Abstract
Proliferative activity, assessed by Ki67 immunohistochemistry (IHC), is an established prognostic and predictive biomarker of breast cancer (BC). However, it remains under-utilized due to lack of standardized robust measurement methodologies and significant intratumor heterogeneity of expression. A recently proposed methodology for IHC biomarker assessment in whole slide images (WSI), based on systematic subsampling of tissue information extracted by digital image analysis (DIA) into hexagonal tiling arrays, enables computation of a comprehensive set of Ki67 indicators, including intratumor variability. In this study, the tiling methodology was applied to assess Ki67 expression in WSI of 152 surgically removed Ki67-stained (on full-face sections) BC specimens and to test which, if any, Ki67 indicators can predict overall survival (OS). Visual Ki67 IHC estimates and conventional clinico-pathologic parameters were also included in the study. Analysis revealed linearly independent intrinsic factors of the Ki67 IHC variance: proliferation (level of expression), disordered texture (entropy), tumor size and Nottingham Prognostic Index, bimodality, and correlation. All visual and DIA-generated indicators of the level of Ki67 expression provided significant cutoff values as single predictors of OS. However, only bimodality indicators (Ashman’s D, in particular) were independent predictors of OS in the context of hormone receptor and HER2 status. From this, we conclude that spatial heterogeneity of proliferative tumor activity, measured by DIA of Ki67 IHC expression and analyzed by the hexagonal tiling approach, can serve as an independent prognostic indicator of OS in BC patients that outperforms the prognostic power of the level of proliferative activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proliferative activity of tumor tissue, commonly measured by Ki67 immunohistochemistry (IHC), remains in the spot light of breast cancer pathology: depending on the tumor type and the clinical setting, Ki67 is a well-established prognostic and a predictive marker [1]. However, its clinical application is hindered by the lack of standardized measurement methodologies and clearly defined cutoffs. After initial recommendation of a cutoff based on gene-expression definition of Luminal A breast cancer from a single reference laboratory [2], it became evident that cutoffs between “high” and “low” values for Ki67 vary between laboratories and should be adjusted to the local practices [3]. Petrelli et al. [4] recently published a systematic review on prognostic value of different cutoff IHC levels of Ki67 in breast cancer, based on meta-analysis of 64,196 patients. It was concluded that Ki67 is an independent prognostic marker for overall survival (OS) in breast cancer patients; the threshold with the greatest prognostic significance remains ill-defined, although a cutoff >25 % was associated with a greater risk of death. However, an evidence-based “optimal” cutoff cannot be achieved without robust measurement techniques; therefore, Ki67 potentially should be used as a continuous biomarker.
Besides the need for accurate measurement of the proportion of the Ki67 labeling index (Ki67 LI), which is simply the proportion of Ki67-positive tumor cell profiles within a defined malignant cell population, the assay is further complicated by its’ intratumor heterogeneity. This involves an additional step of standardized choice of the tissue for evaluation. Detection and evaluation of hotspots of Ki67 expression in the tumor tissue can be performed by conventional microscopy, by review of whole slide images (WSI), or with the assistance of digital image analysis (DIA) tools [5, 6]. However, standardized definitions of hotspots, in terms of their size, shape, and contrast to the surrounding tissue, are needed and do present another challenge for both human and machine-based measurements [7, 8].
A recently proposed methodology for comprehensive Ki67 IHC evaluation in WSI of breast cancer tissue [8] is based on the systematic subsampling of DIA-generated data into a hexagonal tiling (HexT) arrays (honeycomb). It enables computation of a comprehensive set of texture and distribution indicators for Ki67 intratumor variability and has the ability to reveal intrinsic factors behind the Ki67 IHC variance, interpreted as proliferation, entropy, bimodality, and cellularity. It also enables automated detection, quantitative evaluation, and augmented visualization of Ki67 hotspots, based on the upper quintile of the HexT data, conceptualized as “Pareto hotspot”. The methodology was tested on 297 breast cancer WSI; however, the patient follow-up data were not available to test clinical utility of the approach.
