Abstract
Since the introduction of the zebrafish as a model for the study of vertebrate developmental biology, an extensive array of techniques for its experimental manipulation and analysis has been developed. Recently it has become apparent that these powerful methodologies might be deployed in order to elucidate the pathogenesis of human neurodegenerative diseases and to identify candidate therapeutic approaches. In this article, we consider evidence that the zebrafish central nervous system provides an appropriate setting in which to model human neurological disease and we review techniques and resources available for generating transgenic models. We then examine recent publications showing that appropriate phenotypes can be provoked in the zebrafish through transgenic manipulations analogous to genetic abnormalities known to cause human tauopathies, polyglutamine diseases or motor neuron degenerations. These studies show proof of concept that findings in zebrafish models can be applicable to the pathogenic mechanisms underlying human diseases. Consequently, the prospects for providing novel insights into neurodegenerative diseases by exploiting transgenic zebrafish models and discovery-driven approaches seem favorable.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since the initial introduction of the zebrafish as a model organism for the study of vertebrate development, an impressive toolbox of experimental techniques for their experimental manipulation and analysis has been developed. Zebrafish embryos develop externally and are optically transparent, allowing direct observation of development and the deployment of fluorescent reporters and indicators to visualize morphology and physiology of cell groups of interest. Transgenic expression of exogenous genes, and experimental knockdown of endogenous genes, can be carried out using relatively straightforward techniques. Zebrafish breed regularly, produce large clutches of offspring and can be housed practicably in large numbers allowing for large-scale genetic and chemical screens. By exploiting these favorable properties, the zebrafish model has proved a useful means to test hypotheses concerning gene function in development, and has provided numerous novel insights into the molecular basis of embryogenesis through large-scale genetics screens (Solnica-Krezel et al. 1994; Brockerhoff et al. 1995; Driever et al. 1996; Malicki et al. 1996; Amsterdam et al. 1999; Guo et al. 1999). More recently, in vivo chemical modifier screens have been carried out in order to identify new compounds that act specifically on biological pathways of interest (Zon and Peterson 2005; Molina et al. 2009). The ability to house zebrafish larvae in 96-well plates and the use of automated assay end points make the zebrafish uniquely suitable amongst current vertebrate models for high throughput chemical library screening in vivo.
It has been suggested that these powerful techniques might be deployed in order both to elucidate the pathogenesis of human neurodegenerative diseases, and to isolate new treatments. The possibility of exploiting this now well-established model to accelerate the discovery of efficacious treatments for currently fatal neurological conditions is both intriguing and exciting. In this review, we first consider the evidence that the zebrafish brain provides an appropriate setting in which to model human neurological disease. We then review techniques and resources available for generating transgenic models. Finally, we examine recent publications, which provide the first proof-of-concept that appropriate phenotypes can be provoked in the zebrafish through transgenic manipulations analogous to genetic abnormalities known to cause human disease, and occurring via pathways that are conserved and relevant to the disease being modeled.
Is the zebrafish a suitable organism in which to model neurodegeneration?
Zebrafish models of neurodegeneration will be useful for understanding human diseases only if the mechanisms underlying pathogenesis are sufficiently phylogenetically conserved that insights gained in zebrafish models can be applied to the human conditions. There is no way of knowing a priori that this will be the case. However, a variety of convergent lines of evidence suggest that aspects of the zebrafish CNS pertinent to human diseases are conserved with respect to human, offering some support to the prediction that observations made in zebrafish models will be clinically applicable. In the following paragraphs, we examine the degree to which the molecular, cellular and tissue environment of the zebrafish brain mirrors that of the human, particularly with respect to our current knowledge of the pathogenesis of neurodegenerative diseases.
Structure and function of zebrafish CNS regions of relevance to human disease
The zebrafish CNS is organized similarly to that of other vertebrates, and is conventionally divided into the spinal cord, hindbrain, midbrain and forebrain. Zebrafish neuroanatomy, both during development and in the adult, has been reviewed in detail elsewhere (Kimmel 1993; Wullimann et al. 1996). There is a major difference of scale, and some striking differences of structure, between the zebrafish and human CNS. However, key areas of the zebrafish CNS relevant to modeling human diseases show remarkably conserved structure to their human counterparts. For example, similar to human cerebellar cortex, the zebrafish cerebellum has molecular, Purkinje cell and granule cell layers (Fig. 1); cell types present in the each of these laminae are similar to those found in the human brain, show similar inputs and synaptic connections, and express similar genes and specialized markers (Bae et al. 2009). The main difference between zebrafish and mammalian cerebellum is that cell bodies of output projection neurons in the zebrafish cerebellum (eurydendroid cells) are located in the cortex rather than the deep nuclei found in mammals. Other regions of the zebrafish CNS, such as the medulla, hypothalamus, optic tracts and tectum, olfactory system, spinal cord and cranial nerves show easily recognizable structural homology to the relevant areas of the human brain. Interpretation of anatomical homology to human forebrain regions of interest to disease is, however, complicated by both eversion of the developing telencephalon (in contrast to the evagination seen in mammals) and the smaller less well-developed nature of the zebrafish forebrain in comparison with other brain regions. Areas of the zebrafish telencephalon that are thought homologous to regions of the basal ganglia involved in human motor degenerations have been identified through studies of gene expression, neurochemistry and axonal projections (Rink and Wullimann 2001, 2004; Wullimann and Rink 2002). The dorsal nucleus of the ventral telencephalic area, arising from the embryonic subpallium, is thought to be the zebrafish homologue of the mammalian striatum (Rink and Wullimann 2004). Similar to the projection neurons of the mammalian striatum, neurons of the fish ventral telencephalic area are GABAergic, and cells in the region express substance P (Sharma et al. 1989), enkephalin (Reiner and Northcutt 1992), and D1 (Kapsimali et al. 2000) and D2 (Boehmler et al. 2004) dopamine receptors in a variety of different fish species. Furthermore, this area is rich in dopaminergic nerve terminals, derived from a major ascending dopaminergic projection (see below), further corroborating the proposed homology of the region to the striatum. Another division of the dorsal telencephalon is proposed to be homologous to the nucleus basalis of Meynert, on account of prominent choline acetyltransferase-expressing neurons and projections to the dorsal telencephalic area (Mueller et al. 2004; Rink and Wullimann 2004).
Aspects of zebrafish neuroanatomy of relevance to modeling human neurodegenerative diseases. a Shows an epifluorescence micrograph of a sagittal section through the diencephalon of an adult zebrafish brain. The section was labeled using a primary antibody to tyrosine hydroxylase (TH), demonstrating groups of dopaminergic neurons. PP preoptic complex, VT ventral thalamus, PT pretectal nucleus, TP posterior tuberal complex. b, c Show epifluorescence micrographs of sagittal sections through the cerebellum of an adult zebrafish. The sections were labeled with primary antibodies to (b) Zn8 (green; axonal marker) and DAPI (blue; nuclear counterstain) or (c) IP3R1 (red; Purkinje cell marker), in order to demonstrate the laminar arrangement of the zebrafish cerebellum. G granule cell layer, P Purkinje cell layer, M molecular layer. Arrows in c indicate individual Purkinje cell bodies
The dopaminergic system of zebrafish is of significant interest to the development of models of human motor degenerations, because of the central role played by degeneration of dopamine neurons in the clinical expression of akinetic-rigid movement disorders such as Parkinson’s disease. The zebrafish brain contains increasingly well-characterized groups of dopaminergic neurons, located in the olfactory bulbs, telencephalon, pretectal area and ventral diencephalon (Fig. 1) (Rink and Wullimann 2002; Ma 2003; Ryu et al. 2006). Much of our current understanding of the anatomy of the zebrafish dopaminergic system is based on expression patterns of tyrosine hydroxylase (the enzyme catalyzing the rate-limiting step in dopamine biosynthesis). The recent discovery that a gene duplication event in zebrafish resulted in two separate th genes (Candy and Collet 2005) with different expression patterns (Chen et al. 2009c) may necessitate amending the current picture, since it is unclear at present which of the commonly used TH antibodies recognizes one or both isoenzymes. One of the diencephalic groups of TH-immunoreactive neurons, located in the posterior tuberculum of the hypothalamus, sends axonal projections that terminate in the dorsal telencephalic area. This is thought to be the zebrafish homologue of the mammalian nigrostriatal tract (Rink and Wullimann 2001; Fig. 1).
Similar to the situation in mammals, dopaminergic function is important in the regulation of locomotor behavior in zebrafish. Lesions of the dopaminergic system, induced by exposure to the dopamine neuron-specific toxin MPP+ are associated with reduction in spontaneous movement of zebrafish larvae (Bretaud et al. 2004; Lam et al. 2005; McKinley et al. 2005; Sallinen et al. 2009a). Similarly, exposure to agents that antagonize the actions of dopamine at its receptors also leads to decreased spontaneous larval movement (Giacomini et al. 2006; Boehmler et al. 2007). In addition, dopamine function is important in the regulation of adult spontaneous movement. Thus, although systemic exposure to MPTP or 6-OHDA did not seem to produce robust loss of dopamine neurons in the adult zebrafish posterior tuberculum, significant losses of dopamine and noradrenaline were noted, along with a reduction in mean velocity and increase in turn angles during spontaneous swimming (Anichtchik et al. 2004). Interestingly, dopamine also seems to modulate the development of motor behavior during embryogenesis; at 3 days post-fertilization, forebrain dopaminergic function inhibits swimming movements, but this response is lost by 5 days post-fertilization (Thirumalai and Cline 2008). This is the opposite effect to that observed in older animals following manipulation of dopaminergic signaling and the mechanisms are not yet certain. However, this observation raises the possibility that functional abnormalities of dopaminergic neurotransmission provoked in transgenic models of motor disorders may manifest in unexpected ways if the models are evaluated very early during development. It is also recognized that manipulation of other neurotransmitter systems of relevance to human disease can modulate the spontaneous locomotor behavior of larval zebrafish. For example, recent work has shown that inhibition of monoamine oxidase B in larvae resulted in elevated levels of serotonin, but not of other monoamine neurotransmitters, accompanied by dose-dependent reductions in movement of 7-day-old larval zebrafish (Sallinen et al. 2009b). This is compatible with earlier studies showing that serotonin-specific reuptake inhibitors impair locomotor activity in zebrafish (Airhart et al. 2007) and that serotonin stimulates swimming behavior in larval zebrafish (Brustein et al. 2003).
