Abstract
Although lung cancer continues to be the leading cause of cancer-related death, accurate diagnosis followed by personalized treatment is expected to raise the 5-year survival rate. Targeted therapies are now in routine clinical use, in particular for lung adenocarcinoma (ADC). Fibroblast growth factor receptor 1 (FGFR1) has recently emerged as a molecular target, especially in squamous cell/epidermoid carcinoma (SQC) of the lung. This paper evaluates FGFR1 expression and gene copy number in adenocarcinomas, squamous cell carcinomas, pleomorphic carcinomas (PLEOMC) and adenosquamous carcinomas (ADSQC) of the lung and also explores the epithelial-mesenchymal transition (EMT) pathway. We studied 76 lung carcinomas: 34 ADC, 24 SQC, 10 PLEOMC and 8 ADSQC. FGFR1 expression was evaluated by immunohistochemistry and gene amplification by fluorescence in situ hybridization (FISH). Higher FGFR1 protein expression was observed in all tumour types compared to non-tumour tissue. FGFR1 expression was higher in ADC and PLEOMC than in SQC. We found a tendency to higher expression in ADC than in SQC and significantly higher expression in PLEOMC than in other histological subtypes. FISH-based amplification of FGFR1 was identified in 15 (20 %) lung carcinomas: 5 (15 %) ADC, 5 (21 %) SQC, 3 (30 %) PLEOMC and 2 (25 %) ADSQC. Amplification was more frequent in SQC without significant differences. FGFR1 protein is expressed in the majority of lung carcinomas, though it is higher in ADC and PLEOMC (the latter may reflect the importance of FGFR1 control of the EMT pathway). FGFR1 amplification was identified in all types of lung carcinoma. Although FGFR1 is most frequently amplified in SQC, other histological types merit assessment of FGFR1 amplification, in order to select patients that might benefit from targeted therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although lung cancer continues to be the leading cause of cancer-related death, accurate diagnosis followed by personalized treatment is expected to raise the 5-year survival rate, following recent advances in molecular targeted therapy [1–5]. Predictive and targetable oncogenic mutations have mainly been found in adenocarcinomas (ADC) and in tumours of never smokers. Recently, several promising genomic alterations associated with biological pathways have been identified in squamous cell/epidermoid carcinoma (SQC) of the lung, including PIK3CA/AKT1, PTEN and fibroblast growth factor receptor 1 (FGFR1) alterations [6].
FGFRs belong to the super family of receptor tyrosine kinases and are encoded by four genes located on chromosomes 8p11.23-p11.22 (FGFR1), 10q26 (FGFR2), 4p16.3 (FGFR3) and 5q35.2 (FGFR4). They are related to a large family of molecules involved in physiological processes, dysregulation of which may lead to cancer development [7–9]. FGFR1 is a transmembrane tyrosine kinase receptor which promotes cell growth, differentiation and survival by downstream activation of PI3K-AKT, RAS-MEK-MAPK, STAT, Src and PLC signalling pathways [7, 10]. FGFR1 promotes epithelial-mesenchymal transition (EMT) and tissue remodelling by activating these pathways, and upregulation of its expression leads to cell transformation and carcinogenesis [11–15].
FGFR1 is an oncogene that can potentially be targeted by tyrosine kinase inhibitors. It has become the target for therapies now in routine clinical use for lung adenocarcinomas. Recently, comprehensive genomic characterization of SQC has been undertaken to search for potentially targetable mutations [16]. Our study aimed to evaluate FGFR1 expression and gene copy number in ADC, SQC, pleomorphic (PLEOMC) and adenosquamous (ADSQC) carcinomas. We also explored the EMT pathway by studying PLEOMC and vimentin (vim) expression on ADC. ADC were also studied according to TTF1 expression, and SQC according to CK7 expression. The purpose was to identify molecular characteristics of these lung tumour subtypes in order to characterize their potential suitability for targeted therapy.