This study, performed on a different patient cohort (Nottingham, UK), with the HexT methodology applied on another DIA tool, provides further support for the principle of HexT methodology in Ki67 assessment and demonstrates that intratumor heterogeneity, rather than the level of Ki67 expression in the tumor tissue, is an independent predictor of OS in breast cancer patients.
Materials and methods
Study population and tumor characteristics
A series of 182 patients from the Nottingham-Tenovus Primary Breast Carcinoma Series, aged 70 years or less, presenting with primary operable (stages I, II, and IIIa) invasive BC between 1986 and 98 were used. This is a well-characterized consecutive series of patients who were uniformly treated according to locally agreed clinical protocols [9, 10]. All tumors were ≤5 cm diameter on histology of surgical specimens (pT1 and pT2). Women aged over 70 years were not included because of the increased confounding effect of co-morbidities/death from other causes and because the primary treatment protocols for elderly patients often differed from those for younger women. Adjuvant systemic therapies were offered according to the Nottingham Prognostic Index (NPI) [11] and hormone receptor (HR) status. Patients in the moderate I group (NPI 3.41–4.4) with HR-positive tumors were offered hormonal therapy. Patients in the moderate II (NPI 4.41–5.4) and poor (NPI >5.41) groups received hormone therapy for HR-positive tumors and cytotoxic therapy (classical cyclophosphamide, methotrexate, and 5-fluorouracil (CMF)) for HR-negative tumors and if the patient was fit enough to tolerate chemotherapy. Cases were previously stained for Ki67 [12] using full-face tissue sections and scored as previously described; Ki67 LI, established in the previous study, was utilized in the current study for comparison. ER and PR were assessed as previously described [9] and more than 1 % staining was deemed positive. HER2 was assessed by using IHC and dual-color chromogenic in situ hybridization as previously described [13].
This study was approved by the Nottingham Research Ethics Committee 2 under the title “Development of a molecular genetic classification of breast cancer”.
After visual assessment of Ki67 IHC stained slides, five cases were excluded from the study due to the IHC staining quality issues and/or tissue artifacts. In addition, based on DIA-generated and HexT-processed data, minimum sampling requirements for spatial heterogeneity testing were applied (see below), with 152 cases remaining in this study. Clinico-pathological characteristics of the 152 cases are summarized in the Table 1.
Image acquisition and analysis
Digital images were recorded for the study, using a ScanScope XT Slide Scanner (Leica Aperio Technologies, Vista, CA, USA) under ×20 objective magnification (0.5 μm resolution). The DIA was performed on the WSI using the HALO™ Classifier Module/CytoNuclear v1.4 algorithm (Indica Labs, NM, USA) within manually selected region of interest (ROI) enclosing the tumor tissue section. The HALO Classifier was trained to detect tumor cells, while eliminating fibrous and inflammatory stromal compartment, and to enumerate Ki67-positive and negative tumor nuclear profiles in the breast cancer tissue. The tool was not trained to distinguish between invasive carcinoma and non-invasive ductal carcinoma in situ (DCIS); therefore, DCIS component was excluded by manual annotations. Quality of automated tumor and stroma segmentation and Ki67 positivity threshold by the DIA was monitored by visual inspection; reference standard for the accuracy evaluation was not established in this study.