In addition to specialized populations of neurons that show similar neurochemistry and connectivity to neuronal groups involved in human neurodegenerative diseases, the zebrafish CNS has other features that may be relevant to modeling the pathogenesis of these conditions. Glia, including oligodendrocytes (Kirby et al. 2006), cells with astrocyte-like properties (Kawai et al. 2001) and microglia (Peri and Nusslein-Volhard 2008) are present within the brain parenchyma. Since there is increasing evidence that these cell types are critical to pathogenesis in some circumstances (Vila et al. 2001; Mullett and Hinkle 2009), their presence in the zebrafish CNS allows opportunities to study their mechanistic roles in disease. Finally, similar to mammals, the zebrafish CNS has a blood–brain barrier (Jeong et al. 2008), which may be critical for either putative neurotoxins or novel therapeutic agents to gain access to the CNS parenchyma.
One striking difference between mammalian and zebrafish brain is that the zebrafish CNS undergoes continued growth and acquisition of new neurons throughout life, and shows impressive capacity for regeneration of axons and neurons following focal lesions (reviewed in (Becker and Becker 2008)). The ability of zebrafish CNS to reform functional neural circuits after injury to fiber tracts is dependent both on properties of neurons that favor axonal re-growth, and also on the CNS environment, in which glia promote axonal growth and pathfinding, in contrast to the mammalian CNS, which presents an inhibitory environment to axonal regeneration. It is unclear at present whether this capacity for ongoing growth and repair will hamper attempts at modeling degenerative diseases, although the first reports suggest that this is not the case (see below).
Phylogenetic conservation of genes implicated in neurodegeneration
Overall, there is significant genetic similarity between zebrafish and other vertebrates, including mammals. The degree of phylogenetic conservation is perhaps most prominent in pathways governing basic aspects of cellular homeostasis. Interestingly, many of the genes known to cause hereditary neurodegenerative disease have highly conserved homologues in the zebrafish (Table 1); the striking degree of phylogenetic conservation suggests the relevant cellular processes are of fundamental importance to the health and function of neurons and glia throughout vertebrate evolution. The presence of conserved zebrafish homologues of genes involved in human neurodegeneration also argues that it might be possible to provoke neuronal dysfunction or loss through cellular mechanisms similar to those involved in the pathogenesis of human diseases, supporting the notion that zebrafish might be an appropriate model in which to study the functions of the genes and the pathophysiology of the disorders. Orthologues of some human genes are duplicated in the zebrafish genome (Chen et al. 2009a), raising the possibility that resulting sub-functionalisation may help to dissect the functions of the human genes. Orthologues of genes involved in Parkinsonism, Alzheimer’s disease and Huntington’s disease are amongst those identified in zebrafish.
There are currently three well-recognized causes of human autosomal recessive Parkinsonism (this does not include other Mendelian genetic diseases in which Parkinsonism is one component of a complex phenotype): (i) The PRKN gene on chromosome 6q (locus designated PARK2) encodes Parkin, an E3 ubiquitin ligase (Shimura et al. 2000) thought important in the regulation of mitochondrial dynamics (Riparbelli and Callaini 2007) and mitophagy (Narendra et al. 2008); (ii) The DJ-1 gene on chromosome 1p (locus designated PARK7) encodes a protein of unknown function that relocates to mitochondria under conditions of oxidative stress (Bonifati et al. 2003); (iii) The PINK-1 gene, also located on chromosome 1p (locus designated PARK6), encodes phosphatase and tensin homolog-induced putative kinase 1 (PINK-1), a mitochondrially located serine-threonine kinase (Valente et al. 2004) that may be involved in regulation of mitochondrial dynamics (Yang et al. 2008) in a common pathway with Parkin (Clark et al. 2006). With the exception of PARK2-linked juvenile-onset Parkinsonism, cases of clinical Parkinsonism caused by these gene mutations are very uncommon. However, like many of the hereditary diseases discussed in this review, their importance lies in the possibility of understanding the pathogenesis of the common sporadic forms of Parkinsonism through elucidating the pathophysiology in unusual situations where the etiological origins of disease are well-defined.
Zebrafish orthologues of Parkin, DJ-1 and PINK-1 have been described, and their functions investigated in zebrafish by transient knockdown during embryogenesis. This powerful technique is commonly deployed in developmental biology in order to study effects of transient abrogation of gene function during development (Nasevicius and Ekker 2000). Chemically modified morpholino antisense oligonucleotides can be designed to target either translation of a specific mRNA transcript, or prevent splicing of the relevant pre-mRNA, through complementarity to the sequence surrounding the translational initiation codon or splice signals. Morpholino oligonucleotides are stable in vivo, diffuse throughout embryos following microinjection and allow graded suppression of gene expression during the first few days after fertilization. Transient knockdown studies of DJ-1, Parkin and PINK-1 illustrate phylogenetic conservation of function between human and zebrafish.
Zebrafish and human DJ-1 share 83% amino acid identity. Critical residues shown to be affected by pathogenic mutations in humans, and sites of defined biochemical function underlying mitochondrial re-localization and SUMOylation, are completely conserved between human and zebrafish (Bai et al. 2006). DJ-1 is expressed most highly in zebrafish brain and gut (Bai et al. 2006). Within the CNS, DJ-1 is expressed in neurons throughout the brain and spine, including dopaminergic neurons (Bai et al. 2006). The dopaminergic system formed normally in zebrafish embryos after targeting the dj-1 transcript using morpholino antisense oligonucleotides (Bretaud et al. 2007). However, knockdown of dj-1 resulted in an increased sensitivity of dopaminergic neurons to oxidative stress (Bretaud et al. 2007), replicating findings relating to DJ-1 function in cell culture and other animal models. Interestingly, zebrafish embryos subjected to dj-1 knockdown also showed enhanced susceptibility to proteasome inhibition, demonstrating a novel potential link between DJ-1 function and abnormalities of neuronal protein catabolism (Bretaud et al. 2007), which itself has been implicated as a potential pathogenic mechanism in sporadic Parkinson’s disease (McNaught et al. 2003). The increased neuronal death observed in dj-1 knockdown embryos following proteasome inhibition was shown to be apoptotic and p53 dependent; furthermore loss of DJ-1 caused elevation of basal levels of p53, and simultaneous knockdown of dj-1 and the p53 inhibitor mdm2 caused dopamine cell loss without toxin exposure (Bretaud et al. 2007). Subsequently, it was shown that DJ-1 is a transcriptional repressor of p53 in cultured mammalian dopaminergic cells (Fan et al. 2008), indicating that the zebrafish model had provided a new insight into DJ-1 function that was replicable in mammalian cells. Similar to the situation in the mammalian CNS, zebrafish DJ-1 is up-regulated under conditions of oxidative stress (Baulac et al. 2009). Together, these findings suggest that biochemical functions and transcriptional regulation of DJ-1 studies so far are strikingly conserved from zebrafish to mammals and that abnormalities of DJ-1 function can provoke pathological abnormalities that appear relevant to those of Parkinson’s disease.
Zebrafish Parkin shows 62% overall amino acid identity to human Parkin. However, the two RING domains at the C-terminal of the molecule are more highly conserved, showing approximately 90% similarity to the human orthologue (Flinn et al. 2009). Parkin is ubiquitously expressed in zebrafish from early development through adulthood. Morpholino knockdown of Parkin decreased the number of diencephalic dopaminergic neurons, and enhanced their susceptibility to the dopaminergic neurotoxin MPP+, at 3 days post-fertilization (Flinn et al. 2009) This is potentially an important finding, because this is the only animal model of Parkin deficiency in which reduction of dopaminergic neurons similar to that found in human PARK2-linked Parkinsonism has been reported, although it is unclear at present whether this presents a transient developmental delay, a permanent developmental abnormality, or true neurodegeneration. There was no detectable effect of Parkin knockdown on larval swimming behavior at 5 days post-fertilization, the time point at which spontaneous movement is sufficiently developed to make meaningful measurements (Flinn et al. 2009). It is unclear whether the transient knockdown persisted to this time point, and whether the reduction in numbers of dopaminergic neurons reported at 3 days post-fertilization subsequently recovered. However, Parkin knockdown resulted in a quantitative reduction of mitochondrial complex I activity, an abnormality also observed in fibroblasts cultured from patients harboring mutations in parkin (Mortiboys et al. 2008), and reported in sporadic Parkinson’s disease by several different groups. The major findings from this work support the notion that loss of Parkin promotes biochemical and pathological changes in zebrafish that are analogous to those occurring in humans with Parkin mutations, and therefore that the zebrafish could be a representative model for studying the mechanisms underlying neurodegeneration.