Material and methods
Material
Tumour samples were selected from surgical resection specimens of 76 lung carcinomas (34 adenocarcinomas, 24 squamous cell carcinomas, 10 pleomorphic carcinomas and 8 adenosquamous carcinomas). Only samples with more than 75 % of tumour cells were included in this study.
Clinical and pathological characteristics are summarized in Table 1. Median age at diagnosis was 65 years (range 40–87 years); 56 patients were male and 20 were female. Patients included 23 non-smokers, 29 ex-smokers and 24 current smokers. Male gender and smokers were more prevalent in SQC. Male predominance was also found for ADC but with more non-smokers and ex-smokers. A predominance of male smokers was noted for PLEOMC and ADSQC. Stages I and II were more prevalent. Metastases were more frequently diagnosed in lymph nodes.
ADC were classified according to 2015 WHO criteria (main patterns and all present other patterns in the order of their quantitative presence). ADC were subclassified based upon patterns of immunohistochemical marker expression as follows: (1) CK7+/TTF1+/vim+ (14 cases); (2) CK7+/TTF1+/vim− (9 cases); (3) CK7+/TTF1−/vim− (6 cases); and (4) CK7+/TTF−/vim+ (5 cases). ADC were considered as vim positive when the expression was diffusely present in the tumour even though expression was frequently heterogeneous. SQC (24) were subclassified according to CK7 expression: (1) CK7− (17 cases) and (2) CK7+ (7 cases), while all expressed CK5.6. PLEOMC (10) and ADSQC (8) were diagnosed according to the 2015 WHO criteria, and carcinomas with fewer than 10 % fusiform or giant cells were not considered as PLEOMC [17]. PLEOMC showed an adenocarcinoma component in 8 cases and a squamous cell carcinoma component in 2 cases.
Methods
At least two sections of each tumour were independently evaluated by two pathologists. In order to establish tumour subgroups, immunohistochemical staining for the differentiation markers CK7, TTF1, CK5.6 and vim was performed according to existing protocols (see Table 2). For assessment of FGFR1 protein expression, immunohistochemistry was applied. Endogenous peroxidase activity was quenched by incubation in 3 % diluted hydrogen peroxide (15 min). Non-specific binding of primary antibodies was blocked with Ultra V Block (Ultra Vision Kit, TP-125-UB, Lab Vision Corporation, Fremont, CA, USA). A primary antibody against FGFR1 (Polyclonal, Thermo Scientific; dilution 1:75) was applied to the sections, and incubation was 30 min at room temperature. Sections were then washed with phosphate-buffered saline (PBS) (Ultra Vision, TP-125-PB, Lab Vision Corporation, Fremont, CA, USA) and subsequently incubated (15 min) with biotin-labelled secondary antibody (Ultra Vision Kit, TP-125-BN, Lab Vision Corporation, Fremont, CA, USA). Primary antibody binding was visualized using peroxidase-conjugated streptavidin (Ultra Vision Kit, TP-125-HR, Lab Vision Corporation, Fremont, CA, USA) with 3′,3-diaminobenzidine tetrahydrochloride (DAB) (RE7190-K, Novocastra Laboratories Ltd., Newcastle, UK) as chromogen, according to the manufacturer’s instructions. Haematoxylin was used to counterstain the slides, which were then dehydrated and mounted. In parallel, known positive (squamous cell carcinoma with keratin pearls) and negative controls were used.
Evaluation of immunohistochemistry results was performed by two experienced lung pathologists. The intensity of expression was scored in four grades: 0, 1+, 2+ and 3+. The percentage of positive cells was also scored, and a global score was obtained by multiplying intensity by percentage of positive cells. External positive controls (1+, 2+ and 3+) were used.
