β1, α6β1, LFA-1), and other molecules, forming an active tetraspanin web [88]. Latysheva et al. [89] identified Syntenin-1, a PDZ-domain-containing protein, as a new component of TEMs, direct partner of CD63, and referred PDZ-domain of Syntenin-1 specifically binding to C-terminus of CD63. It is supposed that the other tetraspanins also possess the potential PDZ-domain binding sites like CD81, whose C-terminus binds the proteins with PDZ-domain EBP50 (SLC9A3R1) and Sap97 (DLG1) [90]. In general, PDZ-domains containing proteins are very abundant in the cells and participate in many cellular and biological functions, especially mediating the interaction of signal transduction complexes [91].
Seeing that some of the integrins mentioned above have recently been localized in the mouse spermatozoa [85], CD63 could potentially be involved in the sperm–egg interaction by association and may possibly also cooperate with integrins.
Although Cd63 knockout mice appeared viable and fertile with no observable morphological abnormalities in the majority of tissues, this does not completely exclude the role of CD63 in fertilization processes [92]. The same authors suggested that the loss of the CD63 protein could possibly be compensated for, probably by other tetraspanins.
Yoshida et al. [93] suggested that the assurance of the reproduction cycle, certainly in mice, is backed-up by the existence of more than one pathway of sperm–egg fusion (and in all probably also by other processes), that can compensate for the impact of the absence of a single gene and thereby minimize the severity of a malfunction. This could equally be the case of mouse Cd63-knockout, Cd151-knockouts [74], and other tetraspanin-knockouts as well. The significant role in these bypassing mechanisms could involve only tetraspanin proteins; this can be illustrated by recovered fusibility of Cd81-deficient mouse eggs via forced CD9-expression (CD9 mRNA-injection) [70].
The EVs’ function involves discarding unnecessary cellular proteins; transporting proteins and lipids from the cell of origin to the recipient cell; efficient transfer of mRNA and miRNA to selective targets; activation of T cell responses through antigen transport, and suppressing the immune system and enhancing angiogenesis in tumor formation [88, 94]. To date, the functional role of EVs in mammalian reproduction has not been fully clarified. In general, it is proposed that their function is to ensure the intercellular communication in the reproductive tract and many studies documented their participation in processes related to gamete maturation, fertilization, polyspermy prevention, and embryo implantation [3, 95].
EVs (carrying the CD63 molecule) were detected in the seminal plasma of boar [96,97,98] and bull [99], in ovarian follicular fluid in horse [100], cattle [101], and human [102], in uterine luminal fluid of human [103] and sheep [104], in human blastocoel fluid [105] and interestingly, in non-mammalian animals, such as seminiferous tubules and ductuli efferentes of turtle [106, 107] and oviduct of hen [59]. The sperm storage tubules of the hen oviduct are responsible for prolonged spermatozoa storage while maintaining the vitality and fertilizing potential of the sperm. After in vitro insemination, it was observed that there was a decrease of CD63 protein in the sperm storage tubules (SST) cells containing sperm, compared to SST cells without sperm. This could indicate that the CD63-positive EVs secreted by SST cells help mediate a beneficial effect on the sperm stored in the oviduct [108]. Secretion of EVs with CD63 was also observed in human [30] and bovine [57] embryos.
CD63-positive EVs were also suggested to play a role in fertilization, egg implantation, and mother–embryo communication or endometrial embryo cross-talk [104, 109]. At the moment of ejaculation, the seminal fluid carries not only spermatozoa and soluble signaling molecules, but also EVs from the entire male reproductive tract (including epididymosomes and prostasomes) [95]. These components have a complex effect on the female reproductive tract and prime the immune response, thereby affecting the success of egg implantation and female immune response [109].
There are various criteria used to distinguish between different EVs subpopulations. The most common one (also used in the methods of isolation), is the size. Exosomes are smaller EVs of 40–120 nm, while microvesicles are considered to be larger (120–1000 nm) [98]. Regarding the mechanism of formation, microvesicles bud directly out of the plasma membrane, whereas exosomes bud inward into the late endosomes, forming multivesicular bodies inside the cell. When this multivesicular body fuses with the plasma membrane, exosomes are released in a burst [3]. Differences were described between the EVs populations in terms not only of size and mechanisms of formation but also distribution of individual tetraspanins on their surface. Various types of tetraspanin distribution were described in the subpopulations present in porcine seminal plasma by Barranco et al. [98]. While CD9 and CD63 were observed to be mostly expressed on microvesicles, CD81 was more abundant on exosomes [98]. The different expression of individual tetraspanins on EVs surface could reflect their specific roles in the reproductive tract and on fertilization, and also serve as a marker outlining the physiological and pathophysiological processes [97, 98, 110]. These findings could also have practical implications in farm animal breeding, e.g., pigs and cattle, as these EVs were found to bind to the spermatozoa [99] and promote their maturation and long-term viability [107] or to prevent early capacitation [96]. Our preliminary results showed a positive CD63 signal detected by immunohistochemistry, using a polyclonal antibody against CD63 on permeabilized smears of ejaculated frozen-thawed bull sperm. A signal for CD63 was identified in the sperm equatorial region which is commonly associated with the process of sperm–egg interaction and fusion. No significant change was detected after in vitro capacitation of the frozen-thawed sperm, but after inducing an in vitro acrosome reaction, there was either a weak signal or no signal detected. This dynamic change of the tetraspanin CD63 profile on bull sperm could indicate that CD63 plays an unidentified role in mammalian fertilization [111].
Summarized data of presence and localisation of CD9, CD81, CD151, and CD63 in germ cells, gametes, and embryos of mammals
Taken in their totality, tetraspanins were shown to be present in bovine, porcine, mouse, and human gametes and in other mammalian species. However, observations sometimes differed considerably from what could result from a distinct experimental approach and moreover, the obvious species-specific traits of tetraspanin expression and thereby their role in the fertilization process certainly exists. All available data regarding the tetraspanin expression in mammalian gametes and their supposed role is shown in Tables 1, 2.
Other tetraspanins in reproductive tract and gametes
In addition to the tetraspanins, which are known and heavily studied in the reproductive tract and gametes and were described in the previous chapters, this section focused on the tetraspanins less frequently studied in relation to reproduction.
CD82/TSPAN27 was detected in the reproductive tissue of mice, in particular on ciliated epithelia cell and epithelium of epididymis [120]. Risinger et al. [121] described that Cd82−/− mice were normally fertile and have an average litter size. The effect on reproduction has not been demonstrated conclusively. There is no data on the expression of this molecule on gametes. Other tetraspanins such as CD53 was found in transcriptomic profile of human spermatozoa or TSPAN12 has been classified to the transcript group of spermatozoa that were obtained from men whose partners did not achieve pregnancy after intrauterine insemination. The role of this molecule is not clarified; it may possibly be an infertility marker [122]. Expression of four proteins from the tetraspanin family TSPAN1-4 was observed in human reproductive tracts, especially in prostate (TSPAN1) and uterus (TSPAN1-4) [123]. The TSPAN6 gene has been differentially expressed between fertile and infertile individuals. Additionally, TSPAN6 was in the list of transcripts that were down-regulated in the asthenozoospermic infertile group compared to normozoospermic infertile men [124]. Assou et al. [125] reported the whole genome transcriptome of human cumulus oophorus. In addition to CD81, CD63, and CD151 molecules, the other tetraspanin, TM4SF8/Tspan3, has been found to overexpress in cumulus oophorus cells.