Zebrafish PINK1 shares 54% identity with its human orthologue and is expressed ubiquitously from early development (Anichtchik et al. 2008). In the adult zebrafish brain, PINK1 is enriched in periventricular zones and present in some dopaminergic neurons of the diencephalon (Anichtchik et al. 2008). Knockdown of PINK1 resulted in a severe developmental phenotype. A decrease in TH-expressing neurons in the diencephalon at 48 h post-fertilization was observed, along with gross morphological abnormalities including spinal curvature, failure of tail development, and impaired tectal and commissural formation in the brain. Some of these abnormalities were partially rescued by wild-type, but not mutant, human PINK1 over expression. Enhanced free radical production, thought to be a central feature of human Parkinson’s disease pathogenesis, was provoked by pink1 knockdown. These observations suggest that the human and zebrafish proteins share conserved functions. The similarity of the morphological phenotype of pink1 morphants to wnt mutants led to evaluation of the GSK3β pathway, which was found to be activated. Since inhibitors of GSK3β allowed partial phenotypic rescue, it was concluded that abnormalities in the AKT- GSK3β pathway might be relevant to the deleterious effects of PINK-1 deficiency. Studies of GSK3β function have not yet been reported in mammalian PINK1 null models, and so it is currently unclear whether this finding represents a novel insight into human PINK1 pathophysiology.
Zebrafish orthologues of genes implicated in several other human neurodegenerative diseases, including ataxias (Imamura and Kishi 2005; Carlson et al. 2009), Huntington’s disease (Karlovich et al. 1998; Lumsden et al. 2007; Diekmann et al. 2009) and frontotemporal dementia (Cadieux et al. 2005) have also been described. The zebrafish brain expresses homologues of genes involved in Alzheimer’s disease, including APP (Musa et al. 2001), the precursor of Aβ found in senile plaques, and the microtubule associated protein-τ (Chen et al. 2009a), abnormal forms of which accumulate in neurofibrillary tangles. The Aβ domain is conserved between human APP and the two zebrafish paralogues appa and appb. Furthermore, orthologues of β-secretase and components of the γ-secretase complex are expressed in the zebrafish CNS, and zebrafish presenilin-1 can replace human PS1 biochemically for cleavage of APP in vitro (Leimer et al. 1999; Groth et al. 2002, Campbell et al. 2006). These findings suggest that similar enzymatic complexes mediating APP cleavage may be present in both zebrafish and human. However, an equivalent fragment to human Aβ has not yet been reported in the zebrafish CNS, and it is currently unclear whether the details of post-translational processing of APP that are thought central to Alzheimer’s disease pathogenesis are replicated in the zebrafish.
The synuclein gene family may represent an exception to the striking degree of conservation between human genes implicated in neurodegenerative disease and their zebrafish homologues. Mammals express α-, β- and γ-synucleins in the CNS, each encoded by a separate gene (Clayton and George 1998). Deposition of insoluble α-synuclein aggregates is a central feature of Parkinson’s disease and a number of other neurological diseases collectively termed synucleinopathies (Spillantini et al. 1998; Tu et al. 1998). In the pufferfish (Yoshida et al. 2006) and zebrafish (Sun and Gitler 2008; Chen et al. 2009b) genomes, the sncg gene encoding γ-synuclein is duplicated; γ1- and γ2-synucleins are closely related but show different expression patterns. Unlike the pufferfish, however, no evidence of a zebrafish gene encoding an α-synuclein orthologue has yet been found in genomic, cDNA or EST sequences, suggesting that the snca gene may have been deleted from the zebrafish genome, the putative deletion possibly being accommodated by functional redundancy between synuclein family members (Abeliovich et al. 2000).
Together, these studies provide evidence of phylogenetic conservation of selected aspects of structure and function of key brain regions, cell types, genes, proteins and biochemical pathways involved in neurodegeneration in humans. These observations support the argument that mechanisms relevant to neurodegeneration in humans may be conserved to a sufficient degree that appropriate and relevant mechanistic insights can be gained into human diseases, through construction and study of zebrafish models.
It is possible that the zebrafish offers advantages over existing experimental models for studies of neurodegeneration. For example, conservation of brain structure, specialized neuronal and glial cell types, myelin, neurochemical circuits and vertebrate genetics may represent an advantage over insect and nematode models; whereas the applicability of live in vivo imaging and high throughput approaches is unique to zebrafish amongst vertebrate models. It is too early in the development of this new field to be certain whether these features will prove significantly advantageous for this application. However, initial studies raise the exciting possibility of a genetically-tractable in vivo experimental system that is amenable to high throughput screening, and in which aspects of cellular specificity and tissue environment-neuronal interactions are recapitulated.
Tools for generation and analysis of transgenic zebrafish models of neurodegenerative diseases
Transgenic zebrafish are relatively straightforward to produce, and refinements to the methodology have made this a rapid and efficient process. In the following sections, we briefly review techniques for generation of transgenic lines, available zebrafish promoters with utility for generation of transgenic models, and genetic tools for the generation of stable lines expressing transgenes that may be toxic to zebrafish CNS and reduce reproductive potential. We then consider some of the possible limitations of available methodologies for generation and analysis of neurodegenerative disease models.
Techniques for generation of transgenic zebrafish
The first transgenic zebrafish were generated by micro-injecting linearized plasmid DNA into the cytoplasm of one-cell stage embryos (Stuart et al. 1988). This results in concatermerization of DNA, which remains episomal and is distributed in a mosaic manner during subsequent cell divisions. Inefficient integration of concatemers during late cell divisions results in single integration events of many tandem copies of the transgene and significant mosaicism. Consequently, the technique is very inefficient, necessitating screening many fish to identify germline transgenic founders. Two technical advances, meaganucleases and transposons, have substantially improved the efficiency by which foreign DNA can become inserted into the genome for the generation of transgenic models (Fig. 2).
The meganuclease I-sce1 is an intron-encoded endonuclease from saccharomyces cerevisiae (Jacquier and Dujon 1985). I-Sce1 cleaves DNA at an 18 bp sequence-specific recognition site that is not present within the zebrafish genome. Co-injection of I-sce1 with a transgene plasmid, in which the transgene expression cassette is flanked by I-sce1 sites, into zebrafish embryos substantially reduces mosaicism, allowing more efficient examination of the expression pattern of reporter constructs in transient assays, and improving the rate of transmission of the transgene from germline F0 mosaics to their progeny, F1 transgenic founders (Thermes et al. 2002). In addition, the single site of integration usually contains a low number of copies of the transgene (Thermes et al. 2002). The mechanisms underlying these observations are uncertain. I-sce1 shows slow enzymatic turnover, because the monomeric enzyme has high affinity for one of the cleavage products (Grabher and Wittbrodt 2008). I-Sce1 may prevent concatemerisation that usually occurs after microinjection of linearized DNA, by limiting access of ligases to cut ends and by digesting concatemers that do occur. The efficiency of recombination may involve enhanced nuclear import of cleaved DNA and interaction of I-Sce1 with the double strand break repair system. It has been reported that careful optimization of parameters using this technique almost completely eliminates mosaicism in animals with genomic integration, and allows transmission of the transgene from F0 to F1 fish with a near-Mendelian rate (Thermes et al. 2002, Soroldoni et al. 2009). In our hands the rate seldom approaches this level, but is substantially better than linearized plasmid injection. In addition, the single site of integration has resulted in simple Mendelian transmission of a variety of transgenes in subsequent outcrosses, facilitating the generation of double transgenic reporter zebrafish (Bai et al. 2009).
Transposons are mobile DNA elements; type 2 transposons encode an enzyme, transposase, which mediates excision of the transposon from the genome and its re-insertion at another location. Transposons have been used extensively in Drosophila genetics, but their application is species specific and until recently no vertebrate transposons had been identified. The discovery of a transposon, Tol2, in the genome of the freshwater fish medaka (Koga et al. 1996) allowed subsequent development of a powerful genetic tool for zebrafish transgenesis (Kawakami et al. 2000). By deletion of the open reading frame of the transposase from Tol2, a non-autonomous element was generated, which could insert into the genome when the transposase was supplied in trans. By generating a transgene plasmid, in which the transgene expression cassette is flanked by the non-autonomous transposon, and co-injecting the plasmid into zebrafish embryos along with mRNA encoding transposase, highly efficient integration of the transgene into the genome occurs. This approach most frequently yields multiple single copy integration events, although it is possible to select single copy integrants if necessary, by southern blot. The technique is efficient and has been used increasingly since its introduction and subsequent refinement (Kawakami 2004), because it is necessary to inject and screen a smaller number of fish than other techniques in order to identify stable transgenic lines. In addition, the technique yields integrants with sufficient frequency to use for other transgenic approaches, for example gene-trap experiments (Kawakami et al. 2004) (see “Expression of transgenes with adverse effects on viability and reproduction”). Recent studies have sought to enhance the efficiency of synthetic transposons (Mates et al. 2009); it is possible that such customized systems may find applications in zebrafish genetics in the future.