Fluorescence in situ hybridization (FISH) was carried out on tissue sections using a FGFR1 (8p11)/SE 8 (D8Z1) probe (Kreatech, Leica Biosystems). Sections were deparaffinized in xylene (15 min), dehydrated in ethanol (2× for 5 min) and heated in citrate buffer in a pressure cooker for 4 min. They were then washed in 2× sodium citrate (2× SSC) for 5 min, digested in proteinase K (10 min at 37 °C), washed in 2× SSC (5 min), dehydrated in increasing concentrations of ethanol (70, 90 and 100 %) and air-dried. To the slides, 10 μl of probe solution was added and a coverslip was applied, sealed with rubber cement. Slides were denatured at 78 °C (8 min) and hybridized at 37 °C for at least 16 h in a humidified chamber. Post-hybridization washing was in 50 % formamide and PBS at 46 °C (4 min) and 2× SSC (2 min), and counterstaining with 4′,6-diamidino-2-phenylindole (DAPI), and finally sections were air-dried.
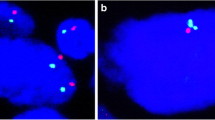
In each section, 100 tumour cells were analysed using a ×100 oil immersion objective on a Nikon 80i fluorescence microscope with appropriate filters. FGFR1 FISH results were classified as high-level amplification when the FGFR1/CEN8 ratio was ≥2.0, with the average number of FGFR1 signals per tumour cell nucleus ≥6 and the percentage of tumour cells containing ≥15 FGFR1 signals or large clusters ≥10 %, or as low-level amplification according to criteria published earlier [18–20]. Two technicians and a pathologist independently scored the results.
Statistical analysis was performed using SPSS statistics 22.0 software for Windows (SPSS, Chicago, USA). Correlations between clinicopathological and histological parameters and FGFR1 status were analysed using χ 2 and Fisher’s exact test. Comparison of FGFR1 protein expression levels between lung carcinomas and non-tumour tissue was done by ANOVA test (when comparing global expression (intensity × % of positive cells)) and by χ2 and Fisher’s exact test (when comparing groups according expression intensity (3+, 2+, 1+, 0)). Correlations of FGFR1 expression/amplification levels among the histological types were assessed using linear (Pearson) correlation. p values <0.05 were considered statistically significant.
Results
Figure 1 illustrates the different histological types based upon HE staining, as well as subtypes according to expression patterns of TTF1, vim and CK7. In non-tumour tissue, FGFR1 expression was seen in respiratory epithelium; pneumocytes and stromal cells showed no or rare expression (Fig. 2). We found significantly higher FGFR1 protein expression in all tumour groups than in non-tumour tissue (Table 3). Significantly higher expression was seen in ADC compared to SQC (p = 0.0232). Significantly higher expression was also observed in PLEOMC compared to SQC (p = 0.0265). No significant differences in expression were found between ADC and ADSQC (p = 0.158), ADC and PLEOMC (p = 0.3190), SQC and ADSQC (p = 0.9651) or ADSQC and PLEOMC (p = 0.1189) (Table 3). We found no differences between FGFR1 expression in SQC with or without CK7 expression (p = 0.2502), ADC with or without TTF1 expression (p = 0.123) or with or without vim expression (p = 0.301) (Table 3).
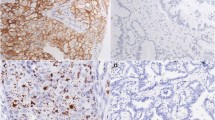
HE-stained tumour samples classified according to histological and subclassified according to immunohistochemical features, namely, expression of TTF1, vim and CK7. a Adenocarcinoma (ADC) vim+/TTF1−, acinar pattern; HE, ×200. b ADC vim−/TTF1−, papillary and micropapillary patterns; HE, ×200. c ADC vim+/TTF1+, micropapillary pattern; HE, ×400. d ADC vim−/TTF1+, solid pattern; HE, ×100. e ADC vim−/TTF1+, cribriform pattern; HE, ×100. f ADC vim−/TTF1−, solid with clear cells; HE, ×200. g ADC vim−/TTF1−, mucinous pattern; HE, ×100. h squamous cell carcinoma (SQC), CK7+; HE, ×100. i SQC, CK7−; HE, ×200. j adenosquamous carcinoma; HE, ×100. k Pleomorphic carcinoma (PLEOMC) with giant cells; HE, ×200. l PLEOMC with fusiform cells; HE, ×200
FGFR1 immunohistochemical expression in lung carcinomas according to histological type and subclassified according to CK7, TTF1 and vim expressions. a Normal respiratory epithelium; FGFR1, ×100. b Adenocarcinoma (ADC) vim−/TTF1+; FGFR1 1+, ×200. c ADC vim+/TTF1−; FGFR1 2+, ×200. d ADC TTF1+/vim+; FGFR1 3+, ×200. e Squamous cell carcinoma (SQC) CK7−; FGFR1 1+, ×400. f SQC CK7−; FGFR1 2+, ×400. g SQC CK7−; FGFR1 3+, ×200. h SQC CK7+; FGFR1 3+, ×400. i Pleomorphic carcinoma (PLEOMC); FGFR1 3+, ×400. j PLEOMC; FGFR1 3+, ×200. k Adenosquamous carcinoma (ADSQC); FGFR1−, ×200. l ADSQC; FGFR1 3+, ×200
When immunohistochemical results were stratified according to expression level 1+ or 2+/3+, a trend was found for higher expression in ADC than in SQC (p = 0.061) and significantly higher expression in PLEOMC than in ADC (p = 0.0212), SQC (p = 0.0004) and ADSQC (p = 0.0015). No differences were observed between the remaining features (Table 4).