Two uroplakin-type proteins, tetraspanins UPIa/TSPAN21 and UPIb/TSPAN20, have been described in mouse and human oocytes, where these proteins form a heterotetramer complex with other uroplakins, UPII and UPIIIa. The UPIa/TSPAN21 was co-localized with molecule CD9 in mouse oocytes [126]. Similarly, tetraspanin uroplakins have been previously described in ovaries of Xenopus laevis [127]. In mice, the rate of fertilization was reduced in the IVF assay with oocytes incubated with an antibody against UPIa and UPIb tetraspanins [126]. The same authors described UPIb/TSPAN20 in the male reproductive tract of mice, in prostate and epididymis. Additionally, mRNA coding of both tetraspanin uroplakins were found in the ovary in a huge amount, while in testis, UPIa and UPIb gene expression has been recorded to a much lesser extent. UPIa/TSPAN21 and UPIb/TSPAN20 were also found in the head of spermatids in mouse testis and mature spermatozoa, mainly associated with hook. The uroplankin-enriched domain is situated in a convex part of acrosome; therefore the uroplakins presumably have no effect on sperm-egg interaction. The role of uroplakins is possibly structural, generating or maintaining the hook arrangement [126].
Schuster et al. [128] created a database (https://spermbase.org/index.php), which is dedicated to the sperm RNA expression profiling for four species (rat, mouse, human, and rabbit). Sperm-borne RNA affected the male germ cell development, fertilization and early development, and epigenetic transgenerational inheritance. Tetraspanins found in the database of sperm-borne RNA sequences (Supplementary Table 1) could be a potential topic for future studies on the participation of these molecules in the reproductive process.
CD9, CD81, CD151, and CD63 in pathologies associated with the reproductive tract
Many studies describe the expression of tetraspanins in relation to tumor stage, tumor type, growth, migration, invasion, and metastasis of tumor cells [13]. These studies have reported association of CD151 with human breast cancer [129, 130] and together with CD63 with cervical cancer, caused by persistent infection with high-risk human papillomaviruses (HPV) in most cases [131]. Several tetraspanins have been studied in connection with prostate cancer [14, 132,133,134]. Changes in CD9 and CD151 expression were observed in relation to the tumor growth and metastasis. Simultaneously, a decrease of CD9 level and an increase of CD151 level could be considered as a prognostic marker [132, 133]. In one of the recent studies, it was observed that the expression of CD151 and CD9 on extracellular vesicles of prostate cells was related to the prostate cancer. The alteration of these molecules on prostate cells changed the proteome of their EVs. These changes resulted in an increase of the invasive and migratory capabilities of the non-tumorigenic population of prostate cells, thus promoting activation of the metastatic potential of these cells [134]. Several studies also suggest the possible involvement of CD63- and CD81-positive vesicles in prostate cancer. Logozzi et al. [135] showed an increased level of EVs expressing both CD81 and prostate-specific antigen in the plasma of prostate cancer patients. However, in a later study, Padda et al. [136] did not confirm these findings but detected CD9-positive EVs.
Tetraspanin CD151 is also associated with a serious pig disease, the porcine viral reproductive and respiratory syndrome (PRRSV). This RNA virus, as the name suggests, causes several reproductive and respiratory diseases [137]. In the case of reproduction, PRRSV is manifested by an increased number of abortions in higher stages of gravidity, with the birth of dead fetuses (50–70%) and premature farrowing birth. In boars, the disease could be manifested by a temporary deterioration of sperm quality, reduction of their number, and altering their motility [138]. CD151 was identified as a key molecule of susceptibility to PRRSV infection. CD151 facilitated virus entry into host cells due to specific interaction with the 3′-untranslated region of PRRSV RNA. The proposed role of CD151 was based on the ability to bind RNA, and possibly in the location of ribonucleoprotein complexes at the site of viral replication [139]. The mechanism of virus entry into cells via the low pH-dependent endocytic pathway [140] and the specific role of CD151 in endocytosis was presumed previously [141]. A similar mechanism of viral infection through cell entry involving CD151 during endocytosis was described for human papillomavirus or cytomegalovirus. Their virions interact with receptors in tetraspanin-enriched microdomains and penetrate cell by fusion at the PM or via endocytic vesicles [142].
CD63 is associated with various pathologies. Its expression was increased in patients with endometriosis, where it might be responsible for survival promotion of ectopic endometrial cells, leading to endometriotic lesions. The expression of CD63 was elevated only in the proliferative phase of the menstrual cycle of patients with endometriosis [143]. According to Menon et al. [144], CD63-positive EVs could serve as biomarkers for possible pregnancy complications, such as pre-term birth or premature rupture of membranes. It is interesting that the topical application of CD63-positive mesenchymal stem cell exosomes to the endometrium of rats with intrauterine adhesions (one of the important causes in human female infertility), promoted endometrial regeneration, fertility restoration, collagen remodeling, and maintained normal uterine structure. This fact can point to the therapeutic effect of EVs not originating from the reproductive tract [145]. In the male reproductive tract, CD63-positive EVs enable correct sperm maturation and prevent pathological events such as early capacitation [96].
Concluding remarks
Tetraspanins, namely, CD9, CD81, CD151, and CD63 play an indispensable, direct or indirect, role in gamete quality, their interaction and fusion during fertilization as well as early embryogenesis. Tetraspanins are also reported to be involved in illnesses such as cancer related to the male and female reproductive tract, and they can serve as potential diagnostic or possibly therapeutic markers. Our review summarizes the present knowledge on CD9, CD81, CD151, and CD63 detection and localization in spermatozoa, oocytes, embryos, as well as their possible function in the reproduction of mammals and reproductive related pathologies. Although we focused only on 4 of 33 tetraspanins expressed in mammals, the role of the other members of this superfamily cannot be excluded without thorough examination.