Promoter elements for driving transgene expression in zebrafish models of neurodegenerative disease
In order to generate transgenic models of neurodegeneration in zebrafish, appropriate cis-acting regulatory elements must be used to drive transgene expression in a suitable temporal and spatial expression pattern, and at sufficient levels to provoke pathology. The first transgenic zebrafish suffered from inactivation of transgenes, possibly because of use of non-zebrafish promoter elements, and the use of zebrafish cis-acting regulatory regions has allowed development of stable lines with reliable expression of transgenes (Higashijima et al. 1997, Long et al. 1997). The details of the desired transgene expression pattern will likely vary between applications and disease models. However, for many genetic and sporadic neurodegenerative diseases in humans, the implicated genes are expressed ubiquitously in the nervous system, resulting in a specific pattern of neuropathology through unknown mechanisms. The possibility of using zebrafish models to understand the selective vulnerability of particular groups of neurons to abnormal protein expression is attractive. For these applications, expression of genes in many different neuronal types, widely distributed throughout the neuraxis, would be desirable. Three such promoter elements have been described. The first is a product of an elegant functional in vivo analysis of the gata2 promoter (Meng et al. 1997). Deletion analysis showed that a fragment of the upstream sequence of gata2, lacking hematopoietic regulatory elements and containing a neuronal enhancer, was able to drive robust expression of GFP in developing neurons. This element was subsequently used to express a Tau-GFP fusion protein in a transient zebrafish model of Tauopathy (Tomasiewicz et al. 2002). Stable transgenic lines using this element to drive transcription have not yet been described, and its activity in the adult CNS is not reported. The second pan-neuronal promoter element reported was derived from the zebrafish huc gene, which is the homologue of the Drosophila elav gene, and encodes an RNA binding protein, HuC/D, commonly used as an early neuronal marker (Kim et al. 1996). A 2.8 kb fragment of the proximal flanking region was sufficient to drive robust pan-neuronal expression in embryos (Park et al. 2000). The promoter has subsequently been used to generate a stable model of Tauopathy (see below). Both of these promoter elements are active in neurons, although the early time points at which they become transcriptionally active are a potential source of concern for neurodegenerative disease model construction: expression of transgenes early in embryogenesis could provoke phenotypes through developmental anomalies rather than neurodegeneration. In view of this concern, genes representing later markers of neuronal differentiation were examined. The eno2 gene encoding the neuron-specific γ-enolase isoenzyme, was identified as a marker of differentiated neurons (Bai et al. 2007). Expression of eno2 was detected at low levels by 24hpf, but the abundance of the mRNA increased substantially in the brain and spinal cord between 60 and 72 h post-fertilization, and expression persisted at high levels into adulthood, in a pan-neuronal pattern. The regulatory region of eno2 is complex; there is an untranslated first exon, and the first intron contains a CpG island that appears important for promoter activity. A 12 kb fragment of the promoter, including the first intron, was active in driving reporter gene expression in neurons throughout the brain and spinal cord from 48 h post-fertilization through adulthood, including neuronal types relevant to neurodegenerative diseases, such as cerebellar Purkinje cells and cholinergic neurons (Fig. 3) (Bai et al. 2007). The eno2 construct was also highly active in the retina and visual pathways (Bai et al. 2009). This element was used to generate a transgenic zebrafish Tauopathy model (Bai et al. 2007) and is currently being used by a number of groups for a variety of applications.
Zebrafish eno2 promoter construct. The pictures show micrographs of Tg(eno2:egfp) zebrafish larvae in order to demonstrate widespread neuronal expression of a GFP transgene under control of the 12 kb eno2 element that was developed for generation of transgenic models of neurodegenerative disease. a The oblique sagittal section is labeled for transgene expression (GFP; green) and a nuclear counterstain (blue: DAPI) to facilitate identification of anatomical landmarks. GFP expression is seen throughout the neuraxis, and is particularly prominent in the retina and optic tectum. Tel telencephalon, TeO optic tectum, CCe cerebellum, MdO medulla oblongata, SC spinal cord, RCGL retinal ganglion cell layer, L ocular lens, RPRL retinal photoreceptor layer, LLG lateral line ganglion. The boxed area marks the approximate region shown at higher magnification in b. b A Z-projection is shown of multiple confocal planes imaged from a live intact transgenic zebrafish, illustrating GFP expression in neurons of the CNS and PNS. LLN lateral line nerve, XIII eighth cranial nerve, otherwise same as a
For particular applications it might be desirable to express transgenes in patterns other than the pan-neuronal pattern described above. A large literature dealing with transgenic approaches to targeting transgene expression to defined subpopulations of neurons is beyond the scope of this review. The dopaminergic system has received particular attention because of its central importance in humans to Parkinsonian movement disorders and other neuropsychiatric diseases. However, several attempts at targeting transgene expression to the dopaminergic system have been only partially successful. Most recently, an 11 kb promoter fragment derived from the zebrafish slc6a3 gene, encoding the dopamine transporter, showed expression in dopamine neurons of the pretectal region, but was not expressed in other dopaminergic groups, and showed ectopic expression in numerous non-dopaminergic neurons (Fig. 4; Bai and Burton 2009). Similar findings have been reported using fragments of the zebrafish th (Meng et al. 2008a) and rat TH (Gao et al. 2005) promoters, although in this case the only dopaminergic cell groups that expressed the promoter were located in the retina. It appears that the transcription of genes whose expression is restricted to neuronal populations that share a common neurochemical phenotype, despite wide anatomical distribution, is complex, and may rely on multiple regulatory elements distributed over a wide area of genomic sequence. An enhancer trap experiment yielded an integration event into the vesicular monoamine transporter gene vmat2, labeling monoaminergic neurons specifically throughout the zebrafish brain (Wen et al. 2008). A similar approach may be necessary in order to generate lines that express transgenes only in dopaminergic neurons, perhaps by Gal4 enhancer trap or similar methodology (see below).
Imaging dopamine neurons in live zebrafish. The image shows an epifluorescence micrograph of a superior view of a live Tg(slc6a3:egfp) zebrafish in which the GFP transgene is expressed under an 11 kb promoter fragment from the gene encoding the dopamine transporter. Dopaminergic neuronal clusters in the pretectal region are seen (green arrows). However, the transgene is also expressed in some non-dopaminergic groups (blue arrows) and does not show expression in other groups of dopaminergic neurons, indicating that some of the relevant regulatory elements lie outside the 11 kb region included in the promoter construct. E eye, otherwise same as Fig. 3
Given the central role played by glial cells in both the physiology of the healthy brain and in the pathogenesis of disease, the description of zebrafish glial promoter elements capable of driving transgene expression in cells with astrocytic (Bernardos and Raymond 2006), oligodendroglial (Yoshida and Macklin 2005) or microglial (Peri and Nusslein-Volhard 2008) properties is potentially of considerable interest. In particular, the zebrafish may provide a powerful system in which to examine non-cell autonomous effects of alterations in glial gene expression that may provoke or prevent neuronal pathology.
Finally, the use of large DNA fragments to drive transgene expression presents several potential advantages. The inclusion of all relevant cis-acting sequences in the construct gives rise to physiological gene expression patterns and levels. Furthermore, dynamic gene expression changes during pathogenesis of the resulting models are more likely to mirror those present during disease progression, through appropriate activation of relevant response elements in the regulatory region. Currently, several techniques are available to enable genetic modification of bacterial artificial chromosomes, allowing large chromosomal loci to be conveniently manipulated and transgenes introduced into the genetic locus contained in the BAC by recombination (Lee et al. 2001a; Yang et al. 2009). The construct is then introduced into the genome by the transgenesis techniques listed above, although this is very inefficient for large BAC transgenes necessitating screening large numbers of fish to find recombinants. However, the BAC transgenic lines that have been reported seem to faithfully recapitulate endogenous gene expression patterns (Kirby et al. 2006; Chen et al. 2007; Sato et al. 2007; Yang et al. 2007; McGraw et al. 2008; Peri and Nusslein-Volhard 2008) and the technique is becoming more widely applied.
Expression of transgenes with adverse effects on viability and reproduction
Expressing transgenes to evoke neurodegeneration by using a promoter that becomes active after only a few days of life has both favorable and potentially troublesome outcomes. Driving disease pathogenesis to occur early in the life of a zebrafish will be critical to exploiting the full potential of the technical toolbox that has arisen from the application of zebrafish models in developmental biology studies. In particular, screening for genetic and chemical modifiers will demand that a discernable and relevant phenotype occurs when the zebrafish are still sufficiently young that the larvae can be accommodated in microtiter plates. Unfortunately, the development of an authentic neurodegenerative phenotype at this young age would likely prevent the zebrafish from reaching sexual maturity, or at least compromise reproductive behavior, making the establishment of stable transgenic lines impossible. One potential way around this issue is to generate stable lines that only express the transgene under particular conditions; the binary Gal4–UAS system successfully employed in Drosophila has recently been adapted for use in zebrafish (Scheer and Campos-Ortega 1999). This scheme entails the generation of ‘driver’ lines that express a chimeric transcription factor containing the DNA binding motif of yeast Gal4 fused to a transactivator domain (Fig. 5). The cognate DNA motif recognized by Gal4 is absent from the zebrafish genome, so the resulting transgenic fish do not express any other transgenes. A second transgenic ‘effector’ line is generated, in which the putative neurodegeneration-causing gene is under transcriptional control of a weak basal promoter linked to multiple copies of the upstream activating sequence (UAS) recognized by Gal4. In the absence of Gal4, the effector line does not express the transgene at appreciable levels. When the driver and effector lines are crossed, however, trans-activation of the UAS enhancer by Gal4 from the driver line results in robust tissue specific expression of the transgene. This system has been successfully used in zebrafish, with modifications. The original Gal4 driver contained the trans-activation domain from the herpes simplex virus virion protein VP16. Although this resulted in strong activation of the UAS promoter, this promiscuous activator is itself toxic, limiting its utility. This problem has been addressed by using attenuated versions of the VP16 trans-activation domain that provoke less toxicity but retain trans-activating activity (Asakawa et al. 2008; Distel et al. 2009). Using these modified Gal4–Vp16 constructs, it has been possible to generate driver lines by random genomic insertion, potentially allowing genetic drivers to mirror expression patterns of endogenous genes, under the control of a full complement of endogenous cis-acting elements. Isolation of useful driver lines for individual applications will likely depend on screening established lines for appropriate expression patterns. This technique could prove extremely useful, since the drivers could be used to express multiple different transgenes in the same pattern for comparison, and standard well-characterized driver lines would reduce the time necessary to generate and isolate transgenic lines with the desired expression patterns. A database of currently available zebrafish Gal4 lines can be accessed at http://www.zfin.org.