FGFR1 amplification by FISH (Fig. 3) was found in 15 cases (20 %). The proportion of FGFR1-amplified cases was not significantly different between tumour types: 5 (15 %) ADC, 5 (21 %) SQC, 3 (30 %) PLEOMC and 2 (25 %) ADSQC (Table 5). Low-level FGFR1 amplification was found in 8 cases (3 ADC, 3 SQC, 1 PLEOMC and 1 ADSQC), and high-level FGFR1 amplification in 7 cases (2 ADC, 2 SQC, 2 PLEOMC and 1 ADSQC). The difference in FISH-positive amplified FGFR1 cases between SQC and ADC was not significant. Of 8 ADSQC, 2 were FGFR1 FISH positive (25 %) and of 10 PLEOMC, 3 (30 %) (Table 5). Although SQC was more often positive than ADC, the difference was not significant (Table 5 ). There were no significant differences in FGFR1 FISH results according to gender or smoking status, neither for ADC nor for SQC, while a tendency was noted for a higher frequency of FGFR1 amplification in ADC of smokers. No correlations were found between FGFR1 immunohistochemical expression and FISH results (Spearman rank correlation p = 0.3323 and linear correlation p = 0.681). All cases of PLEOMC showed 3+ diffuse FGFR1 protein expression, regardless of the epithelial component present. FGFR1 was amplified by FISH in 3 (30 %) cases: 2 with a SQC and one with an ADC component. In ADSQC, no differences between the squamous and adenocarcinoma components were found neither for FGFR1 protein expression nor for FGFR1 FISH results.
Discussion
Gene changes involved in the development of cancer tend to effect three main types of genes: proto-oncogenes, tumour suppressor genes and DNA repair genes. These genes are called drivers of cancer [3–5]. The identification of mutations critical for tumour growth is crucial, as they can be targeted with improvement in survival. Driver events in lung cancer have been identified in ADC in patients who never smoked [3–5]. In SQC, primarily a smoker’s disease, targetable gene changes which would allow more effective targeted therapies have not (yet) been identified [6, 21]. FGFR1 and FGF have recently emerged as driving oncogenes, sufficient to drive tumour growth [22]. Alterations of the FGFR gene have been recognized in gastric, breast, oral squamous cell, ovarian and bladder carcinomas and in SQC of the lung [6, 23, 24]. Several reports have shown that FGFR1-mediated signalling pathways play an essential role in lung carcinogenesis, some suggesting that the activation of the FGF pathway is an early event [6, 21, 25–31]. FGFR1 amplification is associated with a response to FGFR inhibitors and different FGFR1 inhibitors are currently under study [6, 27, 30, 32–37]. Therefore, assessment of FGFR1 gene status is likely to gain in importance.