Computation of heterogeneity parameters from hexagon tiling of the DIA-generated data
DIA results represented by Ki67-negative and Ki67-positive tumor cell nuclei with their X and Y coordinates in the WSI were partitioned into HexT, from which intratumor variance indicators were computed. The process is here described briefly; an in-depth description is available in [8]. Briefly, hexagons of 825 pixel size corresponding to 0.75 mm circular diameter and 0.4421 mm2 area were used in this study. HexT was generated to fit the area of the ROI, and the individual nuclei extracted by DIA were assigned to an appropriate hexagon based on their coordinates. Hexagons containing no nuclear profiles by DIA were regarded as missing data; hexagons containing fewer than 100 nuclear profiles were regarded as insufficiently sampled. A minimum requirement of 20 informative hexagons per tumor was applied in further analyses. Local Ki67% was calculated for each hexagon which was then ranked according to six Ki67 LI intervals: level 0 (0–10 %), level 1 (>10–20 %), level 2 (>20–30 %), level 3 (>30–50 %), level 4 (>50–80 %), and level 5 (>80–100 %). The ranks then formed the basis for the co-occurrence matrix used to compute Haralick texture parameters. Of the 14 classical Haralick parameters [14], 6 were computed from the normalized co-occurrence matrix. Bimodality indicators (Ashman’s D, bimodality index) were calculated based on search of two hidden distributions in a mono-parametric histogram of the HexT data. The search was performed using the algorithm for the Gaussian mixture models, [15] based on the expectation-maximum [16] algorithm.
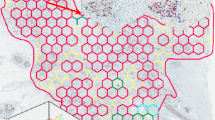
An example of the HexT analysis results is presented in Fig. 1. Local Ki67% values for each hexagon are computed from the DIA data and overlaid on the original WSI with the hexagon color code representing the level of local Ki67%. The HexT data (represented by the local Ki67% values and their coordinates in the WSI) are used to compute texture and distribution indicators for individual tumors. As a result, average Ki67%, obtained by the DIA for each tumor, is supplemented by comprehensive quantitative characteristics of the intratumor variance (heterogeneity) of Ki67 expression (e.g., spatial entropy, bimodality, etc.). In addition, hexagons representing the upper quintile of the local Ki67% distribution indicate approximately 20 % of the tumor tissue area revealing the highest biomarker expression (Pareto hotspot).
An example of two tumors with Ki67 bimodality detected. The color code for the Ki67 % levels in the honeycomb is as follows: dark blue (0-10 %), light blue (10-20 %), green (20-30 %), yellow (30-50 %). The x axis of the histograms corresponds to the Ki67%; the y axis represents a number of hexagons. The first tumor (on the left) was evaluated as Ki67 LI = 47 %, Ki67 Observer Mean = 30 %, Ki67 HexT Mean = 16 %, Ki67 HexT Median = 14 %; the second tumor (on the right), as Ki67 LI = 25 %, Ki67 Observer Mean = 22 %, Ki67 HexT Mean = 17 %, Ki67 HexT Median = 15 %. Both tumors revealed high bimodality (Ashman’s D 3.2 and 2.7) and entropy (2.9 and 3.4). Hotspots were detected by two observers in the first tumor while none of the observers detected a hotspot in the second tumor
Hotspot detection by visual review of the WSI
Four observers (AM, IA, JB, RM) independently reviewed all WSI at low magnification and annotated up to three freeform areas to delineate the Ki67 hotspots in the tumor tissue within the invasive tumor component, if present. These hotspots should be free of (i) tissue folding, (ii) any staining artifacts, (iii) tissue necrosis, and (iv) staining edge effect. Inter-observer agreement of the visual hotspot detection was evaluated. Each observer provided semi-quantitative score of Ki67% in the tumor tissue represented by average Ki67% and hotspot Ki67%, if detected. Final Ki67% score was calculated by substituting the average Ki67% by hotspot Ki67%, if established. Final Ki67% scores of the individual observers were averaged for further analyses (Ki67 Obs Mean).
Statistical analysis
Statistical analysis was performed with SAS 9.4 software. Summary statistics and distribution analyses were performed with significance tests based on the paired sample t test, one-way ANOVA with Bonferoni test for pairwise comparisons. Chi-squared and Fisher’s exact test were used to estimate significant associations in non-parametric statistics. Inter-observer agreement was tested by kappa statistics. Factor analysis was performed using the factoring method of principal component analysis; 5 factors were retained based on a minimum eigenvalue threshold of 1.3, and a general orthomax rotation of the initial factors was performed. Factor analysis of multiple IHC indicators, obtained by DIA, enables extraction of intrinsic factors behind complex interrelationships in the dataset, and has been described previously [8, 17, 18]. Product-limit estimates were used to summarize overall survival data and the log rank test was used for comparing OS distributions. OS was defined as the time from the breast surgery to the patient’s survival at the end of the follow-up period. Cox proportional hazards analysis was used to develop a multiple variable model to predict time to death. A combination of forward, backward, and stepwise procedures was used to arrive at the final model. Continuous variables were dichotomised to predict OS using the web-based tool “Cutoff Finder” [19]. Statistical significance was set at p < 0.05 (two-sided).
