Abstract
Inflammatory bowel diseases (IBDs; both ulcerative colitis [UC] and Crohn’s colitis [CC]) are well-established predisposing pathological conditions for colorectal cancer (CRC) development. In IBDs, both the endoscopy and the histology assessment of CRC precursors (i.e., dysplasia, also defined as intraepithelial neoplasia) are associated with low interobserver consistency, and no reliable dysplasia-specific biomarker is available. The programmed cell death 4 (PDCD4) tumor suppressor gene is involved in sporadic colorectal oncogenesis, but scanty information is available on its involvement in IBD-associated colorectal oncogenesis. One hundred twenty tissue samples representative of active and inactive IBD and of flat dysplasia were obtained from 30 cases of UC and 30 of CC who undergone colectomy. Twenty additional biopsy samples obtained from patients with irritable bowel syndrome acted as normal controls. PDCD4 expression was assessed by immunohistochemistry; the expression of miR-21 (a major PDCD4 regulator) was investigated by quantitative real-time PCR and in situ hybridization in different series of a hundred samples. Tissue specimens from both controls and inactive IBD consistently featured strong PDCD4 nuclear immunostain; conversely, lower PDCD4 nuclear expression was featured by both active IBD and IBD-associated dysplastic lesions. Significant PDCD4 down-regulation distinguished IBD-associated dysplasia (p < 0.001) versus active IBD. In both active IBD and dysplasia, PDCD4 down-regulation was significantly associated with miR-21 up-regulation. PDCD4 nuclear down-regulation (which parallels miR-21 up-regulation) is involved in the molecular pathway of IBD-associated carcinogenesis. PDCD4 nuclear expression may be usefully applied as ancillary maker in the histological assessment of IBD-associated dysplastic lesions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ulcerative colitis and Crohn’s disease are the two major forms of idiopathic inflammatory bowel disease (IBD) [1]. Colorectal cancer (CRC) is a well-established adverse outcome of longstanding IBDs (both ulcerative colitis [UC] and Crohn’s colitis [CC]), accounting for approximately 10–15 % of all IBD deaths [2, 3]. In colitis-associated carcinogenesis, both the histology phenotypes and the molecular events are basically similar to those of sporadic CRCs, but the sequence of the molecular deregulations consistently differ [4, 5].
Secondary prevention of IBD-associated CRC depends on the endoscopy and the histology detection of precancerous lesions (i.e., dysplasia, recently defined as intraepithelial neoplasia). As for endoscopy, IBD-associated dysplasia may arise from both raised (dysplasia-associated lesions or masses) or flat mucosal lesions, which distinction has been proven to rely on subjective criteria [6–8]; moreover, dysplastic lesions can blend with inflammatory abnormalities, which represent an additional confounding gross feature even for experienced endoscopists. As for histology, the reliability of dysplasia assessment is largely affected by the pathologist’s experience and frequently limited by intrinsic characteristics of the biopsy samples (high-grade active inflammation, shrinkage, fragmentation, and “burning”); both situations result in a low rate of interobserver agreement [9]. Because of all these reasons, dysplasia assessment is inconsistent, which prompts to the priority of dysplasia-specific tissue biomarkers.
Programmed cell death 4 (PDCD4) is a newly characterized tumor suppressor gene (TSG) involved in the apoptotic machinery, cell transformation and invasion, and tumor progression by interacting with translation initiation factors eIF4A and eIF4G [10–20]. PDCD4 protein expression is consistently down-regulated in both human cancers and cancer cell lines [19, 21–28]. Different mechanisms are involved in PDCD4 dysregulation; among others, the oncogenic microRNA miR-21 has been shown to specifically target the PDCD4 3′ untranslated region, which negatively regulates PDCD4 expression [21, 25]. In active IBDs, miRNA expression profiling consistently features disease-specific signatures, and miR-21 has been pinpointed as one of the most dysregulated genes [29–31]. PDCD4 involvement in IBD-related carcinogenesis has never been addressed.