We found higher FGFR1 protein expression in tumour tissue compared to non-tumour adjacent parenchyma/bronchial epithelium, which suggests that the FGFR1 pathway might be implicated in lung carcinogenesis and tumour growth, even in the absence of FGFR1 gene copy number increase. Other authors have also found higher protein expression in lung carcinoma compared to non-tumour tissue [26]. Our results suggest that FGFR1 protein overexpression is frequent in lung carcinomas, with stronger expression in ADC and PLEOMC than in SQC. Stronger immunohistochemical expression in PLEOMC than in SQC suggests that overexpression is important in this group of tumours. As some authors have found FGFR1-targeting therapy efficacy in strongly stained (3+) cases, we also stratified our results according to this end-point [38]. We observed strong immunohistochemical expression more frequently in ADC than in SQC and in PLEOMC more frequently than in any other type of carcinoma. No differences were found between ADC and ADSQC, which might reflect the compound morphology and FGFR1 involvement in the carcinogenesis of both.
Some SQC also expressed CK7, representing a group of less differentiated tumours with some features overlapping with ADC but without significant differences in terms of FGFR1 expression and gene copy number, compared to CK7-negative SQC. We found no significant differences between ADC in relation to expression of TTF1 (TTF1-positive vs. TTF1-negative cases). This suggests that TTF1 expression probably has no effect on FGFR1 pathway activation and rather reflects tumour histogenesis. We intended to explore the EMT pathway on PLEOMC and on ADC by studying vim expression. EMT is associated with tumour progression, histological grade, metastatic propensity and probably also drug resistance, including to tyrosine kinase inhibitors (TKIs) [39–42]. EMT in epithelial tumours is characterized by fusiform cells; expression of vim, fibronectin and N-cadherin; and reduced expression of epithelial markers such as E-cadherin. We found 3 (30 %) PLEOMC (tumours with EMT characteristics) and 4 (12 %) vim-positive ADC (tumours with biological EMT behaviour not yet clarified) but only 1 (3 %) vim-negative ADC (tumours without EMT activation) to be FGFR1 FISH positive. More studies on a larger number of samples need to be done in order to clarify potential interactions between EMT and FGFR1 that might have implications for therapy choice, as EMT is associated with drug resistance and poor survival [39, 40, 42].
We found a rate of FGFR1 amplification in SQC similar to that in the literature and a higher rate for ADC. Previously published results are summarized in a supplementary table [6,18,43-49]. We also found FGFR1 amplification in ADC and PLEOMC, not different from the findings in other histological subtypes. Our results show that FGFR1 protein expression and gene copy number gain are frequent events in PLEOMC, regardless of the type of epithelial component (squamous or adenocarcinoma). Overexpression in PLEOMC may reflect implication of FGFR1 in EMT pathways as they are a perfect example of a tumour characterized by EMT. We favour assessment of FGFR1 amplification and protein expression in PLEOMC and ADSQC in order to select patients for targeted therapy. As some authors identified FGFR1 amplification in small-cell carcinomas, these should also be included in FGFR1 amplification screening protocols [20], and the same holds true for large-cell carcinomas, ADSQC and PLEOMC [26, 43, 48].
We found a trend towards more frequent FGFR1 amplification in ADC in smoking patients. This result is controversial since others found no correlations with clinical parameters [45, 48], while other authors reported associations between FGFR1 amplification and male gender and smoker status [38].
We found no correlations between FGFR1 amplification and FGFR1 protein expression, in agreement with an earlier report [47]. This implies that overexpression may not reflect amplification. Other mechanisms might be involved such as slower protein degradation or higher translation. A practical problem in our series might be the small number of cases. For EGFR, activating mutations have positive predictive value for TKI response [50]. However, for HER2 in breast carcinoma, protein expression and gene copy number have predictive value [51–53].
Recently, several studies have shown that FGFR1 gene copy number is a valid predictive biomarker for response to FGFR1-targeting TKIs [6, 21, 32, 38, 44, 54]. Some reports mention absence of response to FGFR1 TKIs in patients with FGFR1 amplification. This might be related to concomitant gene abnormalities like PI3KCA mutation or amplification [55]. Ultimately, an optimal testing algorithm for patient treatment may need to include targets other than FGFR1.