Results
Criteria and results of sampling the DIA-generated data into HexT
The DIA-generated data from 152 WSI were subsampled into HexT, with the minimum requirement of 20 hexagons, each containing at least 100 nuclear profiles, per WSI. The summary statistics of the visual evaluation data, DIA, HexT, and computed indicators is presented in the Online Resource 1.
The tumor area analyzed per WSI ranged from 4 to 164 mm2, with a median of 29 mm2 (overall, over 13 million cells in the tumor area of 6000 mm2 were evaluated). Paired t test revealed significant underestimation bias (−7.4 ± 16.8, p < 0.0001) between the Ki67 Obs Mean score and Ki67% obtained from the WSI DIA; the latter “underestimated” the Ki67 LI (−11.6 ± 30.1, p < 0.0001) established in the previous study [12]; the ICC between the three variables was 0.47 (moderate). Remarkably, the Ki67 Obs Mean score underestimated the Ki67 LI (−19.0 ± 25.0, p < 0.0001), while the latter did not differ (−1.2 ± 31.7, p = 0.63) from the Ki67% 90th Percentile.
Hotspot detection by visual review of the WSI
While reviewing 152 WSI, each of the 4 observers identified respectively 37, 67, 32, and 27 tumors with at least one hotspot. The area of the hotspot annotations provided by the observers varied from 1.5 to 5.7 mm2 (Online Resource 1); accordingly, the relative (to the whole tumor) area of the hotspots varied from 3.3 to 14.6 %. The agreement between the observers (taken pairwise) in detecting at least one hotspot was estimated by kappa coefficients ranging from 0.20 to 0.50. Consequently, hotspots were identified in 33, 23, 12, or 11 tumors by any one, two, three, or all four observers, respectively. Analysis of the actual areas and hotspot overlaps outlined by 2 or more observers in the 46 tumors (as above) revealed that, on average, 24.4, 13.9, and 4.4 % of the hotspot areas coincided between the 2, 3, and all 4 observers, respectively.
Detection of a hotspot by each of the four observers increased their final Ki67% score in average by 14.0 ± 6.1, 14.0 ± 6.2, 13.1 ± 10.1, and 10.2 ± 2 % points, respectively. The ICC between the 4 observers in evaluation of the average Ki67% and final Ki67% was 0.78 (substantial) and 0.81 (almost perfect), respectively. The tumors with hotspots detected by at least two observers were characterized by higher entropy (p < 0.03), higher correlation (p < 0.05), and lower energy (p < 0.02) values but did not differ with regard to the other Haralick or bimodality indicators.
Factor analysis of the comprehensive Ki67 indicators
Factor analysis was performed on 152 patients with a complete set of DIA HexT data along with selected pathology data. The rotated factor pattern of the 5 factors, extracted with eigenvalues of 8.8, 4.2, 2.8, 1.8, and 1.3, respectively, is presented in Fig. 2. Factor 1 was characterized by positive and very similar loadings of the various Ki67% indicators, accompanied by low skewness of the Ki67% distribution in the HexT. Factor 1 therefore was best interpreted as the “proliferation” factor. Factor 2 was characterized by strong positive loadings of the Haralick indicators of “disordered texture” (contrast, dissimilarity, entropy) and negative loadings of energy and homogeneity. Interestingly, standard deviation and interquartile range of the Ki67 distribution contributed equally to both factor 1 and factor 2. Factor 3 was characterized by positive loadings of reflective of tumor sample size evaluated by DIA (total cells and tumor area) and pathology report (tumor size) along with the NPI. Factor 4 was represented by both bimodality indicators, while factor 5 was characterized by the correlation parameter and cellularity (tumor cell density) of the tumor. The factor scores revealed normal distribution; a non-linear relationship between factor 1 and 2 was noted (not shown), consistent with that reported in the previous study [8], namely, higher scores of factor 2 (disordered texture) were noted in the tumors with moderate scores of factor 1 (proliferative activity).