Thus, the aim of this study was to investigate both PDCD4 expression (as assessed by immunohistochemistry) and miR-21 levels/expression (as assessed by real-time PCR and in situ hybridization) in a series of inflammatory and dysplastic IBD surgical specimens in the search of additional diagnostic tools to be used in daily pathology routine.
Materials and methods
Patients
The cases considered in the present study were retrospectively collected from the archives of the Surgical Pathology and Cytopathology Unit at the University of Padua. The institute’s ethical regulations on research conducted on human tissues were followed. A total of 60 consecutive IBD patients [UC = 30; CC = 30] who underwent colectomy (total or sub-total) for histologically proven dysplastic lesions were considered. The surgical specimens were grossly assessed according to a standardized local protocol: the surgical specimens were opened anteriorly, pinned to a cork board, and fixed in an adequate volume of formalin (10 %) for 24 h; subsequently, the specimens were semi-serially sliced into 3–4-mm thick transverse gross sections at intervals of 2 cm.
The assessment of IBD (UC and CC) acute inflammation (a) versus non-active inflammation was based on microscopic criteria, activity being defined as the presence of neutrophils or unequivocal damage of the surface and crypt epithelium typically in conjunction with neutrophils [32, 33].
In all cases, the histology assessment of low-grade dysplasia (LGD) versus high-grade dysplasia (HGD) was done according to the current histology criteria [6, 34]. Original slides (4–6-microns thick) obtained from archival paraffin-embedded tissue samples (H&E) were jointly reassessed by two pathologists (KL and MF); in cases where their opinions differed, a third expert GI pathologist (CM) was involved.
From the 60 considered cases, a total of 120 tissue specimens were selected. The tissue specimens were the representatives of the following histology lesions: (1) non-active IBD (UCna = 20 and CCna = 20 samples); (2) active IBD (UCa = 20 and CCa = 20 samples); (3) flat LGD (UC/LGD = 15 and CC/LGD = 15 samples); and (5) flat HGD (UC/HGD = 5 and CC/HGD = 5 samples).
Twenty additional normal (N) colonic mucosa biopsy samples obtained at colonoscopy from patients with irritable bowel syndrome were also included (Clinica Chirurgica II, University of Padua).
PDCD4 immunohistochemistry
The histological sections were stained with hematoxylin and eosin, and the original diagnosis was histologically confirmed in all cases. Immunohistochemistry (IHC) staining was automatically performed (Ventana Benchmark XT system; Touchstone, AZ, USA) for PDCD4 (catalog #HPA001032; Atlas Antibodies, Stockholm, Sweden; 1:100) [21, 27, 35] according to the manufacturer’s instructions. Sections were lightly counterstained with hematoxylin. Appropriate positive and negative controls were run concurrently. PDCD4 nuclear expression was jointly scored by two pathologists (KL and MF) unaware of the patients’ clinical history. In case of discordance between the two pathologists, the case was reviewed by a third expert gastrointestinal pathologist (CM). As described previously [19, 21], nuclear PDCD4 staining was scored on a four-tiered scale (score 0, no nuclear staining; score 1, ≥1 ≤ 30 %—positive nuclear staining; score 2, >30 < 70 %—positive nuclear staining; score 3, ≥70 %—positive nuclear staining). Cytoplasmic PDCD4 expression was scored by staining intensity (score 0, none; score 1, weak; score 2, intermediate; score 3, strong). In the statistical analysis, only PDCD4 nuclear staining was considered. In all the considered tissue samples, non-epithelial cells (fibroblasts, lymphocytes, smooth muscle cells, and endothelia) always featured PDCD4 nuclear expression and were assumed as positive internal control (not considered in IHC score) [19, 21, 27, 28, 36].