Recently, new scientific data intensified the discussion on the use of FGFR1 messenger RNA (mRNA) and/or protein expression as biomarkers for TKI response. Wynes et al. published data in support of the importance of FGFR1 protein and mRNA expression. They showed that FGFR1 mRNA and protein are frequently expressed in lung cancer and lung cancer cell lines, independent of FGFR1 gene copy number. Both predicted ponatinib response in lung cancer cell lines more accurately than gene copy number, which calls for clinical testing of this approach [56]. A clinical trial assessing response to ponatinib according to FGFR1 mRNA expression and FGFR1 gene copy number or both is under way [56]. Some authors identified a response in tumours with an FGFR1 amplification and strong expression of FGFR1 (3+) [38]. These findings question selection criteria for FGFR1 TKI therapy, and resolving this issue will have clear benefits in patient management and improve our understanding.
Our study has some limitations, such as sample size (76 patients: 34 adenocarcinomas, 24 squamous cell carcinomas, 10 pleomorphic carcinomas, and 8 adenosquamous carcinomas) which might not be sufficient to detect small differences in FGFR1 expression levels between different groups. We cannot exclude effects of other parameters (i.e. age, life habits, diet type, etc.) as confounding factors. A study with a large number of patients will be necessary to confirm our findings. However, our results provide preliminary evidence that FGFR1 expression might have an important role in therapy decision-making in different types of pulmonary carcinoma and needs to be further studied in this multifactorial disease.
In conclusion, we found that FGFR1 protein is expressed in most lung carcinomas, more strongly in ADC and PLEOMC than in SQC. FGFR1 amplification was found (in order of frequency) in PLEOMC, ADC, SQC and ADSQC. We propose that FGFR1 assessment, using both IHC and FISH, should be carried out in lung carcinoma patients to select the best therapy option. Although FGFR1 amplification was observed more frequently in SQC, the other histological types should also be investigated for FGFR1 amplification, in order to select patients that will benefit from targeted therapy.
References
Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. CA Cancer J Clin 64:9–29
Schultheis AM, Bos M, Schmitz K, Wilsberg L, Binot E, et al. (2013) Fibroblast growth factor receptor 1 (FGFR1) amplification is a potential therapeutic target in small-cell lung cancer. Mod Pathol
Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Gainor J, et al. (2011) Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol 12:1004–1012
Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, et al. (2010) Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 362:2380–2388
Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, et al. (2010) Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 11:121–128
Weiss J, Sos ML, Seidel D, Peifer M, Zander T, et al. (2010) Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2:62ra93
Turner N, Grose R (2010) Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 10:116–129
Dailey L, Ambrosetti D, Mansukhani A, Basilico C (2005) Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev 16:233–247
Ribatti D, Vacca A, Rusnati M, Presta M (2007) The discovery of basic fibroblast growth factor/fibroblast growth factor-2 and its role in haematological malignancies. Cytokine Growth Factor Rev 18:327–334
Larsson H, Klint P, Landgren E, Claesson-Welsh L (1999) Fibroblast growth factor receptor-1-mediated endothelial cell proliferation is dependent on the Src homology (SH) 2/SH3 domain-containing adaptor protein Crk. J Biol Chem 274:25726–25734
Bakin AV, Rinehart C, Tomlinson AK, Arteaga CL (2002) p38 mitogen-activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. J Cell Sci 115:3193–3206
Yang Y, Pan X, Lei W, Wang J, Shi J, et al. (2006) Regulation of transforming growth factor-beta 1-induced apoptosis and epithelial-to-mesenchymal transition by protein kinase A and signal transducers and activators of transcription 3. Cancer Res 66:8617–8624