Potential limitations of current techniques; methodologies under development
Although the range of molecular techniques available to the zebrafish geneticist is extensive, there remain some prominent gaps in the current toolbox; ongoing work to address these areas will likely enhance the value of zebrafish models:
First, current zebrafish models of recessive human neurodegenerative diseases are based on transient knockdown during early embryogenesis using morpholino antisense oligonucleotides. It is uncertain whether this provides a representative insight into the pathogenesis of human disease phenotypes that occur during adulthood. Unfortunately, zebrafish germline null alleles cannot currently be generated using similar techniques to those employed highly effectively in mouse studies. This has necessitated use of high-throughput approaches in order to identify suitable mutations from mutagenesis screens. The recent development of engineered zinc-finger nucleases may circumvent this problem and allow targeted introduction of null alleles into genes of interest (Doyon et al. 2008; Meng et al. 2008b). This would potentially permit the study of recessive phenotypes in older zebrafish than is possible using transient techniques. In addition germline null mutants would facilitate the use of high-throughput approaches in larval models with disruption of targeted loci.
Second, there is currently no well-established behavioral measure for neurodegenerative phenotypes in zebrafish, limiting phenotypic analysis to visualization of morphology, histology or gene expression, or demonstration of other biochemical phenotypes. Although these assays are valid, quantifiable end-points, arguably they fail to exploit the advantages of an in vivo model fully. Early neuronal dysfunction prior to cell death may represent a realistic target for neuroprotective drug therapy in humans and could potentially manifest as measurable behavioral abnormalities in animal models of disease, which might therefore represent more suitable assays for drug discovery. In addition, non-invasive behavioral measures could be automated for high-throughput applications. Consequently there is significant interest in establishing reproducible and simple behavioral assays as measures for neurodegenerative phenotypes.
Current transgenic zebrafish models of neurodegenerative disease
Recent publications have described the first proof-of-principle experiments showing that transgenic expression of genes triggering neurodegeneration in humans can provoke relevant phenotypes in zebrafish. Models of human Tauopathies, polyglutamine disorders and motor neuron disease have recently been published.
Tauopathy models
Abnormal forms of the microtubule associated protein Tau are deposited in neurofibrillary tangles in a number of sporadic human neurodegenerative diseases, including Alzheimer’s disease, progressive supranuclear palsy and Pick’s disease (Lee et al. 2001b). In the adult human brain, the pre-mRNA from the MAPT gene encoding Tau is alternatively spliced, giving rise to six protein isoforms that contain either 3 or 4 microtubule binding domains (Goedert et al. 1989). In AD, neurofibrillary tangles contain 3- and 4-reapeat Tau, whereas 4-repeat Tau predominates in the tangles of PSP, and 3-repeat Tau in Pick’s disease (Lee et al. 2001b). In the majority of cases of these diseases, no abnormality has been identified in the MAPT gene. However, some cases of fronto-temporal dementia and Parkinsonism, are caused by MAPT mutations that alter the primary sequence of Tau or the ratio of 3- to 4-repeat isoforms (Hutton et al. 1998). Since individual FTDP17 cases may show clinical and pathological similarity to PSP or Pick’s disease, it is thought that abnormalities of Tau metabolism may be central to the pathogenesis of the sporadic diseases.
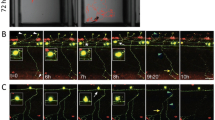
Given the proposed central role of Tau in a number of important neurodegenerative conditions, there has been interest in the construction of zebrafish Tauopathy models. The first publication reported a transient model, in which a Tau-GFP fusion protein was over-expressed in zebrafish larvae using the GATA-2 promoter (Tomasiewicz et al. 2002). The fusion protein was phosphorylated similar to native Tau in vitro and showed an expression pattern in tissue culture suggesting interaction with the cytoskeleton. In zebrafish embryos, neurons expressed the fusion protein in a mosaic pattern, some examples showing fibrillar fluorescence in the cell body and proximal axon, resembling neurofibrillary tangles. The human Tau-GFP fusion was phosphorylated in the zebrafish brain. This initial study validated the use of a GFP fusion protein to monitor evolution of tangle pathology in vivo, and showed that a biochemical change relevant to human disease, phosphorylation, occurs in larval zebrafish. This suggests there is sufficient phylogenetic conservation of endogenous zebrafish kinases to modify the human protein. Stable transgenic zebrafish expressing human 4-repeat Tau were subsequently constructed using the newly described eno2 promoter (Bai et al. 2007). The phenotype of these transgenic fish has not yet been fully reported. The initial report showed evidence of refractile Tau accumulations within neuronal cell bodies and proximal axons, resembling neurofibrillary tangles. These accumulations were present in neurons throughout the brain, including regions of pathological relevance to PSP, such as the optic tectum. More recently, the Gal4–UAS system has been exploited in order to generate a Tauopathy model that shows a larval phenotype, with potential application to high throughput screening. Expression of the FTDP-17 Tau mutant P301L was driven from a novel bidirectional UAS promoter, allowing simultaneous expression of a separate red fluorescent protein in Tau-expressing cells (Paquet et al. 2009). The high levels of mutant Tau expression provoked by the huc:gal4-vp16 driver were sufficient to induce a transient motor phenotype during embryogenesis, caused by a motor axonal outgrowth delay. At later time points, the Tau mutant caused enhanced cell death and protein aggregation in the spinal cord. In addition, rapid progression from early to late pathological Tau phosphorylation was seen over the first few days of life; the phosphorylation of Tau was reduced by application of GSK3β inhibitors, suggesting that the model may be used to identify other similar pharmacological inhibitors from chemical libraries. Unfortunately, loss of promoter activity prevented the examination of later pathological changes, and so it is unclear whether the phenotype was progressive and age-dependent, or transient. In addition, the huc promoter fragment used in this model only induced robust transgene expression in the spinal cord, which is not a prominent site of Tauopathy changes in human disease. However, this valuable study showed the utility of the Gal4–UAS system for modeling neurodegeneration in transgenic zebrafish and demonstrated evidence that biochemical changes characteristic of Tauopathy, including an orderly acquisition of abnormal phospho-epitopes and conformers, can be recapitulated in larval zebrafish.
Polyglutamine models
A number of autosomal dominant neurodegenerative diseases, including Huntington’s disease (HDCRG 1993) and several of the spinocerebellar ataxias (Orr et al. 1993, Kawaguchi et al. 1994; Imbert et al. 1996), are caused by pathological expansion of a tandem trinucleotide CAG repeat in the relevant gene, resulting in an elongated stretch of glutamine residues in the resulting protein. It is thought that the mechanism of pathogenesis involves a toxic gain of function mediated by the expanded polyglutamine tract, rather than loss of function of the affected gene (Landles and Bates 2004; Zoghbi and Orr 2009). Since this general pathogenic mechanism may be shared by these diseases, a polyQ toxicity model in zebrafish would present a possible means to elucidate pathogenesis and perhaps isolate a common treatment for the whole group of conditions. In the first report of a zebrafish polyQ model, transient expression of GFP-polyQ fusion proteins was achieved by microinjection of plasmids, encoding the fluorescent fusion with polyQ tracts of differing lengths, under transcriptional control of a strong viral promoter (Miller et al. 2005). In human polyQ diseases, there is correlation between the length of the polyQ expansion and the severity of the phenotype, as measured by age of onset or rate of clinical progression. Expression of GFP-polyQ fusion proteins in zebrafish caused a decrease in embryo length and loss of tissue differentiation, resulting in gross morphological deficits and reduced viability. Although this acute response does not reflect the chronic neurological diseases seen in patients with polyQ expansion mutations, significant over-expression of these artificial proteins would be expected to provoke acute and severe phenotypes. Importantly, however, the model recapitulated two key features of polyQ diseases: first, there was correlation between the polyQ repeat length and the severity of the morphological phenotype. Second, GFP-positive inclusion bodies were formed, suggesting the formation of aggregates dependent on the polyQ tract (Miller et al. 2005). C-terminal Hsp70 (heat shock protein 70)-interacting protein (CHIP), which functions both as a co-chaperone and ubiquitin ligase, was shown to suppress aggregation of the PolyQ-GFP fusion, and the resulting toxicity, in this transient zebrafish model. The role of CHIP was then confirmed in a chronic mammalian model of Huntington’s disease: N171-Q82 mice, in which the prion promoter drives expression of a Huntingtin fragment with a pathologically expanded polyQ tract, develop a neurobehavioral phenotype consisting of abnormal clasping movements, gait disturbance and tremor, associated with inclusion body neuropathology and loss of DARRP-32 immunoreactivity in the striatum. This phenotype was exacerbated when the mice were bred onto a CHIP haplo-insufficient background, resulting in accelerated clinical deterioration and premature death (Miller et al. 2005). These novel findings indicate that the acute, transient zebrafish model was predictive of at least one key biochemical event underlying the pathogenesis of the chronic mammalian model, demonstrating that an appropriate mechanistic insight into disease pathogenesis was gained by studying the zebrafish model. A similar transient study, using mRNA injection to express GFP-polyQ(4, 25 or 102) confirmed that the expanded polyQ tract induced aggregation of the GFP fusion reporter in vivo (Schiffer et al. 2007). Time lapse photomicrography allowed direct visualization of the polyQ fusion protein being depleted from the cytoplasm as it was incorporated into growing aggregates. Interestingly, visualization of apoptotic cells relative to aggregates showed an unexpected dissociation, suggesting that aggregation was cytoprotective and that the toxic species may be pre-fibrillar GFP-polyQ (Schiffer et al. 2007). This report was remarkable for the first use of a zebrafish polyQ model to test possible chemical inhibitors of polyQ aggregation in vivo; some differences in the anti-aggregate activity of compounds were seen between cell culture and zebrafish and it is possible that the in vivo setting of the zebrafish model will provide a more representative environment for identification of compounds with relevant properties. A more recent study used a cell culture model in order to screen for enhancers of autophagy that might be efficacious in clearing aggregated Huntingtin and other substrates from cells (Williams et al. 2008). The indentified compounds were then subjected to verification in a novel stable transgenic zebrafish line, expressing a GFP-Huntingtin71Q fusion protein under control of the rhodopsin promoter, leading to aggregation of the fusion protein and loss of rod outer segments and rhodopsin expression from the retina. Several of the compounds identified as reducing aggregation in the cell culture model also prevented formation of aggregates in the zebrafish model, providing validation of the cell culture system, and suggesting that zebrafish models might be useful in the future for primary screens of therapeutic compounds.