Quantitative real-time polymerase chain reaction
Formalin-fixed paraffin-embedded biopsy samples were deparaffinized with xylene at 50 °C for 3 min. Total RNA extraction was done using the RecoverAll kit (Ambion Inc, Austin, TX, USA) according to the manufacturer’s instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis was performed using the GeneAmp PCR 9700 thermocycler (Applied Biosystems, Foster City, CA, USA), and gene expression levels were quantified using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems), as previously described [37]. Primers’ sequences for PDCD4 were forward 5′-GGCCTCCAAGGAGTAAGACC-3′ and reverse 5′-AGGGGTCTACATGGCAACTG-3′. GAPDH was used as the internal control gene (forward 5′-AAGGGAAGGTTGCTGGATAGG-3′; reverse 5′-CACATCCACCTCCTCCACATC-3′). The NCodeTM miRNA qRT-PCR method (Invitrogen, Carlsbad, CA, USA) was used to detect and quantify mature miR-21 (primer sequence: 5′-CGGTAGCTTATCAGACTGATGTTGA-3′) according to the manufacturer’s instructions. Normalization was done with the small nuclear RNA U6B (Invitrogen). PCR reactions were run in triplicate, including no-template controls. The data were analyzed using the comparative CT method. Additional tissue samples obtained from 20 N, 20 CCa, 15 CC/LGD, 5 CC/HGD, 20 UCa, 15 UC/LGD, and 5 UC/HGD were considered for the qRT-PCR study.
In situ hybridization
To confirm qRT-PCR data, miR-21 expression was further investigated by in situ hybridization (ISH). ISH was performed using the GenPoint™ Catalyzed Signal Amplification System (DakoCytomation) according to the manufacturer’s protocol. Briefly, slides were incubated at 60 °C for 30 min and deparaffinized as previously described [26, 37]. Sections were treated with Proteinase K (DakoCytomation) for 30 min at room temperature, rinsed several times with dH2O, and immersed in 95 % ethanol for 10 s before air-drying. The slides were prehybridized at 49–56 °C for 1 h with mRNA ISH buffer (Ambion) before overnight incubation at 49–56 °C in buffer containing the 5′-biotin-labeled miR-21 miRCURY™ LNA detection probe (Exiqon, Woburn, MA, USA) or the scrambled negative control probe (U6, Exiqon) at a final concentration of 200 nM. The slides were washed in both Tris-buffered saline with Tween (TBST) and GenPoint™ stringent wash solution (54 °C for 30 min), then exposed to H2O2 blocking solution (DakoCytomation) for 20 min, and then further blocked in a blocking buffer (DakoCytomation) for 30 min before they were exposed to primary streptavidin–horseradish peroxidase (HRP) antibody, biotinyl tyramide, secondary streptavidin–HRP antibody, and DAB chromogen solutions, following the manufacturer’s protocol. The slides were then briefly counterstained in hematoxylin and rinsed with TBST and water before mounting. Out of the initial series of biopsy samples, a total of 5 N, 5 UCa, 5 CCa, 5 UC/LGD, and 5 CC/LGD were considered for the ISH study.
Statistical analysis
Differences and correlations between groups were tested by applying the modified Kruskal–Wallis nonparametric test for trend, the chi-square test, Pearson’s correlation, and the t test. P values <0.05 were considered significant. All statistical assessments were performed with STATA 8.0 software (Stata Corporation, College Station, TX, USA).
Results
PDCD4 protein is down-regulated in active UC/CC and in IBD-associated dysplastic lesions
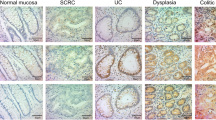
Normal mucosa (i.e., non-IBD tissue samples) consistently featured strong PDCD4 nuclear expression coexisting with moderate/strong cytoplasm immunostain (Fig. 1; Table 1).
PDCD4 is down-regulated in IBD carcinogenesis models. Immunohistochemical scores of nuclear PDCD4 in the different histological lesions (white—score 0, light gray—score 1, dark gray—score 2, black—score 3). Nuclear PDCD4 staining was significantly lower in active disease (UCa and CCa) and associated dysplasia (LGD and HGD) specimens than in normal tissues (p < 0.001)
Overall, in both UC and CC, PDCD4 nuclear expression decreased progressively and significantly from inactive IBD to IBD-associated intraepithelial neoplastic lesions (Kruskal–Wallis: p < 0.001; Figs. 1 and 2; Table 1). Inactive IBDs (both UC and CC) retained PDCD4 strong nuclear expression (score 3, never featured by the crypts’ proliferative compartment) and moderate cytoplasm immunostain (Fig. 2).