Zuo W, Chen YG (2009) Specific activation of mitogen-activated protein kinase by transforming growth factor-beta receptors in lipid rafts is required for epithelial cell plasticity. Mol Biol Cell 20:1020–1029
Ellenrieder V, Hendler SF, Boeck W, Seufferlein T, Menke A, et al. (2001) Transforming growth factor beta1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res 61:4222–4228
Arbeit JM, Olson DC, Hanahan D (1996) Upregulation of fibroblast growth factors and their receptors during multi-stage epidermal carcinogenesis in K14-HPV16 transgenic mice. Oncogene 13:1847–1857
Cancer Genome Atlas Research N (2012) Comprehensive genomic characterization of squamous cell lung cancers. Nature 489:519–525
Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG (2015) WHO classification of tumours of the lung, pleura, thymus and heart. IARC, Lyon, 412 p
Schildhaus HU, Heukamp LC, Merkelbach-Bruse S, Riesner K, Schmitz K, et al. (2012) Definition of a fluorescence in-situ hybridization score identifies high- and low-level FGFR1 amplification types in squamous cell lung cancer. Mod Pathol 25:1473–1480
Kim HR, Kim DJ, Kang DR, Lee JG, Lim SM, et al. (2013) Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol 31:731–737
Schultheis AM, Bos M, Schmitz K, Wilsberg L, Binot E, et al. (2014) Fibroblast growth factor receptor 1 (FGFR1) amplification is a potential therapeutic target in small-cell lung cancer. Mod Pathol 27:214–221
Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, et al. (2011) Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One 6:e20351
Ware KE, Hinz TK, Kleczko E, Singleton KR, Marek LA, et al. (2013) A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis 2:e39
Yoshimura N, Sano H, Hashiramoto A, Yamada R, Nakajima H, et al. (1998) The expression and localization of fibroblast growth factor-1 (FGF-1) and FGF receptor-1 (FGFR-1) in human breast cancer. Clin Immunol Immunopathol 89:28–34
Wesche J, Haglund K, Haugsten EM (2011) Fibroblast growth factors and their receptors in cancer. Biochem J 437:199–213
Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, et al. (2009) SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 41:1238–1242
Behrens C, Lin HY, Lee JJ, Raso MG, Hong WK, et al. (2008) Immunohistochemical expression of basic fibroblast growth factor and fibroblast growth factor receptors 1 and 2 in the pathogenesis of lung cancer. Clin Cancer Res 14:6014–6022
Berger W, Setinek U, Mohr T, Kindas-Mugge I, Vetterlein M, et al. (1999) Evidence for a role of FGF-2 and FGF receptors in the proliferation of non-small cell lung cancer cells. Int J Cancer 83:415–423
Donnem T, Al-Shibli K, Al-Saad S, Busund LT, Bremnes RM (2009) Prognostic impact of fibroblast growth factor 2 in non-small cell lung cancer: coexpression with VEGFR-3 and PDGF-B predicts poor survival. J Thorac Oncol 4:578–585
Fischer H, Taylor N, Allerstorfer S, Grusch M, Sonvilla G, et al. (2008) Fibroblast growth factor receptor-mediated signals contribute to the malignant phenotype of non-small cell lung cancer cells: therapeutic implications and synergism with epidermal growth factor receptor inhibition. Mol Cancer Ther 7:3408–3419
Kuhn H, Kopff C, Konrad J, Riedel A, Gessner C, et al. (2004) Influence of basic fibroblast growth factor on the proliferation of non-small cell lung cancer cell lines. Lung Cancer 44:167–174
Volm M, Koomagi R, Mattern J, Stammler G (1997) Prognostic value of basic fibroblast growth factor and its receptor (FGFR-1) in patients with non-small cell lung carcinomas. Eur J Cancer 33:691–693
Ren M, Hong M, Liu G, Wang H, Patel V, et al. (2013) Novel FGFR inhibitor ponatinib suppresses the growth of non-small cell lung cancer cells overexpressing FGFR1. Oncol Rep 29:2181–2190
O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, et al. (2009) AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell 16:401–412
Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, et al. (2009) Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol 75:196–207
(2013) ClinicalTrials.gov.
