Abstract
Genes of the twist family encode bHLH transcription factors known to be involved in the regulation and differentiation of early mesoderm. Here, we report our characterization of Hau-twist, a twist homolog from the leech Helobdella austinensis, a tractable lophotrochozoan representative. Hau-twist was expressed in segmental founder cells of the mesodermal lineage, in subsets of cells within the mesodermal lineage of the germinal plate, in circumferential muscle fibers of a provisional integument during segmentation and organogenesis stages and on the ventral side of the developing proboscis. Thus, consistent with other systems, our results suggest that twist gene of the leech Helobdella might function in mesoderm differentiation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genes in the evolutionarily conserved twist gene family encode basic helix-loop-helix (bHLH) transcription factors (O’Rourke et al. 2002; Dill et al. 2007). Structurally, these proteins are composed of two domains: a basic domain in charge of mediating specific DNA binding and a helix-loop-helix (HLH) domain containing two alpha amphipathic helices (separated by a non-conserved loop region) that allow Twist proteins to homo-dimerize and/or hetero-dimerize (Hamamori et al. 1997; Sablitzky 2005).
The eponymous Twist was discovered in Drosophila (Simpson 1983), where it is required for both the specification of the mesodermal germ layer (Leptin 1991) and for the later differentiation of specific mesodermal derivatives (Baylies and Bate 1996). Through its role as one of several repressors of E-cadherin, it can also regulate the epithelial-mesenchymal transitions (EMTs), that are important in the progression of many carcinomas (Kang and Massagué 2004). Homologs of twist have been found in developing mesoderm of numerous animal species from all three of the bilaterian super-phyla: Ecdysozoa, Deuterostomia, and Lophotrochozoa. Therefore, twist family genes should be useful for comparative studies to unravel the evolutionary history of mesoderm formation in Bilateria (Dill et al. 2007).
A general question is the extent to which the specification of this third germ layer can be tied to the evolution of mesoderm-specific genes (Price and Patel 2008). In contrast to the results from Drosophila, the expression of twist in most species is observed only in subsets of mesodermal cells, after germ layer specification has already occurred, leading to the conclusion that the ancestral role for the twist gene product was in mesoderm differentiation rather than in mesoderm specification. Within Lophotrochozoa, for example, a twist ortholog in Patella (Pv-twist) is involved in early differentiation of a subset of mesoderm (Nederbragt et al. 2002), and the expression of two twist paralogs in Capitella (CapI-twist1 and CapI-twist2) is correlated with later stages of mesoderm differentiation and not during mesoderm specification. These findings provide support to the notion that the function of mesoderm specification is unique to insects, while the function in mesoderm differentiation represents a trait of ancestral bilaterians (Dill et al. 2007).
To extend our knowledge about the role of twist genes in mesoderm development in lophotrochozoans, we characterized the expression pattern of a twist homolog Hau-twist throughout development in the leech species Helobdella austinensis.
Materials and methods
Embryos
Embryos were obtained from a laboratory colony of the species H. austinensis (Kutschera et al. 2013) as described elsewhere (Gline et al. 2011; Weisblat and Kuo 2014).
Gene identification, cloning, and in vitro transcription
Following previous gene discovery strategies (Kuo and Weisblat 2011), putative Twist bHLH protein domains encoded by the annotated Helobdella robusta genome (DOE, Joint Genome Institute) were identified by a Pfam search, using accession number PF00010. This search identified 68 unique loci. TBLASTN search of the genome assembly and BLASTP search of the pool of proteins deduced from these gene models suggest that of these 68 loci, only 2 (JGI Protein ID 155122 and JGI Protein ID 176734) encode bona fide Twist class proteins. The genomic DNA coding sequences are nearly identical (>97%) between the recently described H. austinensis (Kutschera et al. 2013) and H. robusta. Genes isolated from H. austinensis are denoted with a “Hau-” prefix (Kuo and Weisblat 2011). Based on the sequences of conserved areas in gene models, specific primers (twist forward: 5′ TCA-CCA-TCT-TTT-CCA-ACT-ACG-TCA-TCC-TCT 3′, twist reverse: 5′ CAT-CCT-CCA-AAC-AAA-CGC-GTA-CCC-CAA 3′) were designed against the twist gene model (JGI Protein ID 155122), which corresponded to Hro-twist (Soto et al. 1997), and then amplified from complementary DNA (cDNA) prepared from the closely related H. austinensis. The amplified cDNA fragments were subcloned into pGEM-T vector (Promega) and sequenced to ascertain their identities. These plasmids were then used as templates for synthesizing riboprobe for in situ hybridization.