Rotated factor loadings, also with inclusion of the mean values of observers (Ki67 Obs Mean), and Ki67 LI, are presented in the Online Resource 2. The factor pattern remained essentially the same; however, lower loadings of the visual scoring data on the factor 1 values could be noted.
Associations between the Ki67 indicators and pathology characteristics of the tumors
Associations of the tumor Ki67 indicators and the factor scores with relevant tumor characteristics were explored by ANOVA. In particular, the histological grade (G) was associated with higher factor 1 (p < 0.0001) and factor 3 (p < 0.0001) scores as well the corresponding primary variables. Triple negative tumors revealed higher factor 1 scores compared to the HR positive tumors (p < 0.05). Triple negative tumors and HER2 positive tumors revealed higher NPI compared to the HR positive tumors (p < 0.05). Factor 2, 4, and 5 scores did not reveal significant associations.
Predictors of the overall survival of the patients
Mean duration of follow-up after the surgery was 143.4 ± 71.4 months (range 5 to 248 months, median 156). Seventy-nine patients died during the follow-up period. The G, tumor stage, axillary nodal stage (N), NPI category, and patient’s age group did not predict the OS by product-limit analysis.
Several multivariable models were developed to account simultaneously for the comprehensive Ki67 indicators and other characteristics of the tumors to predict OS (Table 2). Model nos. 1 and 2 were derived from a dataset consisting of patient’s age, pathological parameters, and the comprehensive Ki67 indicators, as primary variables or factor scores. Both models revealed independent prognostic value of worse OS for Ki67 bimodality indicators (Ashman’s D or factor 4 scores) in the context of HR and HER2 positivity. Model nos. 3 and 4 were obtained by adding the therapy modes to the dataset, where chemotherapy predicted better OS (of note, chemotherapy was not a significant single predictor of OS). Remarkably, none of the Ki67 indicators of the level of proliferative activity (Ki67 LI, Ki67 Obs Mean, Ki67 WSI. or Ki67 HexT Mean, median, percentiles) could be verified as significant independent predictors of OS in this dataset.
The Ki67 indicators and factor scores were dichotomised using the web-based tool “Cutoff Finder” [19] and were analyzed using Kaplan–Meier estimates and log rank tests. Many indicators allowed significant dichotomization of the patients into the prognostic subgroups (Fig. 3). Bimodality of Ki67 intratumor expression, represented by factor 4 scores (p = 0.0081) and Ashman’s D (p = 0.0017) but not bimodality index (p = 0.07, not shown), provided significant cutoff values to predict OS. The level of proliferative activity, represented by broad range of indicators (factor 1 scores, Ki67 HexT Mean, median, percentiles, Ki67 LI, Ki67 Obs Mean, positive cell density) served as significant single predictor as well. Importantly, other indicators of Ki67% distribution (standard deviation, interquartile range, skewness) and spatial heterogeneity (factor 5 scores, correlation, entropy) also provided significant cutoff values. Factor 2 (disordered texture) did not reach statistical significance (p = 0.14), while factor 3 (tumor size and NPI) provided highly significant cutoff (p < 0.0001, not shown).
Cutoff values for the Ki67 indicators as single predictors of overall survival. Overall survival probability plots for the Ki67 indicators are presented. The patients were stratified by the cutoff values obtained by the “Cutoff Finder” [19]
Discussion
Our study reveals that spatial heterogeneity of proliferative tumor activity, measured by DIA of Ki67 IHC expression and analyzed by the HexT approach, can serve as an independent prognostic indicator of OS in breast cancer patients that outperforms the prognostic power of the level of proliferative activity.