Representative PDCD4 immunostaining of normal colonic tissues, UC and CC inactive/active disease, and UC/CC-associated dysplasia. a In native mucosa, colocytes featured strong nuclear and moderate/strong cytoplasm stain. b Non-active UC maintains strong nuclear and weak cytoplasm immunostain. Note the nuclear PDCD4 loss in the proliferative basal compartment of the glands. c–d Active CC and active UC tissues showed a significant down-regulation of PDCD4 nuclear expression. Note the presence of cryptic abscess in active UC (d). e A minority of active UC specimens throughout featured a strong cytoplasmic expression with no nuclear immunoreaction. f Almost complete nuclear PDCD4 loss in CC-associated dysplastic lesion. Original magnifications ×10, ×20, and ×40
Tissue specimens obtained from active IBD (both UC and CC) consistently showed PDCD4 nuclear down-regulation (scores 2/3 = 10/20 and 12/20, respectively; Figs. 1 and 2c, d; Table 2). As for the cytoplasm immunostain, active lesions retained weak to moderate positivity. Five cases of active UC throughout featured a strong cytoplasmic expression coexisting with no nuclear immunoreaction (Fig. 2e).
Dysplasia (low and high grade, from both UC and CC cases) showed null or weak nuclear immunostain (scores 0/1 = 17/20, for both) associated to weak to moderate cytoplasm protein expression (Figs. 1 and 2f; Table 2).
Of note, by merging UC and CC cases, a significant lower PDCD4 nuclear expression distinguished LGD from active IBD (nuclear scores 0/1 assessed in 24/30 cases (80 %) of dysplasia versus 18/40 (45 %) cases of active IBD; chi-square: p = 0.003).
miR-21 is overexpressed in active CC/UC and associated dysplastic lesions
A significant miR-21 up-regulation (Fig. 3) was found in samples of active UC (with a 6.3-fold increase over normal colonic mucosa samples; t test: p < 0.001), active CC (with a 5.8-fold increase; t test: p < 0.001), UC/LGD (with a sevenfold increase; t test: p < 0.001), CC/LGD (with a sixfold increase; t test: p < 0.001), UC/HGD (with a 7.3-fold increase; t test: p < 0.001), and CC/HGD (with a 6.1-fold increase; t test: p < 0.001) (Fig. 3a). Of note, there was a significantly higher expression of miR-21 in UC/LGD and UC/HGD tissue samples compared with active UC (t test: p = 0.025 and p = 0.021; Fig. 3a). No significant difference was observed between LGD and HGD lesions in both UC and CD.
miR-21 is overexpressed in UC and CC active disease and in UC/CC-associated dysplasia. a Altered miR-21 expression between active UC/CC and UC/dysplasia and CC/dysplasia (CT; active disease and dysplasia) and normal tissue (NCT) in IBD patients by qRT-PCR analysis. miR-21 was significantly up-regulated in active UC and CC, and IBD-associated dysplastic lesions (p < 0.001). Note the significantly higher expression of miR-21 in UC/LGD and UC/HGD when compared to active UC (values are mean ± SD). Representative examples of miR-21 overexpression by epithelial cells in active UC (b) and UC/LGD (c), and active CC (d) and CC/LGD (e). The presence of miR-21 is revealed by a granular brown stain (original magnifications, ×20 and ×40)
Discussion
Colorectal cancer is an adverse late outcome of both ulcerative and Crohn’s colitis [3]. Dysplasia (currently named as intraepithelial neoplasia) is the major risk factor for IBD-associated CRC, but its low interobserver agreement brought about efforts to identify ancillary biomarkers of cancer-prone histology.
The PDCD4 tumor suppressor gene is consistently down-regulated in sporadic CRC, but its value as marker of IBD-associated oncogenesis is still unfathomed [19, 25, 38]. By exploring the IHC expression of PDCD4 in a series of UC and CC (with and without coexisting dysplastic lesions), two main results were established: (1) normal mucosa and inactive IBD feature strong PDCD4 nuclear immunostain (associated with moderate/strong cytoplasm expression) (Fig. 2) and (2) both active IBDs and IBD-associated dysplasia consistently show PDCD4 nuclear down-regulation (paralleling with weak/moderate cytoplasm stain) (Fig. 1).