Change history
20 June 2020
A Correction to this paper has been published: https://doi.org/10.1007/s00430-020-00686-y
References
Rubinstein E, Ziyyat A, Wolf JP, Le Naour F, Boucheix C (2006) The molecular players of sperm–egg fusion in mammals. Semin Cell Dev Biol 17:254–263. https://doi.org/10.1016/j.semcdb.2006.02.012
Barraud-Lange V, Boucheix C (2013) The role of tetraspanin complexes in egg-sperm fusion. In: Berditchevski F, Rubinstein E (eds) Tetraspanins proteins and cell regulation 9. Springer, Dordrecht, pp 203–231
Machtinger R, Laurent LC, Baccarelli AA (2016) Extracellular vesicles: roles in gamete maturation, fertilization and embryo implantation. Hum Reprod Update 22:182–193. https://doi.org/10.1093/humupd/dmv055
Hemler ME (2001) Specific tetraspanin functions. J Cell Biol 155:1103–1108. https://doi.org/10.1083/jcb.200108061
Berditchevski F, Odintsova E, Sawada S, Gilbert E (2002) Expression of the palmitoylation-deficient CD151 weakens the association of α3β1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J Biol Chem 277:36991–37000. https://doi.org/10.1074/jbc.M205265200
Charrin S, Manié S, Oualid M, Billard M, Boucheix C, Rubinstein E (2002) Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett 516:139–144. https://doi.org/10.1016/S0014-5793(02)02522-X
Yang X, Claas C, Kraeft SK, Chen LB, Wang Z, Kreidberg JA, Hemler ME (2002) Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol Biol Cell 13:767–781. https://doi.org/10.1091/mbc.01-05-0275
Boucheix C, Rubinstein E (2001) Tetraspanins. Cell Mol Life Sci CMLS 58:1189–1205. https://doi.org/10.1007/PL00000933
Huang S, Yuan S, Dong M, Su J, Yu C, Shen Y, Xie X, Yu Y, Yu X, Chen S, Zhang S, Pontarotti P, Xu A (2005) The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics 86:674–684. https://doi.org/10.1016/j.ygeno.2005.08.004
van Spriel AB, Figdor CG (2010) The role of tetraspanins in the pathogenesis of infectious diseases. Microbes Infect 12:106–112. https://doi.org/10.1016/j.micinf.2009.11.001
Martin F, Roth DM, Jans DA, Pouton CW, Partridge LJ, Monk PN, Moseley GW (2005) Tetraspanins in viral infections: a fundamental role in viral biology? J Virol 79:10839–10851. https://doi.org/10.1128/JVI.79.17.10839-10851.2005
Florin L, Lang T (2018) Tetraspanin assemblies in virus infection. Front Immunol 9:1140. https://doi.org/10.3389/fimmu.2018.01140
Hemler ME (2014) Tetraspanin proteins promote multiple cancer stages. Nat Rev Cancer 14:49–60. https://doi.org/10.1038/nrc3640
Detchokul S, Williams ED, Parker MW, Frauman AG (2014) Tetraspanins as regulators of the tumour microenvironment: implications for metastasis and therapeutic strategies. Br J Pharmacol 171:5462–5490. https://doi.org/10.1111/bph.12260
Yanagimachi R (1994) Mammalian fertilization. In: Knobil E, Neill JD (eds) The physiology of reproduction. Raven Press, New York, pp 189–317
Jin M, Fujiwara E, Kakiuchi Y, Okabe M, Satouh Y, Baba SA, Chiba K, Hirohashi N (2011) Most fertilizing mouse spermatozoa begin their acrosome reaction before contact with the zona pellucida during in vitro fertilization. Proc Natl Acad Sci USA 108:4892–4896. https://doi.org/10.1073/pnas.1018202108
Austin CR (1951) Observations on the penetration of the sperm into the mammalian egg. Aust J Biol Sci 4:581–596. https://doi.org/10.1071/bi9510581
Chang MC (1951) Fertilizing capacity of spermatozoa deposited into the fallopian tubes. Nature 168:697–698. https://doi.org/10.1038/168697b0
Chang H, Suarez SS (2011) Two distinct Ca2+ signaling pathways modulate sperm flagellar beating patterns in mice. Biol Reprod 85:296–305. https://doi.org/10.1095/biolreprod.110.089789
Florman HM, Fissore RA (2015) Fertilization in Mammals. In: Plant TM, Zeleznik AJ (eds) Knobil and Neill’s physiology of reproduction, 4th edn. Academic Press, San Diego, pp 149–196
Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, Suzuki K, Kosai K, Inoue K, Ogura A, Okabe M, Mekada E (2000) Requirement of CD9 on the egg plasma membrane for fertilization. Science 287:321–324. https://doi.org/10.1126/science.287.5451.321
Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C (2000) Severely reduced female fertility in CD9-deficient mice. Science 287:319–321. https://doi.org/10.1126/science.287.5451.319
Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, Tada N, Miyazaki S, Kudo A (2000) The gamete fusion process is defective in eggs of CD9-deficient mice. Nat Genet 24:279–282. https://doi.org/10.1038/73502
Kaji K, Oda S, Miyazaki S, Kudo A (2002) Infertility of CD9-deficient mouse eggs is reversed by mouse CD9, human CD9, or mouse CD81; polyadenylated mRNA injection developed for molecular analysis of sperm–egg fusion. Dev Biol 247:327–334. https://doi.org/10.1006/dbio.2002.0694
Zhu GZ, Miller BJ, Boucheix C, Rubinstein E, Liu CC, Hynes RO, Myles DG, Primakoff P (2002) Residues SFQ (173–175) in the large extracellular loop of CD9 are required for gamete fusion. Development 129:1995–2002
Chen MS, Tung KSK, Coonrod SA, Takahashi Y, Bigler D, Chang A, Yamashita Y, Kincade PW, Herr JC, Whiteet JM (1999) Role of the integrin-associated protein CD9 in binding between sperm ADAM 2 and the egg integrin α6β1: Implications for murine fertilization. Proc Natl Acad Sci 96:11830–11835. https://doi.org/10.1073/pnas.96.21.11830
Runge KE, Evans JE, He ZY, Gupta S, McDonald KL, Stahlberg H, Primakoff P, Mylesa DG (2007) Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Dev Biol 304:317–325. https://doi.org/10.1016/j.ydbio.2006.12.041
Coskun S, Elnour A, Hellani A, Gaafar T (2003) CD9 is expressed on human oocytes. Fertil Steril 80:268. https://doi.org/10.1016/S0015-0282(03)01678-9
Ziyyat A, Rubinstein E, Monier-Gavelle F, Barraud V, Kulski O, Prenant M, Boucheix C, Bomsel M, Wolf JP (2006) CD9 controls the formation of clusters that contain tetraspanins and the integrin α6β1, which are involved in human and mouse gamete fusion. J Cell Sci 119:416–424. https://doi.org/10.1242/jcs.02730