A broad set of Ki67 IHC parameters, representing the level of proliferation, pattern of distribution in the tissue, bimodality, and texture indicators were tested in prognostic models along with conventional clinico-pathologic characteristics of the breast cancer patients. Remarkably, although all visual and machine-generated indicators of the level of Ki67 expression in this study provided significant cutoff values as single predictors of OS, only bimodality indicators (Ashman’s D, in particular) served as the independent OS predictors in the context of HR and HER2 status. Other indicators of Ki67 spatial heterogeneity—entropy, energy, and correlation—were also significant as single predictors; however, they were “out-powered” in the multivariate Cox regression model. It is likely that the Ashman’s D bimodality is a more sensitive indicator of intratumor heterogeneity than the parameters computed from the covariance matrix based on Ki67% ranks (Ashman’s D is computed from histograms, searching for two Gaussian functions, taking into account the distance between the spikes and the thickness of each peak). In addition, bimodality indicators do not account for and are independent of spatial peculiarities of the individual tumor samples (size, shape, continuity of growth, etc.), which may cause “noise” in the indicators computed from the covariance matrix. Therefore, although our study presents an independent prognostic value of bimodality indicators, our findings can be indicative of the role of intratumor heterogeneity of the proliferative activity in general.
Clinical utility of Ki67 IHC as prognostic and predictive factor in breast and other cancers is greatly obscured by the lack of standardized measurement methodologies. Our data are in line with this notion: we found significant bias between Ki67% measured as microscope-based Ki67 LI in the previous study [12], DIA, and visual evaluation of the WSI on computer monitor. Furthermore, our study revealed that four observers showed rather low agreement in visual detection of at least one hotspot (kappa ranging from 0.2 to 0.5). Moreover, the size and shape of the hotspots and their spatial overlap varied greatly between the cases and observers. Without proper definitions and standardization of the hotspot detection, the efforts to find hotspots with subsequent evaluation of Ki67 expression may be another source of variation for Ki67 LI measurement. It is also remarkable that the tumors, with at least two observers detecting at least one hotspot, revealed significantly higher entropy, higher correlation, and lower energy; however, they were not associated with the bimodality indicators. Employment of DIA technologies may be useful in automated detection and standardization of the size and other characteristics of the hotspots. In particular, the HexT approach enables automated evaluation of hotspots of desired absolute or relative size; for example, the Pareto hotspot would always reflect Ki67% in 20 % of the “hottest tumor” area with the 90th percentile representing the median value in this subsampled tumor tissue [7].
Our finding that bimodality of Ki67% distribution in the tumor tissue may be a more important prognostic factor of OS than the level of Ki67% itself is somewhat unexpected but may have great practical impact. While efforts to standardize Ki67 LI measurements and to define clinically valid cutoffs can lead to consensus and recommendations [4], DIA tools can be calibrated [20]; it is challenging to ensure the analytic accuracy of the IHC test where high precision of quantification is required. Biological variability of the tumors and tissue processing variation may interfere with the DIA-based approaches. In that regard, the bimodality and other tissue heterogeneity indicators may prove to be more robust and less sensitive to these variations.
Our data suggest that variability of intratumor proliferative activity may be a fundamental feature of tumor aggressiveness affecting the final outcome of the disease, even more important than the average level of proliferation in tumor tissue. At least, it is an independent factor of the disease behavior. It is intriguing that in our study, the level of proliferation (factor 1) and tumor size/NPI (factor 3) were associated with the histological grade (G), while the heterogeneity indicators (factor 2, 4, and 5 scores) did not reveal significant clinico-pathologic associations, except the impact on OS. It should also be taken into account that the factors extracted from the comprehensive Ki67 IHC dataset are linearly independent by definition. Moreover, we noted non-linear relationship between factors 1 and 2, comparable to that reported previously in another patient cohort [8]: higher factor 2 scores (disordered texture) were noted in tumors with moderate scores of factor 1 (proliferative activity). It can be interpreted that intratumor spatial heterogeneity of the proliferative activity is a potential feature of tumors in the mid-scale of proliferative activity and represents an independent factor of unfavorable disease outcome.