ALS model
Familial amyotrophic lateral sclerosis is uncommon, but a fifth of such cases arise from mutations in the gene encoding superoxide dismutase (SOD) (Deng et al. 1993; Rosen et al. 1993). The mutations are thought to provoke degeneration of upper and lower motor neurons by a gain of function mechanism (Turner and Talbot 2008), although the details remain uncertain. In order to evaluate the validity of a zebrafish model of ALS, a recent study used mRNA microinjection to effect transient over-expression of SOD mutants (Lemmens et al. 2007). The microinjected animals showed normal morphology and normal development of Mauthner neurons, Rohan-Beard sensory neurons and lateral line sensory neurons, despite robust expression of the mutant SOD protein. However, a motor axonopathy was observed, manifest as shortened length and abnormal branching, suggesting that, similar to the human diseases, ubiquitous expression of the mutant protein had evoked motor neuron-specific neuropathology (Lemmens et al. 2007). This important finding is the first example of pathology specific to relevant neuronal populations being provoked by ubiquitous expression of a pathogenic protein in a zebrafish, suggesting that this model may be useful to elucidate the mechanisms underlying specific vulnerability of motor neurons to this mutation. The pathological axonal changes were rescued by simultaneous over-expression of VEGF and were exacerbated by morpholino knockdown of VEGF expression (Lemmens et al. 2007). These findings are similar to those observed in SOD transgenic mice, and suggest that at least some of the biochemical mechanisms underlying the axonopathy in the zebrafish model may be shared with a mammalian model.
Together, these emerging lines of evidence indicate that relevant phenotypic abnormalities, mediated by conserved mechanisms, can be provoked in zebrafish models by transgenic expression of mutant human proteins involved in the pathogenesis of neurodegenerative diseases. These findings are therefore encouraging that mechanistic insights and putative interventions identified in zebrafish models will be applicable to the human diseases.
Conclusions
The use of transgenic zebrafish models for the analysis of human neurodegenerative diseases provides the possibility of exploiting powerful imaging and screening methods to yield novel insights into pathogenesis, new treatment targets, and lead therapeutic compounds. Despite some prominent differences between the nervous systems of zebrafish and humans, the zebrafish CNS shows similar basic organization to that of other vertebrates, and contains neuronal and glial cell types of specific relevance to neurodegenerative diseases. Importantly, there appears to be a significant amount of phylogenetic conservation of many of the genes involved in neurodegenerative disease and their related biochemical pathways. Recent proof of concept studies have shown that transgenic expression of mutated human genes, or synthetic constructs mimicking pathogenic mutations, provokes neurological abnormalities in the zebrafish CNS that can be detected and measured. Several key aspects of the morphological and biochemical features of these models are shared with the relevant human diseases. Consequently, the outlook for application of transgenic zebrafish models to study disease pathogenesis seems excellent. Several questions, which will be critical to exploiting the model fully for clinically relevant applications, are currently unanswered. A complete picture of the initial models is only just emerging, and it is possible that these early positive results are not yet fully representative of the degree to which zebrafish models can yield insights into human diseases. For example, it is uncertain whether biochemical abnormalities underlying the phenotypes detected in larval models at early time points following fertilization accurately mirror those of chronic human disease. Some aspects of rapid disease pathogenesis in the developing zebrafish brain may represent a temporally compressed version of the chronically progressive changes seen in patients. However, it will be important to understand any ways in which the diseases and models diverge mechanistically, in order to provide representative assays for high throughput screens. The next steps in development of this approach will involve further development of stable transgenic lines using different strategies to express pathogenic genes, followed by detailed analysis of the resulting models, selection of those that seem most representative of the relevant disease and then adaptation into formats suitable for high-throughput screening. Optimization of both the models and the assay outputs will be critical to enhance the prospects for generating valuable results from screens. The possibilities for accelerating progress in this important area of neurology are very exciting; consequently the results of the first genetic and chemical screens in zebrafish models of neurodegeneration are awaited with much anticipation.
References
Abeliovich A, Schmitz Y, Farinas I, Choi-Lundberg D, Ho WH, Castillo PE, Shinsky N, Verdugo JM, Armanini M, Ryan A, Hynes M, Phillips H, Sulzer D, Rosenthal A (2000) Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron 25:239–252
Airhart MJ, Lee DH, Wilson TD, Miller BE, Miller MN, Skalko RG (2007) Movement disorders and neurochemical changes in zebrafish larvae after bath exposure to fluoxetine (PROZAC). Neurotoxicol Teratol 29:652–664
Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N (1999) A large-scale insertional mutagenesis screen in zebrafish. Genes Dev 13:2713–2724
Anichtchik OV, Kaslin J, Peitsaro N, Scheinin M, Panula P (2004) Neurochemical and behavioural changes in zebrafish Danio rerio after systemic administration of 6-hydroxydopamine and 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine. J Neurochem 88:443–453
Anichtchik O, Diekmann H, Fleming A, Roach A, Goldsmith P, Rubinsztein DC (2008) Loss of PINK1 function affects development and results in neurodegeneration in zebrafish. J Neurosci 28:8199–8207
Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K (2008) Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci USA 105:1255–1260
Bae YK, Kani S, Shimizu T, Tanabe K, Nojima H, Kimura Y, Higashijima S, Hibi M (2009) Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Dev Biol 330:406–426
Bai Q, Burton EA (2009) Cis-acting elements responsible for dopaminergic neuron-specific expression of zebrafish slc6a3 (dopamine transporter) in vivo are located remote from the transcriptional start site. Neuroscience 164:1138–1151
Bai Q, Mullett SJ, Garver JA, Hinkle DA, Burton EA (2006) Zebrafish DJ-1 is evolutionarily conserved and expressed in dopaminergic neurons. Brain Res 1113:33–44
Bai Q, Garver JA, Hukriede NA, Burton EA (2007) Generation of a transgenic zebrafish model of Tauopathy using a novel promoter element derived from the zebrafish eno2 gene. Nucleic Acids Res 35:6501–6516
Bai Q, Wei X, Burton EA (2009) Expression of a 12-kb promoter element derived from the zebrafish enolase-2 gene in the zebrafish visual system. Neurosci Lett 449:252–257
Baulac S, Lu H, Strahle J, Yang T, Goldberg MS, Shen J, Schlossmacher MG, Lemere CA, Lu Q, Xia W (2009) Increased DJ-1 expression under oxidative stress and in Alzheimer’s disease brains. Mol Neurodegener 4:12
Becker CG, Becker T (2008) Adult zebrafish as a model for successful central nervous system regeneration. Restor Neurol Neurosci 26:71–80
Bernardos RL, Raymond PA (2006) GFAP transgenic zebrafish. Gene Expr Patterns 6:1007–1013
Boehmler W, Obrecht-Pflumio S, Canfield V, Thisse C, Thisse B, Levenson R (2004) Evolution and expression of D2 and D3 dopamine receptor genes in zebrafish. Dev Dyn 230:481–493
Boehmler W, Carr T, Thisse C, Thisse B, Canfield VA, Levenson R (2007) D4 Dopamine receptor genes of zebrafish and effects of the antipsychotic clozapine on larval swimming behaviour. Genes Brain Behav 6:155–166
Bonifati V, Rizzu P, Squitieri F, Krieger E, Vanacore N, van Swieten JC, Brice A, van Duijn CM, Oostra B, Meco G, Heutink P (2003) DJ-1(PARK7), a novel gene for autosomal recessive, early onset Parkinsonism. Neurol Sci 24:159–160
Bretaud S, Lee S, Guo S (2004) Sensitivity of zebrafish to environmental toxins implicated in Parkinson’s disease. Neurotoxicol Teratol 26:857–864
Bretaud S, Allen C, Ingham PW, Bandmann O (2007) p53-dependent neuronal cell death in a DJ-1-deficient zebrafish model of Parkinson’s disease. J Neurochem 100:1626–1635
Brockerhoff SE, Hurley JB, Janssen-Bienhold U, Neuhauss SC, Driever W, Dowling JE (1995) A behavioral screen for isolating zebrafish mutants with visual system defects. Proc Natl Acad Sci USA 92:10545–10549