Giacomini E, Vago R, Sanchez AM, Podini P, Zarovni N, Murdica V, Rizzo R, Bortolotti D, Candiani M, Viganò P (2017) Secretome of in vitro cultured human embryos contains extracellular vesicles that are uptaken by the maternal side. Sci Rep 7:1–13. https://doi.org/10.1038/s41598-017-05549-w
Vyas P, Balakier H, Librach CL (2019) Ultrastructural identification of CD9 positive extracellular vesicles released from human embryos and transported through the zona pellucida. Syst Biol Reprod Med. https://doi.org/10.1080/19396368.2019.1619858
Li YH, Hou Y, Ma W, Yuan JX, Zhang D, Sun QY, Wang WH (2004) Localization of CD9 in pig oocytes and its effects on sperm–egg interaction. Reproduction 127:151–157. https://doi.org/10.1530/rep.1.00006
Airuungowa WJ, Uyhan U, Shi Z, Zamgaa O, Enkhmaart YW, Wuri L, Cui Y, Nasanochir N, Feng JS (2016) Localization of CD9 on sheep oocytes and early embryos. Int J Clin Exp Me 9:7996–8004
Pan Y, Wang M, Baloch AR, Zhang Q, Wang J, Ma R, Xu G, Kashif J, Wang L, Fan J, Cui Y, Set Y (2019) FGF10 enhances yak oocyte fertilization competence and subsequent blastocyst quality and regulates the levels of CD9, CD81, DNMT1, and DNMT3B. J Cell Physiol 234:17677–17689. https://doi.org/10.1002/jcp.28394
Zhou GB, Liu GS, Meng QG, Liu Y, Hou YP, Wang XX, Li N, Zhu SE (2009) Tetraspanin CD9 in bovine oocytes and its role in rertilization. J Reprod Dev 55:305–308. https://doi.org/10.1262/jrd.20099
Jankovicova J, Secova P, Manaskova-Postlerova P, Simonik O, Frolikova M, Chmelikova E, Horovska L, Michalkova K, Dvorakova-Hortova K, Antalikova J (2019) Detection of CD9 and CD81 tetraspanins in bovine and porcine oocytes and embryos. Int J Biol Macromol 123:931–938. https://doi.org/10.1016/j.ijbiomac.2018.11.161
Dusinský R, Simon M, Nouzovská D (1988) Preparation of monoclonal antibodies against cell surface antigens in cattle. Vet Med (Praha) 33:135–142
Zuidscherwoude M, Göttfert F, Dunlock VME, Figdor CG, van den Bogaart G, van Spriel AB (2015) The tetraspanin web revisited by super-resolution microscopy. Sci Rep 5:1–18. https://doi.org/10.1038/srep12201
Dominguez R (2004) Actin-binding proteins—a unifying hypothesis. Trends Biochem Sci 29:572–578. https://doi.org/10.1016/j.tibs.2004.09.004
Macaulay AD, Gilbert I, Scantland S, Fournier E, Ashkar F, Bastien A, Saadi HAS, Gagné D, Sirard MA, Khandjian ÉW, Richard FJ, Hyttel P, Robert C (2016) Cumulus cell transcripts transit to the bovine oocyte in preparation for maturation. Biol Reprod 94:16. https://doi.org/10.1095/biolreprod.114.127571
Barraud-Lange V, Naud-Barriant N, Bomsel M, Wolf JP, Ziyyat A (2007) Transfer of oocyte membrane fragments to fertilizing spermatozoa. FASEB J 21:3446–3449. https://doi.org/10.1096/fj.06-8035hyp
Miyado K, Yoshida K, Yamagata K, Sakakibara K, Okabe M, Wang X, Miyamoto K, Akutsu H, Kondo T, Takahashi Y, Ban T, Ito Ch, Toshimori K, Nakamura A, Ito M, Miyado M, Mekada E, Umezawa A (2008) The fusing ability of sperm is bestowed by CD9-containing vesicles released from eggs in mice. Proc Natl Acad Sci 105:12921–12926. https://doi.org/10.1073/pnas.0710608105
Barraud-Lange V, Chalas Boissonnas C, Serres C, Auer J, Schmitt A, Lefèvre B, Wolf JP, Ziyyat A (2012) Membrane transfer from oocyte to sperm occurs in two CD9-independent ways that do not supply the fertilising ability of Cd9-deleted oocytes. Reprod Camb Engl 144:53–66. https://doi.org/10.1530/REP-12-0040
Miller BJ, Georges-Labouesse E, Primakoff P, Myles DG (2000) Normal fertilization occurs with eggs lacking the integrin α6β1 and is Cd9-dependent. J Cell Biol 149:1289–1296. https://doi.org/10.1083/jcb.149.6.1289
Takahashi Y, Bigler D, Ito Y, White JM (2001) Sequence-specific interaction between the disintegrin domain of mouse ADAM 3 and murine eggs: role of beta1 integrin-associated proteins CD9, CD81, and CD98. Mol Biol Cell 12:809–820. https://doi.org/10.1091/mbc.12.4.809
Evans JP, Schultz RM, Kopf GS (1997) Characterization of the binding of recombinant mouse sperm fertilin α subunit to mouse eggs: evidence for function as a cell adhesion molecule in sperm–egg binding. Dev Biol 187:94–106. https://doi.org/10.1006/dbio.1997.8612
Evans JP, Kopf GS, Schultz RM (1997) Characterization of the binding of recombinant mouse sperm fertilin β subunit to mouse rggs: evidence for adhesive activity via an egg β1 integrin-mediated interaction. Dev Biol 187:79–93. https://doi.org/10.1006/dbio.1997.8611
Yamagata K, Nakanishi T, Ikawa M, Yamaguchi R, Moss SB, Okabe M et al (2002) Sperm from the calmegin-deficient mouse have normal abilities for binding and fusion to the egg plasma membrane. Dev Biol 250:348–357. https://doi.org/10.1006/dbio.2002.0803
Stein KK, Primakoff P, Myles D (2004) Sperm-egg fusion: events at the plasma membrane. J Cell Sci 117:6269–6274. https://doi.org/10.1242/jcs.01598
Jégou A, Ziyyat A, Barraud-Lange V, Perez E, Wolf JP, Pincet F, Gourier Ch (2011) CD9 tetraspanin generates fusion competent sites on the egg membrane for mammalian fertilization. Proc Natl Acad Sci 108:10946–10951. https://doi.org/10.1073/pnas.1017400108
Chalbi M, Barraud-Lange V, Ravaux B, Howan K, Rodriguez N, Soule P, Ndzoudi A, Boucheix C, Rubinstein E, Wolf JP, Ziyyat A, Perez E, Pincet F, Gourier Ch (2014) Binding of sperm protein Izumo1 and its egg receptor Juno drives CD9 accumulation in the intercellular contact area prior to fusion during mammalian fertilization. Development 141:3732–3739. https://doi.org/10.1242/dev.111534
Inoue N, Ikawa M, Isotani A, Okabe M (2005) The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 434:234–238. https://doi.org/10.1038/nature03362
Ravaux B, Favier S, Perez E, Gourier C (2018) Egg CD9 protein tides correlated with sperm oscillations tune the gamete fusion ability in mammal. J Mol Cell Biol 10:494–502. https://doi.org/10.1093/jmcb/mjy005