The reasons behind these (nuclear/cytoplasm) immunophenotypes are only partially understood. Similar patterns of PDCD4 expression have already been reported in both sporadic colorectal [20, 38] and in Barrett’s carcinogenesis [21]. In vitro studies have shown that, under normal growth conditions, PDCD4 is located mainly in the nucleus, moving to the cytoplasm on serum deprivation in vitro [10]. The PDCD4 nuclear overexpression may identify a proliferative “attitude” shared by both “reparative” reactions (to inflammation) and epithelial dedifferentiation. Such observation is consistent with the absence of any PDCD4 nuclear stain in the proliferative compartment of normal colonic crypts. In this study, the nuclear expression found in active IBD and associated dysplasia supports active inflammatory disease as the “cancerization field” of the IBD-associated carcinogenesis [39, 40].
Because of the histological similarity between hyperplastic/regenerative lesions (coexisting with active inflammation) and low-grade intraepithelial lesions, PDCD4 nuclear expression was specifically compared in such gray-zone histology phenotypes. In both UC and CC, the nuclear protein down-regulation significantly prevailed in associated dysplasia versus active IBDs. PDCD4 nuclear loss could be used in combination with other dysplasia-specific IHC markers when active inflammation acts as confounding variable in the histology assessment. In fact, several other specific markers have been proposed for the prediction of colorectal cancer development and dysplasia assessment in the IBD context [41]. Among the others, p53 and ki-67 IHC assessments are the most widely used in routine practice to differentiate dysplastic epithelia within inflamed colonic mucosa [40, 41]. However, mutations and loss of heterozygosity of p53 and cell proliferation (as assessed by ki-67 labeling index) are early events in IBD mucosa, often occurring before dysplasia is detected. In this respect, PDCD4 down-regulation did not show a superior diagnostic performance by comparing it to previously described IHC markers. These data further underlines the absence of a real dysplasia-specific marker and the need of a multi-marker immunohistochemical panel (in which PDCD4 could be reasonabily included) to identifiy colorectal dysplasia in inflamed mucosa in IBD patients.
In esophageal squamous carcinoma cell lines, Hiyoshi and colleagues recently demonstrated that PDCD4 is regulated by miR-21 and such interplay affects both cancer cell proliferation and invasion [43]. In the CRC setting, evidence has been provided on the inverse relationship between miR-21 and PDCD4, suggesting that miR-21 post-transcriptionally modulates PDCD4 via mRNA degradation [39]. The present in vivo study supports the miR-21 regulatory activity on PDCD4 and consistently demonstrates a significant up-regulation of miR-21 in both active IBD (UC and CC) and UC/CC-associated dysplasia. MicroRNA expression profiling in active IBD has been addressed by various studies, which featured both a disease-specific miRNA signature [29–31, 44] and a miRNA role in the onset/relapse of active inflammation [44]. Some miRNAs have been already indicated as specific markers of IBD-related dysplasia [45, 46], and the different expressions of miR-21 in active ulcerative colitis versus UC dysplasia further support the specific role of the oncomir in UC-associated oncogenesis.
In conclusion, PDCD4 is involved in IBD-associated colon carcinogenesis. Nuclear down-regulation of PDCD4 expression is featured by both active IBD and IBD-associated intraepithelial neoplasia; in histology doubtful cases (differential diagnosis between regenerative and dysplastic lesions), nuclear protein down-regulation can be considered as supporting information in favor of a neoplastic intraepithelial lesion. miR-21 up-regulation significantly parallels the PDCD4 down-regulation supporting the current knowledge that the oncomiR post-transcriptionally modulates the TSG activity via mRNA degradation.