Last but not least, our current study provides additional proof of principle for the HexT approach [3]: while the first study was based on different DIA tool (Aperio/Leica) and different patient cohort, the HexT approach generated essentially the same factor pattern and clinico-pathologic associations of the comprehensive Ki67 indicators (the patient follow-up data was not available in the previous study). This demonstrates that the HexT approach, which has been successfully applied in many aspects in image processing and machine vision related fields [21], has its potential in the field of microscopy images and can serve as a tool for comprehensive IHC test. Likely, this approach can be applied to any IHC or other tissue-based biomarkers where intra-tissue heterogeneity needs to be assessed [22–25].
In summary, intratumor heterogeneity of proliferative activity, represented by bimodality indicators, is an independent prognostic factor of worse OS in breast cancer patients, while the level of proliferative activity was significant only in univariate prognostic models. Our study supports the concept that assessment of IHC staining, based on the “honeycomb subsampling” of DIA data, enables comprehensive and efficient methodology for tissue-based biomarker testing. Furthermore, our findings indicate that tissue-based biomarker assessment should take into account intra-tissue heterogeneity aspects to reveal invisible aspects of disease, benefiting from methodologies enabled by digital pathology.
References
Esposito A, Criscitiello C, Curigliano G (2015) Highlights from the 14(th) St Gallen International Breast Cancer Conference 2015 in Vienna: Dealing with classification, prognostication, and prediction refinement to personalize the treatment of patients with early breast cancer Ecancermedicalscience 9:518. doi:10.3332/ecancer.2015.518
Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, Perou CM, Ellis MJ, Nielsen TO (2009) Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101:736–750. doi:10.1093/jnci/djp082
Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, HJ S (2013) Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer. Ann Oncol 24:2206–2223. doi:10.1093/annonc/mdt303
Petrelli F, Viale G, Cabiddu M, Barni S (2015) Prognostic value of different cut-off levels of Ki-67 in breast cancer: a systematic review and meta-analysis of 64,196 patients. Breast Cancer Res Treat 153:477–491. doi:10.1007/s10549-015-3559-0
Lu H, Papathomas TG, van Zessen D, Palli I, de Krijger RR, van der Spek PJ, Dinjens W, Stubbs AP (2014) Automated Selection of Hotspots (ASH): enhanced automated segmentation and adaptive step finding for Ki67 hotspot detection in adrenal cortical cancer Diagn Pathol 9:216. doi:10.1186/s13000–014-0216-6
Romero Q, Bendahl PO, Ferno M, Grabau D, Borgquist S (2014) A novel model for Ki67 assessment in breast cancer. Diagn Pathol 9:118. doi:10.1186/1746-1596-9-118
Christgen M, von Ahsen S, Christgen H, Länger F, Kreipe H (2015) The region of interest (ROI) size impacts on Ki67 quantification by computer-assisted image analysis in breast cancer Human Pathology
Plancoulaine B, Laurinaviciene A, Herlin P, Besusparis J, Meskauskas R, Baltrusaityte I, Iqbal Y, Laurinavicius A (2015) A methodology for comprehensive breast cancer Ki67 labeling index with intra-tumor heterogeneity appraisal based on hexagonal tiling of digital image analysis data Virchows Arch. doi:10.1007/s00428-015-1865-x
Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, IO E (2005) High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cdna expression analyses. Int J Cancer 116:340–350. doi:10.1002/ijc.21004