Glazar AI, Evans JP (2009) Immunoglobulin superfamily member IgSF8 (EWI-2) and CD9 in fertilisation: Evidence of distinct functions for CD9 and a CD9-associated protein in mammalian sperm-egg interaction. Reprod Fertil Dev 21:293–303. https://doi.org/10.1071/rd08158
Sala-Valdés M, Ailane N, Greco C, Rubinstein E, Boucheix C (2012) Targeting tetraspanins in cancer. Expert Opin Ther Targets 16:985–997. https://doi.org/10.1517/14728222.2012.712688
Umeda R, Satouh Y, Takemoto M, Nakadu-Nakura Y, Liu K, Yokoyama T, Shirouzu M, Iwata S, Nomura N, Sato K, Ikawa M, Nishizawa T, Nureki O (2020) Structural insights into tetraspanin CD9 function. Nat Commun 11:1–11. https://doi.org/10.1038/s41467-020-15459-7
Mellisho EA, Velásquez AE, Nuñez MJ, Cabezas JG, Cueto JA, Fader C, Castro FO, Rodríguez-Álvarezet L (2017) Identification and characteristics of extracellular vesicles from bovine blastocysts produced in vitro. PLoS ONE 12:e0178306. https://doi.org/10.1371/journal.pone.0178306
Marin D, Scott RT (2018) Extracellular vesicles: a promising tool for assessment of embryonic competence. Curr Opin Obstet Gynecol 30:171–178. https://doi.org/10.1097/GCO.0000000000000458
Kanatsu-Shinohara M, Toyokuni S, Shinohara T (2004) CD9 is a surface marker on mouse and rat male germline stem cells. Biol Reprod 70:70–75. https://doi.org/10.1095/biolreprod.103.020867
Kierszenbaum AL, Rosselot C, Rivkin E, Tres LL (2006) Role of integrins, tetraspanins, and ADAM proteins during the development of apoptotic bodies by spermatogenic cells. Mol Reprod Dev 73:906–917. https://doi.org/10.1002/mrd.20470
Ito C, Yamatoya K, Yoshida K, Maekawa M, Miyado K, Toshimori K (2010) Tetraspanin family protein CD9 in the mouse sperm: unique localization, appearance, behavior and fate during fertilization. Cell Tissue Res 340:583–594. https://doi.org/10.1007/s00441-010-0967-7
Frolikova M, Manaskova-Postlerova P, Cerny J, Jankovicova J, Simonik O, Pohlova A, Secova P, Antalikova J, Dvorakova-Hortova K (2018) CD9 and CD81 interactions and their structural modelling in sperm prior to fertilization. Int J Mol Sci 19:1236. https://doi.org/10.3390/ijms19041236
Antalíková J, Jankovičová J, Simon M, Cupperová P, Michalková K, Horovská Ľ (2015) Localization of CD9 molecule on bull spermatozoa: its involvement in the sperm–egg interaction. Reprod Domest Anim 50:423–430. https://doi.org/10.1111/rda.12508
Charrin S, Naour FL, Oualid M, Billard M, Faure G, Hanash SM, Boucheix C, Rubinstein E (2001) The major CD9 and CD81 molecular partner identification and characterization of complexes. J Biol Chem 276:14329–14337. https://doi.org/10.1074/jbc.M011297200
Sala-Valdés M, Ursa Á, Charrin S, Rubinstein E, Hemler ME, Sánchez-Madrid F, Yáñez-Mó M (2006) EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with Ezrin-Radixin-Moesin proteins. J Biol Chem 281:19665–19675. https://doi.org/10.1074/jbc.M602116200
Wang L, Chen W, Zhao C, Huo R, Guo XJ, Lin M, Huang XY, Mao YD, Zuo-Min Zhou ZM, Sha JH (2010) The role of ezrin-associated protein network in human sperm capacitation. Asian J Androl 12:667–676. https://doi.org/10.1038/aja.2010.79
Salvolini E, Buldreghini E, Lucarini G, Vignini A, Lenzi A, Di Primo R, Balercia G (2013) Involvement of sperm plasma membrane and cytoskeletal proteins in human male infertility. Fertil Steril 99:697–704. https://doi.org/10.1016/j.fertnstert.2012.10.042
The UniProt Consortium (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47:D506–D515. https://doi.org/10.1093/nar/gky1049
Rubinstein E, Ziyyat A, Prenant M, Wrobel E, Wolf JF, Levy S, Le Naour F, Boucheix C (2006) Reduced fertility of female mice lacking CD81. Dev Biol 290:351–358. https://doi.org/10.1016/j.ydbio.2005.11.031
Ohnami N, Nakamura A, Miyado M, Sato M, Kawano N, Yoshida K, Harada Y, Takezawa Y, Kanai S, Ono Ch, Takahashi Y, Kimura K, Shida T, Miyado K, Umezawa A (2012) CD81 and CD9 work independently as extracellular components upon fusion of sperm and oocyte. Biol Open 1:640–647. https://doi.org/10.1242/bio.20121420
Jankovicova J, Frolikova M, Sebkova N, Simon M, Cupperova P, Lipcseyova D, Michalkova K, Horovska L, Sedlacek R, Stopka P, Dvorakova-Hortova AJK (2016) Characterization of tetraspanin protein CD81 in mouse spermatozoa and bovine gametes. Reproduction 152:785–793. https://doi.org/10.1530/REP-16-0304
Zimmerman B, Kelly B, McMillan BJ, Seegar TCM, Dror RO, Kruse AC, Blacklow SC (2016) Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell 167:1041–1051.e11. https://doi.org/10.1016/j.cell.2016.09.056
Gupta S, Primakoff P, Myles DG (2009) Can the presence of wild-type oocytes during insemination rescue the fusion defect of CD9 null oocytes? Mol Reprod Dev 76:602. https://doi.org/10.1002/mrd.21040
Wright MD, Geary SM, Fitter S, Moseley GW, Lau LM, Sheng KCh, Apostolopoulos V, Stanley EG, Jackson DE, Ashman LK (2004) Characterization of mice lacking the tetraspanin superfamily member CD151. Mol Cell Biol 24:5978–5988. https://doi.org/10.1128/MCB.24.13.5978-5988.2004
Talbot P, Dandekar P (2003) Perivitelline space: does it play a role in blocking polyspermy in mammals? Microsc Res Tech 61:349–357. https://doi.org/10.1002/jemt.10348
Campbell S, Swann HR, Aplin JD, Seif MW, Kimber SJ, Elstein M (1995) CD44 is expressed throughout pre-implantation human embryo development. Hum Reprod Oxf Engl 10:425–430. https://doi.org/10.1093/oxfordjournals.humrep.a135955
Jones PH, Bishop LA, Watt FM (1996) Functional significance of CD9 association with beta 1 integrins in human epidermal keratinocytes. Cell Adhes Commun 4:297–305. https://doi.org/10.3109/15419069609010773
Berditchevski F, Gilbert E, Griffiths MR, Fitter S, Ashman L, Jenner SJ (2001) Analysis of the CD151 α3β1 integrin and CD151 tetraspanin interactions by mutagenesis. J Biol Chem 276:41165–41174. https://doi.org/10.1074/jbc.M104041200