Abbreviations
- IHC:
-
Immunohistochemistry
- IBD:
-
Inflammatory bowel disease
- PDCD4:
-
Programmed cell death 4
- UC:
-
Ulcerative colitis
- CC:
-
Crohn’s colitis
References
Fiocchi C (1998) Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology 115:182–205
Crohn BB (1925) The sigmoidoscopic picture of chronic ulcerative colitis (non-specific). Am J Med Sci 170:220–228
Munkholm P (2003) The incidence and prevalence of colorectal cancer in inflammatory bowel disease. Aliment Pharmacol Ther 18(S2):1–5
Feagins LA, Souza RF, Spechler SJ (2009) Carcinogenesis in IBD: potential targets for the prevention of colorectal cancer. Nat Rev Gastroenterol Hepatol 6(5):297–305
Goel GA, Kandiel A, Achkar JP et al (2011) Molecular pathways underlying IBD-associated colorectal neoplasia: therapeutic implications. Am J Gastroenterol 106(4):719–730
Hamilton SR, Bosman FT, Boffetta P et al (2010) Carcinoma of the colon and rectum. In: Bosman FT, Carneiro F, Hruban R, Theise ND (eds) WHO classification of tumours of the digestive system, 4th edn. International Agency for Research on Cancer, Lyon, France, pp 134–182
Neumann H, Vieth M, Langner C et al (2011) Cancer risk in IBD: how to diagnose and how to manage DALM and ALM. World J Gastroenterol 17(27):3184–3191
Eaden J, Abrams K, McKay H et al (2001) Inter-observer variation between general and specialist gastrointestinal pathologists when grading dysplasia in ulcerative colitis. J Pathol 194(2):152–157
Eaden J (2004) Colorectal carcinoma and inflammatory bowel disease. Aliment Pharmacol Ther 20(S4):24–30
Bohm M, Sawicka K, Siebrasse JP et al (2003) The transformation suppressor protein Pdcd4 shuttles between nucleus and cytoplasm and binds RNA. Oncogene 22:4905–4910
Yang HS, Cho MH, Zakowicz H et al (2004) A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol Cell Biol 24:3894–3906
Afonja O, Juste D, Das S et al (2004) Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene 23:8135–8145
Bitomsky N, Bohm M, Klempnauer KH (2004) Transformation suppressor protein Pdcd4 interferes with JNK-mediated phosphorylation of c-Jun and recruitment of the coactivator p300 by c-Jun. Oncogene 23:7484–7493
Zakowicz H, Yang HS, Stark C et al (2005) Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI. RNA 11:261–274
Jansen AP, Camalier CE, Colburn NH (2005) Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res 65:6034–6041
Dorrello NV, Peschiaroli A, Guardavaccaro D et al (2006) S6K1-and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314:467–471
Bitomsky N, Wethkamp N, Marikkannu R et al (2008) SiRNA-mediated knockdown of Pdcd4 expression causes upregulation of p21(Waf1/Cip1) expression. Oncogene 27:4820–4829
Carayol N, Katsoulidis E, Sassano A et al (2008) Suppression of programmed cell death 4 (PDCD4) protein expression by BCRABL-regulated engagement of the mTOR/p70 S6 kinase pathway. J Biol Chem 283:8601–8610
Mudduluru G, Medved F, Grobholz R et al (2007) Loss of programmed cell death 4 expression marks adenoma–carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer 110:1697–1707
Allgayer H (2010) Pdcd4, a colon cancer prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol 73:185–191
Fassan M, Pizzi M, Battaglia G et al (2010) Programmed cell death 4 (PDCD4) expression during multistep Barrett’s carcinogenesis. J Clin Pathol 63:692–696
Chen Y, Knosel T, Kristiansen G et al (2003) Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol 200:640–646
Zhang H, Ozaki I, Mizuta T et al (2006) Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene 25:6101–6112
Wang Q, Sun Z, Yang HS (2008) Downregulation of tumor suppressor Pdcd4 promotes invasion and activates both betacatenin/Tcf and AP-1-dependent transcription in colon carcinoma cells. Oncogene 27:1527–1535
Baffa R, Fassan M, Volinia S et al (2009) MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol 219:214–221
Yamamichi N, Shimomura R, Inada K et al (2009) Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res 15:4009–4016