Hilberg F, Roth GJ, Krssak M, Kautschitsch S, Sommergruber W, et al. (2008) BIBF 1120: triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy. Cancer Res 68:4774–4782
Reck M, Kaiser R, Eschbach C, Stefanic M, Love J, et al. (2011) A phase II double-blind study to investigate efficacy and safety of two doses of the triple angiokinase inhibitor BIBF 1120 in patients with relapsed advanced non-small-cell lung cancer. Ann Oncol 22:1374–1381
Zhang J, Zhang L, Su X, Li M, Xie L, et al. (2012) Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res 18:6658–6667
Chung JH, Rho JK, Xu X, Lee JS, Yoon HI, et al. (2011) Clinical and molecular evidences of epithelial to mesenchymal transition in acquired resistance to EGFR-TKIs. Lung Cancer 73:176–182
Grygielewicz P, Dymek B, Bujak A, Gunerka P, Stanczak A, et al. (2014) Epithelial-mesenchymal transition confers resistance to selective FGFR inhibitors in SNU-16 gastric cancer cells. Gastric Cancer
Kim SH, Kim JM, Shin MH, Kim CW, Huang SM, et al. (2012) Correlation of epithelial-mesenchymal transition markers with clinicopathologic parameters in adenocarcinomas and squamous cell carcinoma of the lung. Histol Histopathol 27:581–591
Xiao D, He J (2010) Epithelial mesenchymal transition and lung cancer. J Thorac Dis 2:154–159
Cihoric N, Savic S, Schneider S, Ackermann I, Bichsel-Naef M, et al. (2014) Prognostic role of FGFR1 amplification in early-stage non-small cell lung cancer. Br J Cancer 110:2914–2922
Goke F, Franzen A, Menon R, Goltz D, Kirsten R, et al. (2012) Rationale for treatment of metastatic squamous cell carcinoma of the lung using fibroblast growth factor receptor inhibitors. Chest 142:1020–1026
Kohler LH, Mireskandari M, Knosel T, Altendorf-Hofmann A, Kunze A, et al. (2012) FGFR1 expression and gene copy numbers in human lung cancer. Virchows Arch 461:49–57
Sasaki H, Shitara M, Yokota K, Hikosaka Y, Moriyama S, et al. (2012) Increased FGFR1 copy number in lung squamous cell carcinomas. Mol Med Rep 5:725–728
Seo AN, Jin Y, Lee HJ, Sun PL, Kim H, et al. (2014) FGFR1 amplification is associated with poor prognosis and smoking in non-small-cell lung cancer. Virchows Arch
Tran TN, Selinger CI, Kohonen-Corish MR, McCaughan BC, Kennedy CW, et al. (2013) Fibroblast growth factor receptor 1 (FGFR1) copy number is an independent prognostic factor in non-small cell lung cancer. Lung Cancer 81:462–467
Yang W, Yao YW, Zeng JL, Liang WJ, Wang L, et al. (2014) Prognostic value of FGFR1 gene copy number in patients with non-small cell lung cancer: a meta-analysis. J Thorac Dis 6:803–809
Sequist LV, Bell DW, Lynch TJ, Haber DA (2007) Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol 25:587–595
Slamon DJ (1990) Studies of the HER-2/neu proto-oncogene in human breast cancer. Cancer Investig 8:253
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, et al. (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235:177–182
Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, et al. (1989) Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244:707–712
Gozgit JM, Wong MJ, Moran L, Wardwell S, Mohemmad QK, et al. (2012) Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther 11:690–699
Popper HH, Ryska A, Timar J, Olszewski W (2014) Molecular testing in lung cancer in the era of precision medicine. Transl Lung Cancer Res 3:291–300
Wynes MW, Hinz TK, Gao D, Martini M, Marek LA, et al. (2014) FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res 20:3299–3309
Acknowledgments
The authors recognize Portuguese Leica Biosystems services and CIMAGO—Research Center for Environment, Genetics and Oncobiology and Centro de Pneumologia for their support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Sousa, V., Reis, D., Silva, M. et al. Amplification of FGFR1 gene and expression of FGFR1 protein is found in different histological types of lung carcinoma. Virchows Arch 469, 173–182 (2016). https://doi.org/10.1007/s00428-016-1954-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-016-1954-5