Brustein E, Chong M, Holmqvist B, Drapeau P (2003) Serotonin patterns locomotor network activity in the developing zebrafish by modulating quiescent periods. J Neurobiol 57:303–322
Cadieux B, Chitramuthu BP, Baranowski D, Bennett HP (2005) The zebrafish progranulin gene family and antisense transcripts. BMC Genom 6:156
Campbell WA, Yang H, Zetterberg H, Baulac S, Sears JA, Liu T, Wong ST, Zhong TP, Xia W (2006) Zebrafish lacking Alzheimer presenilin enhancer 2 (Pen-2) demonstrate excessive p53-dependent apoptosis and neuronal loss. J Neurochem 96:1423–1440
Candy J, Collet C (2005) Two tyrosine hydroxylase genes in teleosts. Biochim Biophys Acta 1727:35–44
Carlson KM, Melcher L, Lai S, Zoghbi HY, Clark HB, Orr HT (2009) Characterization of the zebrafish atxn1/axh gene family. J Neurogenet 23:313–323
Chen YH, Wang YH, Chang MY, Lin CY, Weng CW, Westerfield M, Tsai HJ (2007) Multiple upstream modules regulate zebrafish myf5 expression. BMC Dev Biol 7:1
Chen M, Martins RN, Lardelli M (2009a) Complex splicing and neural expression of duplicated tau genes in zebrafish embryos. J Alzheimers Dis 18(2):305–317
Chen YC, Cheng CH, Chen GD, Hung CC, Yang CH, Hwang SP, Kawakami K, Wu BK, Huang CJ (2009b) Recapitulation of zebrafish sncga expression pattern and labeling the habenular complex in transgenic zebrafish using green fluorescent protein reporter gene. Dev Dyn 238:746–754
Chen YC, Priyadarshini M, Panula P (2009c) Complementary developmental expression of the two tyrosine hydroxylase transcripts in zebrafish. Histochem Cell Biol 132(4):375–381
Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441:1162–1166
Clayton DF, George JM (1998) The synucleins: a family of proteins involved in synaptic function, plasticity, neurodegeneration and disease. Trends Neurosci 21:249–254
Deng HX, Hentati A, Tainer JA, Iqbal Z, Cayabyab A, Hung WY, Getzoff ED, Hu P, Herzfeldt B, Roos RP et al (1993) Amyotrophic lateral sclerosis and structural defects in Cu, Zn superoxide dismutase. Science 261:1047–1051
Diekmann H, Anichtchik O, Fleming A, Futter M, Goldsmith P, Roach A, Rubinsztein DC (2009) Decreased BDNF levels are a major contributor to the embryonic phenotype of huntingtin knockdown zebrafish. J Neurosci 29:1343–1349
Distel M, Wullimann MF, Koster RW (2009) Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc Natl Acad Sci USA 106:13365–13370
Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL (2008) Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol 26:702–708
Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C (1996) A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123:37–46
Fan J, Ren H, Jia N, Fei E, Zhou T, Jiang P, Wu M, Wang G (2008) DJ-1 decreases Bax expression through repressing p53 transcriptional activity. J Biol Chem 283:4022–4030
Flinn L, Mortiboys H, Volkmann K, Koster RW, Ingham PW, Bandmann O (2009) Complex I deficiency and dopaminergic neuronal cell loss in parkin-deficient zebrafish (Danio rerio). Brain 132:1613–1623
Gao Y, Li P, Li L (2005) Transgenic zebrafish that express tyrosine hydroxylase promoter in inner retinal cells. Dev Dyn 233:921–929
Giacomini NJ, Rose B, Kobayashi K, Guo S (2006) Antipsychotics produce locomotor impairment in larval zebrafish. Neurotoxicol Teratol 28:245–250
Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA (1989) Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 3:519–526
Grabher C, Wittbrodt J (2008) Recent advances in meganuclease-and transposon-mediated transgenesis of medaka and zebrafish. Methods Mol Biol 461:521–539
Groth C, Nornes S, McCarty R, Tamme R, Lardelli M (2002) Identification of a second presenilin gene in zebrafish with similarity to the human Alzheimer’s disease gene presenilin2. Dev Genes Evol 212:486–490
Guo S, Wilson SW, Cooke S, Chitnis AB, Driever W, Rosenthal A (1999) Mutations in the zebrafish unmask shared regulatory pathways controlling the development of catecholaminergic neurons. Dev Biol 208:473–487
Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G (1997) High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol 192:289–299
Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Heutink P et al (1998) Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393:702–705
Imamura S, Kishi S (2005) Molecular cloning and functional characterization of zebrafish ATM. Int J Biochem Cell Biol 37:1105–1116
Imbert G, Saudou F, Yvert G, Devys D, Trottier Y, Garnier JM, Weber C, Mandel JL, Cancel G, Abbas N, Durr A, Didierjean O, Stevanin G, Agid Y, Brice A (1996) Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet 14:285–291
Jacquier A, Dujon B (1985) An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell 41:383–394
Jeong JY, Kwon HB, Ahn JC, Kang D, Kwon SH, Park JA, Kim KW (2008) Functional and developmental analysis of the blood–brain barrier in zebrafish. Brain Res Bull 75:619–628
Kapsimali M, Vidal B, Gonzalez A, Dufour S, Vernier P (2000) Distribution of the mRNA encoding the four dopamine D(1) receptor subtypes in the brain of the european eel (Anguilla anguilla): comparative approach to the function of D(1) receptors in vertebrates. J Comp Neurol 419:320–343
Karlovich CA, John RM, Ramirez L, Stainier DY, Myers RM (1998) Characterization of the Huntington’s disease (HD) gene homologue in the zebrafish Danio rerio. Gene 217:117–125
Kawaguchi Y, Okamoto T, Taniwaki M, Aizawa M, Inoue M, Katayama S, Kawakami H, Nakamura S, Nishimura M, Akiguchi I et al (1994) CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet 8:221–228
Kawai H, Arata N, Nakayasu H (2001) Three-dimensional distribution of astrocytes in zebrafish spinal cord. Glia 36:406–413
Kawakami K (2004) Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol 77:201–222
Kawakami K, Shima A, Kawakami N (2000) Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci USA 97:11403–11408
Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M (2004) A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell 7:133–144
Kim CH, Ueshima E, Muraoka O, Tanaka H, Yeo SY, Huh TL, Miki N (1996) Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci Lett 216:109–112
Kimmel CB (1993) Patterning the brain of the zebrafish embryo. Annu Rev Neurosci 16:707–732
Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, Appel B (2006) In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci 9:1506–1511
Koga A, Suzuki M, Inagaki H, Bessho Y, Hori H (1996) Transposable element in fish. Nature 383:30
Lam CS, Korzh V, Strahle U (2005) Zebrafish embryos are susceptible to the dopaminergic neurotoxin MPTP. Eur J Neurosci 21:1758–1762
Landles C, Bates GP (2004) Huntingtin and the molecular pathogenesis of Huntington’s disease. Fourth in molecular medicine review series. EMBO Rep 5:958–963
Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG (2001a) A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56–65
Lee VM, Goedert M, Trojanowski JQ (2001b) Neurodegenerative tauopathies. Annu Rev Neurosci 24:1121–1159
Leimer U, Lun K, Romig H, Walter J, Grunberg J, Brand M, Haass C (1999) Zebrafish (Danio rerio) presenilin promotes aberrant amyloid beta-peptide production and requires a critical aspartate residue for its function in amyloidogenesis. Biochemistry 38:13602–13609
Lemmens R, Van Hoecke A, Hersmus N, Geelen V, D’Hollander I, Thijs V, Van Den Bosch L, Carmeliet P, Robberecht W (2007) Overexpression of mutant superoxide dismutase 1 causes a motor axonopathy in the zebrafish. Hum Mol Genet 16:2359–2365
Long Q, Meng A, Wang H, Jessen JR, Farrell MJ, Lin S (1997) GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development 124:4105–4111
Lumsden AL, Henshall TL, Dayan S, Lardelli MT, Richards RI (2007) Huntingtin-deficient zebrafish exhibit defects in iron utilization and development. Hum Mol Genet 16:1905–1920
Ma PM (2003) Catecholaminergic systems in the zebrafish. IV. Organization and projection pattern of dopaminergic neurons in the diencephalon. J Comp Neurol 460:13–37
Malicki J, Neuhauss SC, Schier AF, Solnica-Krezel L, Stemple DL, Stainier DY, Abdelilah S, Zwartkruis F, Rangini Z, Driever W (1996) Mutations affecting development of the zebrafish retina. Development 123:263–273
Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, Ma L, Samara-Kuko E, Gysemans C, Pryputniewicz D, Miskey C, Fletcher B, Vandendriessche T, Ivics Z, Izsvak Z (2009) Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 41:753–761
McGraw HF, Nechiporuk A, Raible DW (2008) Zebrafish dorsal root ganglia neural precursor cells adopt a glial fate in the absence of neurogenin1. J Neurosci 28:12558–12569