Fassan M, Cagol M, Pennelli G et al (2010) Programmed cell death 4 (PDCD4) protein in esophageal cancer. Oncol Rep 24:135–139
Goke R, Barth P, Schmidt A et al (2004) Programmed cell death protein 4 suppresses CDK1/cdc2 via induction of p21 (Waf1/Cip1). Am J Physiol Cell Physiol 287:C1541–C1546
Dalal SR, Kwon JH (2010) The role of microRNA in inflammatory bowel disease. Gastroenterol Hepatol (NY) 6:714–722
Wu F, Zhang S, Dassopoulos T, Harris ML et al (2010) Identification of microRNAs associated with ileal and colonic Crohn’s disease. Inflamm Bowel Dis 16:1729–1738
Okubo M, Tahara T, Shibita T et al (2011) Association study of common genetic variants in pre-microRNAs in patients with ulcerative colitis. J Clin Immunol 31:69–73
Goldman H (1994) Interpretation of large intestinal mucosal biopsy specimens. Hum Pathol 25(11):1150–1159
Cornaggia M, Leutner M, Mescoli C et al (2011) Chronic idiopathic inflammatory bowel diseases: the histology report. Dig Liver Dis 43(4):293–303
Rugge M, Correa P, Dixon MF (2000) Padova classification gastric dysplasia: the Padova international classification. Am J Surg Pathol 24(2):167–176, Review
Rugge M, Fassan M, Clemente R et al (2008) Bronchopulmonary carcinoid: phenotype and long-term outcome in a single institution series of Italian patients. Clin Cancer Res 14:149–154
Yoshinaga H, Matsuhashi S, Fujiyama C et al (1999) Novel human PDCD4 (H731) gene expressed in proliferative cells is expressed in the small duct epithelial cells of the breast as revealed by an anti-H731 antibody. Pathol Int 49:1067–1077
Fassan M, Volinia S, Palatini J et al (2011) MicroRNA expression profiling in human Barrett’s carcinogenesis. Int J Cancer 129(7):1661–1670
Fassan M, Pizzi M, Giacomelli L et al (2011) PDCD4 nuclear loss inversely correlates with miR-21 levels in colon carcinogenesis. Virchows Arch 458(4):413–419
Chang KH, Miller N, Kheirelseid EA et al (2011) MicroRNA-21 and PDCD4 expression in colorectal cancer. Eur J Surg Oncol 37(7):597–603
Yu Y, Kanwar SS, Patel BB et al (2012) MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis 33(1):68–76
Itzkowitz SH (2006) Molecular biology of dysplasia and cancer in inflammatory bowel disease. Gastroenterol Clin North Am 35(3):553–571
Gerrits MM, Chen M, Theeuwes M et al (2011) Biomarker-based prediction of inflammatory bowel disease-related colorectal cancer: a case–control study. Cell Oncol (Dordr) 34(2):107–117
Hiyoshi Y, Kamohara H, Karashima R et al (2009) Micro-RNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res 15(6):1915–1922
Gerrits MM, Chen M, Theeuwes M et al (2011) Biomarker-based prediction of inflammatory bowel disease-related colorectal cancer: a case–control study. Cell Oncol 34:107–117
Rutter M, Saunders B, Wilkinson K et al (2004) Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology 126:451–459
Brest P, Lapaquette P, Souidi M et al (2011) A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet 43(3):242–245
Acknowledgments
We wish to acknowledge the continuous support of the “G. Berlucchi” and the “G.B. Morgagni” Foundations. This work was partially supported by an AIRC Regional grant (2008). We also thank Dr. Cristiano Lanza, Dr. Vincenza Guzzardo, and Dr. Vanni Lazzarin for their excellent technical support. All authors of this research paper participated directly in the planning and execution of the study and in the analysis of the results.
Conflict of interest
The authors have no competing interests to declare.
Author information
Authors and Affiliations
Corresponding author
Additional information
Kathrin Ludwig and Matteo Fassan contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ludwig, K., Fassan, M., Mescoli, C. et al. PDCD4/miR-21 dysregulation in inflammatory bowel disease-associated carcinogenesis. Virchows Arch 462, 57–63 (2013). https://doi.org/10.1007/s00428-012-1345-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-012-1345-5