Yauch RL, Berditchevski F, Harler MB, Reichner J, Hemler ME (1998) Highly stoichiometric, stable, and specific association of integrin α3β1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol Biol Cell 9:2751–2765
Yauch RL, Kazarov AR, Desai B, Lee RT, Hemler ME (2000) Direct extracellular contact between integrin α3β1 and TM4SF protein CD151. J Biol Chem 275:9230–9238. https://doi.org/10.1074/jbc.275.13.9230
Serru V, Le Naour F, Billard M, Azorsa DO, Lanza F, Boucheix C, Rubinstein E (1999) Selective tetraspan-integrin complexes (CD81/alpha4beta1, CD151/alpha3beta1, CD151/alpha6beta1) under conditions disrupting tetraspan interactions. Biochem J 340:103–111
Stipp CS, Kolesnikova TV, Hemler ME (2003) EWI-2 regulates α3β1 integrin–dependent cell functions on laminin-5. J Cell Biol 163:1167–1177. https://doi.org/10.1083/jcb.200309113
Charrin S, Manié S, Billard M, Ashman L, Gerlier D, Boucheix C, Rubinstein E (2003) Multiple levels of interactions within the tetraspanin web. Biochem Biophys Res Commun 304:107–112. https://doi.org/10.1016/S0006-291X(03)00545-X
Sabetian S, Shamsir MS, Naser MA (2014) Functional features and protein network of human sperm-egg interaction. Syst Biol Reprod Med 60:329–337. https://doi.org/10.3109/19396368.2014.955896
Frolikova M, Valaskova E, Cerny J, Lumeau A, Sebkova N, Palenikova V, Sanchez-Hernandez N, Pohlova A, Manaskova-Postlerova P, Dvorakova-Hortova K (2019) Addressing the compartmentalization of specific integrin heterodimers in mouse sperm. Int J Mol Sci 20:1004. https://doi.org/10.3390/ijms20051004
Jankovicova J, Frolikova M, Palenikova V, Valaskova E, Cerny J, Secova P, Bartokova M, Horovska L, Manaskova-Postlerova P, Antalikovaet J, Komrskova K (2020) Expression and distribution of CD151 as a partner of alpha6 integrin in male germ cells. Sci Rep 10:1–12. https://doi.org/10.1038/s41598-020-61334-2
Kierszenbaum AL, Rivkin E, Tres LL (2007) Molecular biology of sperm head shaping. Soc Reprod Fertil Suppl 65:33–43
Pols MS, Klumperman J (2009) Trafficking and function of the tetraspanin CD63. Exp Cell Res 315:1584–1592. https://doi.org/10.1016/j.yexcr.2008.09.020
Latysheva N, Muratov G, Rajesh S, Padgett M, Hotchin NA, Overduin M, Berditchevski F (2006) Syntenin-1 is a new component of tetraspanin-enriched microdomains: mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol Cell Biol 26:7707–7718. https://doi.org/10.1128/MCB.00849-06
Pan Y, Brown C, Wang X, Geisert EE (2007) The developmental regulation of CD81 in the rat retina. Mol Vis 13:181–189
Lee H-J, Zheng JJ (2010) PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal 8:8. https://doi.org/10.1186/1478-811X-8-8
Schröder J, Lüllmann-Rauch R, Himmerkus N, Pleines I, Nieswandt B, Orinska Z, Koch-Nolte F, Schröder B, Bleich M, Saftig P (2009) Deficiency of the tetraspanin CD63 associated with kidney pathology but normal lysosomal function. Mol Cell Biol 29:1083–1094. https://doi.org/10.1128/MCB.01163-08
Yoshida K, Kawano N, Harada Y, Miyado K (2014) Role of CD9 in sperm–egg fusion and virus-induced cell fusion in mammals. In: Sawada H, Inoue N, Iwano M (eds) Sexual reproduction in animals and plants. Springer, Tokyo, pp 383–391
Rana S, Yue S, Stadel D, Zöller M (2012) Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int J Biochem Cell Biol 44:1574–1584. https://doi.org/10.1016/j.biocel.2012.06.018
Sullivan R, Saez F (2013) Epididymosomes, prostasomes, and liposomes: their roles in mammalian male reproductive physiology. Reproduction 146:R21–R35. https://doi.org/10.1530/REP-13-0058
Du J, Shen J, Wang Y, Pan C, Pang W, Diao H, Dong W (2016) Boar seminal plasma exosomes maintain sperm function by infiltrating into the sperm membrane. Oncotarget 7:58832–58847. https://doi.org/10.18632/oncotarget.11315
Alvarez-Rodriguez M, Ljunggren SA, Karlsson H, Rodriguez-Martinez H (2019) Exosomes in specific fractions of the boar ejaculate contain CD44: a marker for epididymosomes? Theriogenology 140:143–152. https://doi.org/10.1016/j.theriogenology.2019.08.023
Barranco I, Padilla L, Parrilla I, Álvarez-Barrientos A, Pérez-Patiño C, Peña FJ, Martínez EA, Rodriguez-Martínez H, Jordi Rocaet J (2019) Extracellular vesicles isolated from porcine seminal plasma exhibit different tetraspanin expression profiles. Sci Rep. https://doi.org/10.1038/s41598-019-48095-3
Pagano N, Kosior MA, Gasparrini B, Longobardi V, De Canditiis C, Albero G, Deregibus MC, Bosi G, Idda A, Consiglioet AL (2020) 148 Bull spermatozoa uptake of extracellular vesicles from bovine seminal plasma. Reprod Fertil Dev 32:200–200. https://doi.org/10.1071/RDv32n2Ab148
da Silveira JC, Veeramachaneni DNR, Winger QA, Carnevale EM, Bouma GJ (2012) Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle. Biol Reprod 86:71. https://doi.org/10.1095/biolreprod.111.093252
Sohel MH, Hoelker M, Noferesti SS, Salilew-Wondim D, Tholen E, Looft Ch, Rings F, Uddin MJ, Spencere TE, Schellander K, Tesfaye D (2013) Exosomal and non-exosomal transport of extra-cellular microRNAs in follicular fluid: implications for bovine oocyte developmental competence. PLoS ONE 8:e78505. https://doi.org/10.1371/journal.pone.0078505
Santonocito M, Vento M, Guglielmino MR, Battaglia R, Wahlgren J, Ragusa M, Barbagallo D, Borzi P, Rizzari S, Maugeri M, Scollo P, Tatone C, Valadi H, Purrello M, Di Pietro C (2014) Molecular characterization of exosomes and their microRNA cargo in human follicular fluid: bioinformatic analysis reveals that exosomal microRNAs control pathways involved in follicular maturation. Fertil Steril 102:1751–1761.e1. https://doi.org/10.1016/j.fertnstert.2014.08.005
Ng YH, Rome S, Jalabert A, Forterre A, Singh H, Hincks CL, Salamonsen LA (2013) Endometrial exosomes/microvesicles in the uterine microenvironment: a new paradigm for embryo-endometrial cross talk at implantation. PLoS ONE 8:e58502. https://doi.org/10.1371/journal.pone.0058502