Rakha EA, El-Sayed ME, Lee AHS, Elston CW, Grainge MJ, Hodi Z, Blamey RW, IO E (2008) Prognostic significance of Nottingham histologic grade in invasive breast carcinoma. Journal of Clinical Oncology 26:3153–3158. doi:10.1200/Jco.2007.15.5986
Galea MH, Blamey RW, Elston CE, IO E (1992) The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat 22:207–219
Aleskandarany MA, Green AR, Rakha EA, Mohammed RA, Elsheikh SE, Powe DG, Paish EC, Macmillan RD, Chan S, Ahmed SI, IO E (2010) Growth fraction as a predictor of response to chemotherapy in node-negative breast cancer. International Journal of Cancer 126:1761–1769. doi:10.1002/ijc.24860
Barros FF, Abdel-Fatah TM, Moseley P, Nolan CC, Durham AC, Rakha EA, Chan S, Ellis IO, AR G (2014) Characterisation of HER heterodimers in breast cancer using in situ proximity ligation assay. Breast Cancer Res Treat 144:273–285. doi:10.1007/s10549-014-2871-4
Haralick RM, Shanmugan K, Distein I (1973) Textural features for image classification IEEE transactions on systems. Man, and Cybernetics SMC-3:610–621
Xuan GR, Zhang W, Chai PQ (2001) EM algorithms of Gaussian Mixture Model and Hidden Markov Model Ieee Image Proc:145–148
Dempster A, Land NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society 39:1–38
Laurinavicius A, Laurinaviciene A, Ostapenko V, Dasevicius D, Jarmalaite S, Lazutka J (2012) Immunohistochemistry profiles of breast ductal carcinoma: factor analysis of digital image analysis data. Diagn Pathol 7:27. doi:10.1186/1746-1596-7-27
Laurinavicius A, Green AR, Laurinaviciene A, Smailyte G, Ostapenko V, Meskauskas R, Ellis IO (2015) Ki67/SATB1 ratio is an independent prognostic factor of overall survival in patients with early hormone receptor-positive invasive ductal breast carcinoma Oncotarget. doi: 10.18632/oncotarget.5838
Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, Denkert C (2012) Cutoff finder: a comprehensive and straightforward web application enabling rapid biomarker cutoff optimization. PLoS One 7:e51862. doi:10.1371/journal.pone.0051862
Laurinavicius A, Plancoulaine B, Laurinaviciene A, Herlin P, Meskauskas R, Baltrusaityte I, Besusparis J, Dasevicius D, Elie N, Iqbal Y, Bor C, Ellis IO (2014) A methodology to ensure and improve accuracy of Ki67 labelling index estimation by automated digital image analysis in breast cancer tissue Breast Cancer Res 16:R35. doi:10.1186/bcr3639
Her I (1995) Geometric transformations on the hexagonal grid. IEEE Trans Image Process 4:1213–1222. doi:10.1109/83.413166
Potts SJ, Krueger JS, Landis ND, Eberhard DA, Young GD, Schmechel SC, Lange H (2012) Evaluating tumor heterogeneity in immunohistochemistry-stained breast cancer tissue Lab Invest 92:1342–1357. doi:10.1038/labinvest.2012.91
Faratian D, Christiansen J, Gustavson M, Jones C, Scott C, Um I, Harrison DJ (2011) Heterogeneity mapping of protein expression in tumors using quantitative immunofluorescence J Vis Exp:e3334. doi:10.3791/3334
Dodd LG, Kerns BJ, Dodge RK, Layfield LJ (1997) Intratumoral heterogeneity in primary breast carcinoma: study of concurrent parameters. J Surg Oncol 64:280–287 discussion 287-288
Brown JR, DiGiovanna MP, Killelea B, Lannin DR, Rimm DL (2014) Quantitative assessment Ki-67 score for prediction of response to neoadjuvant chemotherapy in breast cancer. Lab Investig 94:98–106. doi:10.1038/labinvest.2013.128
Acknowledgments
This research is funded by the European Social Fund under the Global Grant measure, Grant #VP1-3.1-SMM-07-K-03-051.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Laurinavicius, A., Plancoulaine, B., Rasmusson, A. et al. Bimodality of intratumor Ki67 expression is an independent prognostic factor of overall survival in patients with invasive breast carcinoma. Virchows Arch 468, 493–502 (2016). https://doi.org/10.1007/s00428-016-1907-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-016-1907-z