McKinley ET, Baranowski TC, Blavo DO, Cato C, Doan TN, Rubinstein AL (2005) Neuroprotection of MPTP-induced toxicity in zebrafish dopaminergic neurons. Brain Res Mol Brain Res 141:128–137
McNaught KS, Belizaire R, Isacson O, Jenner P, Olanow CW (2003) Altered proteasomal function in sporadic Parkinson’s disease. Exp Neurol 179:38–46
Meng A, Tang H, Ong BA, Farrell MJ, Lin S (1997) Promoter analysis in living zebrafish embryos identifies a cis-acting motif required for neuronal expression of GATA-2. Proc Natl Acad Sci USA 94:6267–6272
Meng S, Ryu S, Zhao B, Zhang DQ, Driever W, McMahon DG (2008a) Targeting retinal dopaminergic neurons in tyrosine hydroxylase-driven green fluorescent protein transgenic zebrafish. Mol Vis 14:2475–2483
Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA (2008b) Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol 26:695–701
Miller VM, Nelson RF, Gouvion CM, Williams A, Rodriguez-Lebron E, Harper SQ, Davidson BL, Rebagliati MR, Paulson HL (2005) CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J Neurosci 25:9152–9161
Molina G, Vogt A, Bakan A, Dai W, Queiroz de Oliveira P, Znosko W, Smithgall TE, Bahar I, Lazo JS, Day BW, Tsang M (2009) Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat Chem Biol 5:680–687
Mortiboys H, Thomas KJ et al (2008) Mitochondrial function and morphology are impaired in parkin-mutant fibroblasts. Ann Neurol 64(5):555–565
Mueller T, Vernier P, Wullimann MF (2004) The adult central nervous cholinergic system of a neurogenetic model animal, the zebrafish Danio rerio. Brain Res 1011:156–169
Mullett SJ, Hinkle DA (2009) DJ-1 knock-down in astrocytes impairs astrocyte-mediated neuroprotection against rotenone. Neurobiol Dis 33:28–36
Musa A, Lehrach H, Russo VA (2001) Distinct expression patterns of two zebrafish homologues of the human APP gene during embryonic development. Dev Genes Evol 211:563–567
Narendra D, Tanaka A, Suen DF, Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183:795–803
Nasevicius A, Ekker SC (2000) Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet 26:216–220
Orr HT, Chung MY, Banfi S, Kwiatkowski TJ Jr, Servadio A, Beaudet AL, McCall AE, Duvick LA, Ranum LP, Zoghbi HY (1993) Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet 4:221–226
Paquet D, Bhat R, Sydow A, Mandelkow EM, Berg S, Hellberg S, Falting J, Distel M, Koster RW, Schmid B, Haass C (2009) A zebrafish model of tauopathy allows in vivo imaging of neuronal cell death and drug evaluation. J Clin Invest 119:1382–1395
Park HC, Kim CH, Bae YK, Yeo SY, Kim SH, Hong SK, Shin J, Yoo KW, Hibi M, Hirano T, Miki N, Chitnis AB, Huh TL (2000) Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol 227:279–293
Peri F, Nusslein-Volhard C (2008) Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell 133:916–927
Reiner A, Northcutt RG (1992) An immunohistochemical study of the telencephalon of the senegal bichir (Polypterus senegalus). J Comp Neurol 319:359–386
RG HDC (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72:971–983
Rink E, Wullimann MF (2001) The teleostean (zebrafish) dopaminergic system ascending to the subpallium (striatum) is located in the basal diencephalon (posterior tuberculum). Brain Res 889:316–330
Rink E, Wullimann MF (2002) Development of the catecholaminergic system in the early zebrafish brain: an immunohistochemical study. Brain Res Dev Brain Res 137:89–100
Rink E, Wullimann MF (2004) Connections of the ventral telencephalon (subpallium) in the zebrafish (Danio rerio). Brain Res 1011:206–220
Riparbelli MG, Callaini G (2007) The Drosophila parkin homologue is required for normal mitochondrial dynamics during spermiogenesis. Dev Biol 303:108–120
Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62
Ryu S, Holzschuh J, Mahler J, Driever W (2006) Genetic analysis of dopaminergic system development in zebrafish. J Neural Transm Suppl (70):61–66
Sallinen V, Torkko V, Sundvik M, Reenila I, Khrustalyov D, Kaslin J, Panula P (2009a) MPTP and MPP+ target specific aminergic cell populations in larval zebrafish. J Neurochem 108(3):719–731
Sallinen V, Sundvik M, Reenila I, Peitsaro N, Khrustalyov D, Anichtchik O, Toleikyte G, Kaslin J, Panula P (2009b) Hyperserotonergic phenotype after monoamine oxidase inhibition in larval zebrafish. J Neurochem 109:403–415
Sato Y, Miyasaka N, Yoshihara Y (2007) Hierarchical regulation of odorant receptor gene choice and subsequent axonal projection of olfactory sensory neurons in zebrafish. J Neurosci 27:1606–1615
Scheer N, Campos-Ortega JA (1999) Use of the Gal4–UAS technique for targeted gene expression in the zebrafish. Mech Dev 80:153–158
Schiffer NW, Broadley SA, Hirschberger T, Tavan P, Kretzschmar HA, Giese A, Haass C, Hartl FU, Schmid B (2007) Identification of anti-prion compounds as efficient inhibitors of polyglutamine protein aggregation in a zebrafish model. J Biol Chem 282:9195–9203
Sharma SC, Berthoud VM, Breckwoldt R (1989) Distribution of substance P-like immunoreactivity in the goldfish brain. J Comp Neurol 279:104–116
Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 25:302–305
Solnica-Krezel L, Schier AF, Driever W (1994) Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics 136:1401–1420
Soroldoni D, Hogan BM, Oates AC (2009) Simple and efficient transgenesis with meganuclease constructs in zebrafish. Methods Mol Biol 546:117–130
Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M (1998) alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc Natl Acad Sci USA 95:6469–6473
Stuart GW, McMurray JV, Westerfield M (1988) Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development 103:403–412
Sun Z, Gitler AD (2008) Discovery and characterization of three novel synuclein genes in zebrafish. Dev Dyn 237:2490–2495
Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS (2002) I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev 118:91–98
Thirumalai V, Cline HT (2008) Endogenous dopamine suppresses initiation of swimming in prefeeding zebrafish larvae. J Neurophysiol 100:1635–1648
Tomasiewicz HG, Flaherty DB, Soria JP, Wood JG (2002) Transgenic zebrafish model of neurodegeneration. J Neurosci Res 70:734–745
Tu PH, Galvin JE, Baba M, Giasson B, Tomita T, Leight S, Nakajo S, Iwatsubo T, Trojanowski JQ, Lee VM (1998) Glial cytoplasmic inclusions in white matter oligodendrocytes of multiple system atrophy brains contain insoluble alpha-synuclein. Ann Neurol 44:415–422
Turner BJ, Talbot K (2008) Transgenics, toxicity and therapeutics in rodent models of mutant SOD1-mediated familial ALS. Prog Neurobiol 85:94–134
Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW (2004) Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304:1158–1160
Vila M, Jackson-Lewis V, Guegan C, Wu DC, Teismann P, Choi DK, Tieu K, Przedborski S (2001) The role of glial cells in Parkinson’s disease. Curr Opin Neurol 14:483–489
Wen L, Wei W, Gu W, Huang P, Ren X, Zhang Z, Zhu Z, Lin S, Zhang B (2008) Visualization of monoaminergic neurons and neurotoxicity of MPTP in live transgenic zebrafish. Dev Biol 314:84–92
Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, O’Kane CJ, Floto RA, Rubinsztein DC (2008) Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol 4:295–305
Wullimann MF, Rink E (2002) The teleostean forebrain: a comparative and developmental view based on early proliferation, Pax6 activity and catecholaminergic organization. Brain Res Bull 57:363–370
Wullimann MF, Rupp B, Reichert H (1996) Neuroanatomy of the zebrafish brain. Birkhauser-Verlag, Berlin
Yang Z, Jiang H, Zhao F, Shankar DB, Sakamoto KM, Zhang MQ, Lin S (2007) A highly conserved regulatory element controls hematopoietic expression of GATA-2 in zebrafish. BMC Dev Biol 7:97
Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B (2008) Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci USA 105:7070–7075
Yang Z, Jiang H, Lin S (2009) Bacterial artificial chromosome transgenesis for zebrafish. Methods Mol Biol 546:103–116
Yoshida M, Macklin WB (2005) Oligodendrocyte development and myelination in GFP-transgenic zebrafish. J Neurosci Res 81:1–8
Yoshida H, Craxton M, Jakes R, Zibaee S, Tavare R, Fraser G, Serpell LC, Davletov B, Crowther RA, Goedert M (2006) Synuclein proteins of the pufferfish Fugu rubripes: sequences and functional characterization. Biochemistry 45:2599–2607
Zoghbi HY, Orr HT (2009) Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. J Biol Chem 284:7425–7429
Zon LI, Peterson RT (2005) In vivo drug discovery in the zebrafish. Nat Rev Drug Discov 4:35–44
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sager, J.J., Bai, Q. & Burton, E.A. Transgenic zebrafish models of neurodegenerative diseases. Brain Struct Funct 214, 285–302 (2010). https://doi.org/10.1007/s00429-009-0237-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-009-0237-1