Burns G, Brooks K, Wildung M, Navakanitworakul R, Christenson LK, Spencer TE (2014) Extracellular vesicles in luminal fluid of the ovine uterus. PLoS ONE 9:e90913. https://doi.org/10.1371/journal.pone.0090913
Battaglia R, Palini S, Vento ME, La Ferlita A, Lo Faro MJ, Caroppo E, Borzì P, Falzone L, Barbagallo D, Ragusa M, Scalia M, D’Amato G, Scollo P, Musumeci P, Purrello M, Gravotta E, Di Pietro C (2019) Identification of extracellular vesicles and characterization of miRNA expression profiles in human blastocoel fluid. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-018-36452-7
Tarique I, Haseeb A, Bai X, Li W, Yang P, Huang Y, Yang S, Xu M, Zhang Y, Vistro WA, Fazlani SA, Chen Q (2019) Cellular evidence of CD63-enriched exosomes and multivesicular bodies within the seminiferous tubule during the spermatogenesis of turtles. Microsc Microanal 26:148–156. https://doi.org/10.1017/S1431927619015149
Tarique I, Liu Y, Bai X, Haseeb A, Yang P, Huang Y, Qu W, Wu R, Vistro WA, Chen Q (2019) Characterization of extracellular vesicles from cilia and epithelial cells of ductuli efferentes in a turtle (Pelodiscus sinensis). Animals 9:888. https://doi.org/10.3390/ani9110888
Huang A, Isobe N, Yoshimura Y (2017) Changes in localization and density of CD63-positive exosome-like substances in the hen oviduct with artificial insemination and their effect on sperm viability. Theriogenology 101:135–143. https://doi.org/10.1016/j.theriogenology.2017.06.028
Robertson SA, Sharkey DJ (2016) Seminal fluid and fertility in women. Fertil Steril 106:511–519. https://doi.org/10.1016/j.fertnstert.2016.07.1101
Caballero JN, Frenette G, Belleannée C, Sullivan R (2013) CD9-positive microvesicles mediate the transfer of molecules to bovine spermatozoa during epididymal maturation. PLoS ONE 8:e65364. https://doi.org/10.1371/journal.pone.0065364
Jankovičová J, Bartóková M, Horovská Ľ et al (2019) Poster: detection of cluster of differentiation molecule 63 in bull sperm. Reprod Dom Anim 54:110–111
Liu WM, Cao YJ, Yang YJ, Hu Z, Duan EK (2006) Tetraspanin CD9 regulates invasion during mouse embryo implantation. J Mol Endocrinol 36:121–130. https://doi.org/10.1677/jme.1.01910
Yubero N, Jiménez-Marín Á, Lucena C, Barbancho M, Garrido JJ (2010) Immunohistochemical distribution of the tetraspanin CD9 in normal porcine tissues. Mol Biol Rep 38:1021–1028. https://doi.org/10.1007/s11033-010-0198-8
Saadeldin IM, Kim SJ, Choi YB, Lee BC (2014) Improvement of cloned embryos development by co-culturing with parthenotes: a possible role of exosomes/microvesicles for embryos paracrine communication. Cell Reprogr 16:223–234. https://doi.org/10.1089/cell.2014.0003
Zhou GB, Zeng Y, Meng QG, Liu Y, Dai YP, Zhu SE, Bunch TD, Hou YP (2013) Decreased expression of CD9 in bovine oocytes after cryopreservation and the relationship to fertilization capacity. Mol Reprod Dev 80:451–459. https://doi.org/10.1002/mrd.22181
Qu P, Qing S, Liu R, Qin H, Wang W, Qiao F, Ge H, Liu J, Zhang Y, Cui W, Wang Y (2017) Effects of embryo-derived exosomes on the development of bovine cloned embryos. PLoS ONE 12:e0174535. https://doi.org/10.1371/journal.pone.0174535
Tanigawa M, Miyamoto K, Kobayashi S, Sato M, Akutsu H, Okabe M, Mekada E, Sakakibara K, Miyado M, Umezawa A, Miyado K (2008) Possible involvement of CD81 in acrosome reaction of sperm in mice. Mol Reprod Dev 75:150–155. https://doi.org/10.1002/mrd.20709
Tres LL, Kierszenbaum AL (2005) The ADAM-integrin-tetraspanin complex in fetal and postnatal testicular cords. Birth Defects Res Part C Embryo Today Rev 75:130–141. https://doi.org/10.1002/bdrc.20041
Kaewmala K, Uddin MJ, Cinar MU, Große-Brinkhaus Ch, Jonas E, Tesfaye D, Phatsara Ch, Tholen E, Looft Ch, Schellander K (2011) Association study and expression analysis of CD9 as candidate gene for boar sperm quality and fertility traits. Anim Reprod Sci 125:170–179. https://doi.org/10.1016/j.anireprosci.2011.02.017
Custer MC, Risinger JI, Hoover S, Simpson RM, Patterson T, Barrett JC (2006) Characterization of an antibody that can detect the Kai1/CD82 murine metastasis suppressor. Prostate 66:567–577. https://doi.org/10.1002/pros.20386
Risinger JI, Custer M, Feigenbaum L, Simpson RM, Hoover SB, Webster JD, Chandramouli GVR, Tessarollo L, Barrett JC (2014) Normal viability of Kai1/Cd82 deficient mice. Mol Carcinog 53:610–624. https://doi.org/10.1002/mc.22009
García-Herrero S, Meseguer M, Martínez-Conejero JA, José Remohí J, Pellicer A, Garrido N (2010) The transcriptome of spermatozoa used in homologous intrauterine insemination varies considerably between samples that achieve pregnancy and those that do not. Fertil Steril 94:1360–1373. https://doi.org/10.1016/j.fertnstert.2009.07.1671
Todd SC, Doctor VS, Levy S (1998) Sequences and expression of six new members of the tetraspanin/TM4SF family. Biochim Biophys Acta 1399:101–104. https://doi.org/10.1016/s0167-4781(98)00087-6
Bansal SK, Gupta N, Sankhwar SN, Rajender S (2015) Differential genes expression between fertile and infertile spermatozoa revealed by transcriptome analysis. PLoS ONE 10:e0127007. https://doi.org/10.1371/journal.pone.0127007
Assou S, Anahory T, Pantesco V, Le Carrour T, Pellestor F, Klein B, Reyftmann L, Dechaud H, De Vos J, Hamamah S (2006) The human cumulus–oocyte complex gene-expression profile. Hum Reprod 21:1705–1719. https://doi.org/10.1093/humrep/del065
Liao Y, Chang HC, Liang FX, Chung PJ, Wei Y, Nguyen TP, Zhou G, Talebian S, Lewis C, Krey LC, Fang-Ming Deng FM, Wong TW, Chicote JU, Grifo JA, Keefe DL, Shapiro E, Lepor H, Wu XR, DeSalle R, Garcia-España A, Kim SY, Sun TT (2018) Uroplakins play conserved roles in egg fertilization and acquired additional urothelial functions during mammalian divergence. Mol Biol Cell 29:3128–3143. https://doi.org/10.1091/mbc.E18-08-0496
Garcia-España A, Chung P-J, Zhao X, Lee A, Pellicer A, Yu J, Sun TT, DeSalle R (2006) Origin of the tetraspanin uroplakins and their co-evolution with associated proteins: implications for uroplakin structure and function. Mol Phylogenet Evol 41:355–367. https://doi.org/10.1016/j.ympev.2006.04.023
Schuster A, Tang C, Xie Y, Ortogero N, Yuan S, Yan W (2016) SpermBase: a database for sperm-borne RNA contents. Biol Reprod 95:1–12. https://doi.org/10.1095/biolreprod.116.142190
Yang XH, Richardson AL, Torres-Arzayus MI, Zhou P, Sharma Ch, Kazarov AR, Andzelm SJL, Brown M, Hemler ME (2008) CD151 accelerates breast cancer by regulating α6 integrin function, signaling, and molecular organization. Cancer Res 68:3204–3213. https://doi.org/10.1158/0008-5472.CAN-07-2949
Sadej R, Romanska H, Baldwin G, Gkirtzimanaki K, Novitskaya V, Filer AD, Krcova Z, Kusinska R, Ehrmann J, Buckley ChD, Kordek R, Potemski P, Eliopoulos AG, Lalani EN, Berditchevski F (2009) CD151 regulates tumorigenesis by modulating the communication between tumor cells and endothelium. Mol Cancer Res 7:787–798. https://doi.org/10.1158/1541-7786.MCR-08-0574
Scheffer KD, Berditchevski F, Florin L (2014) The tetraspanin CD151 in papillomavirus infection. Viruses 6:893–908. https://doi.org/10.3390/v6020893
Ang J, Lijovic M, Ashman LK, Ang J, Lijovic M, Ashman LK, Kan K, Frauman AG (2004) CD151 protein expression predicts the clinical outcome of low-grade primary prostate cancer better than histologic grading: a new prognostic indicator? Cancer Epidemiol Prev Biomark 13:1717–1721
Wang JC, Bégin LR, Bérubé NG, Chevalier S, Aprikian AG, Gourdeau H, Chevrette M (2007) Down-regulation of CD9 expression during prostate carcinoma progression is associated with CD9 mRNA modifications. Clin Cancer Res 13:2354–2361. https://doi.org/10.1158/1078-0432.CCR-06-1692
Brzozowski JS, Bond DR, Jankowski H et al (2018) Extracellular vesicles with altered tetraspanin CD9 and CD151 levels confer increased prostate cell motility and invasion. Sci Rep 8:1–13. https://doi.org/10.1038/s41598-018-27180-z
Logozzi M, Angelini DF, Iessi E, Mizzoni D, Raimo RD, Federici C, Lugini L, Borsellino G, Gentilucci A, Pierella F, Marzio V, Sciarra A, Battistini L, Fais S (2017) Increased PSA expression on prostate cancer exosomes in in vitro condition and in cancer patients. Cancer Lett 403:318–329. https://doi.org/10.1016/j.canlet.2017.06.036
Padda RS, Deng FK, Brett SI, Biggs CN, Durfee PN, Brinker ChJ, Williams KC, Leong HS (2019) Nanoscale flow cytometry to distinguish subpopulations of prostate extracellular vesicles in patient plasma. Prostate 79:592–603. https://doi.org/10.1002/pros.23764
Goyal SM (1993) Porcine reproductive and respiratory syndrome. J Vet Diagn Investig Off Publ Am Assoc Vet Lab Diagn Inc 5:656–664. https://doi.org/10.1177/104063879300500435
de Jong M, Cromvijk W, Cromvijk P (1991) The new pig dis-ease-epidemiology and production losses in the Netherlands. Comm Eur Commun 1:9–19
Shanmukhappa K, Kim JK, Kapil S (2007) Role of CD151, a tetraspanin, in porcine reproductive and respiratory syndrome virus infection. Virol J 4:62. https://doi.org/10.1186/1743-422X-4-62
Kreutz LC, Ackermann MR (1996) Porcine reproductive and respiratory syndrome virus enters cells through a low pH-dependent endocytic pathway. Virus Res 42:137–147. https://doi.org/10.1016/0168-1702(96)01313-5
Sincock PM, Fitter S, Parton RG, Berndt MC, Gamble JR, Ashman LK (1999) PETA-3/CD151, a member of the transmembrane 4 superfamily, is localised to the plasma membrane and endocytic system of endothelial cells, associates with multiple integrins and modulates cell function. J Cell Sci 112:833–844
Fast LA, Lieber D, Lang T, Florin L (2017) Tetraspanins in infections by human cytomegalo- and papillomaviruses. Biochem Soc Trans 45:489–497. https://doi.org/10.1042/BST20160295
Gomes VA, de Moraes BC, Rosa-e-Silva JC, de Paz CCP, Ferriani RA, Meola J (2018) The apoptotic, angiogenic and cell proliferation genes CD63, S100A6 e GNB2L1 are altered in patients with endometriosis. Rev Bras Ginecol E Obstetrícia RBGO Gynecol Obstet 40:606–613. https://doi.org/10.1055/s-0038-1673364
Menon R, Dixon CL, Sheller-Miller S, Fortunato SJ, Saade GR, Palma C, Lai A, Guanzon D, Salomon C (2019) Quantitative proteomics by SWATH-MS of maternal plasma exosomes determine pathways associated with term and preterm birth. Endocrinology 160:639–650. https://doi.org/10.1210/en.2018-00820
Zhao S, Qi W, Zheng J, Tian Y, Qi X, Kong D, Zhang J, Huang X (2020) Exosomes derived from adipose mesenchymal stem cells restore functional endometrium in a rat model of intrauterine adhesions. Reprod Sci. https://doi.org/10.1007/s43032-019-00112-6
Acknowledgements
This work was supported by the Scientific Grant Agency of the Ministry of Education, Science, Research, and Sport of the Slovak Republic and the Slovak Academy of Sciences (VEGA-2/0027/20), by the Slovak Research and Development Agency (APVV-15-0196), bilateral project SAS-CAS (18-17); by the Internal Grant Agency of Czech University of Life Sciences in Prague (SV18-08-21230); by the CellFit COST Action CA16119, Ministry of Education, Youth and Sports, Czech Republic INTER-COST LTC 18059; by the Grant Agency of the Czech Republic No. GA-18-11275S; by the project BIOCEV (CZ.1.05/1.1.00/02.0109) from the ERDF; and by the Institutional support of the Institute of Biotechnology RVO: 86652036.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Edited by Charlotte M de Winde.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Special Issue on Tetraspanins in Infection and Immunity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jankovičová, J., Neuerová, Z., Sečová, P. et al. Tetraspanins in mammalian reproduction: spermatozoa, oocytes and embryos. Med Microbiol Immunol 209, 407–425 (2020). https://doi.org/10.1007/s00430-020-00676-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00430-020-00676-0