Alignment and phylogenetic analyses
A Twist data set was built with twist homologs from representatives of the major metazoan clades with a total of 20 amino acid sequences (the basic helix-loop-helix domains). For this purpose, Twist protein sequences were retrieved from the UniProt database. Alignment was carried out using the ClustalW software (http://clustalw.genome.jp). Phylogenetic analysis was performed by the maximum likelihood method using the MEGA 6.0 program (http://www.megasoftware.net). Bootstrap analysis was performed with 1000 replicates.
Developmental RT-PCR
Total RNA samples were prepared from H. austinensis embryos at selected stages (Weisblat and Huang 2001) with RNA Wiz (Ambion) according to the manufacturer’s instructions, using 40 embryos for each stage. The total RNA obtained (3 μg) was reverse transcribed using a first-strand cDNA synthesis kit (BD Biosciences, Palo Alto, CA). The PCR mixture (50 ml) contained 10× Taq buffer, 0.3 U Taq polymerase (PerkinElmer), 2.5 mM of dNTPs, 5 pmol of each set of primers, and 50 ng of cDNA from the selected stages as template (Fig. 2 (A)).
To amplify Hau-twist1-specific fragments, a pair of PCR primers was designed as follows: twist1 forward: 5′-TCA-CCA-TCT-TTT-CCA-ACT-ACG-TCA-TCC-TCT-3′, twist1 reverse: 5′-TTC-ATT-CTC-CTT-CAG-TTG-CGA-ATA-CAT-GAA-3′; this primer set yielded a fragment of 852 bp for Hau-twist1. The PCR reactions were performed under the following cycling conditions: an initial denaturation at 94 °C for 5 min, followed by 25–35 cycles of denaturation at 94 °C for 30 s, and elongation at 72 °C for 1 min, with a final elongation step at 72 °C for 10 min. A 10 ml aliquot of each PCR reaction was removed after 25 cycles, while the remaining material underwent five more cycles of amplification. The 18S rRNA sequence was used as an internal standard (QuantumRNA, Ambion). The extent of amplification was chosen empirically to avoid saturation of the amplified bands. To quantify PCR products, each sample was run in a 1.5% agarose gel and stained with ethidium bromide. Band intensity was measured with an AlphaImager (Alpha Innotech Corp.) using the Alphaease (v3.3b) program.
In situ hybridization
In situ hybridization (ISH) was performed as previously described (Cho et al. 2010), using a 1671 bp probe for Hau-twist1. The embryos were incubated at 37 °C for 10 min in 20 mg/ml Pronase E in PBT (PBS plus 0.1% Tween 20, pH 7.4), then rinsed three times in PBT at room temperature for 20 min. Embryos were then post fixed for 20 min in 4% paraformaldehyde and finally rinsed twice in PBT. Pre-hybridization was carried out at 67 °C for 30 min in hybridization buffer (50% deionized formamide, 5× SSC, 1× Denhardt’s solution, 0.1% CHAPS, 100 mg/ml heparin, 0.1% Tween 20, 100 mg/ml tRNA). The pre-hybridization buffer was replaced with fresh hybridization buffer containing 2 ng/ml of the Hau-twist1 probe, and embryos were incubated at 67 °C overnight. Embryos were washed, then incubated at room temperature for 1.5 h in 1% blocking reagents (Roche) in PBT, then incubated at 4 °C for 16 h with 1/1000 anti-DIG/AP antibody (Roche) in 1% blocking reagents. After incubation, the color reaction was carried out using nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indoyl-phosphate (Roche) by standard procedures. Stained embryos were dehydrated in ethanol, mounted in Fluoromount-G (Southern Biotech), and imaged using brightfield microscopy.
For fluorescent whole mount in situ hybridization (FWMISH), we used the NEN tyramide signal amplification (TSA) Plus kit (PerkinElmer, Wellesley, MA). The protocol was identical to that in Cho et al. (2010), where embryos were rinsed once in 1× PBTw and blocked for 2 h at room temperature in a solution of 0.1 M Tris-HCl pH 7.5, 0.15 M NaCl, and 0.1% Tween 20 (TNT) containing 1% NEN TSA blocking reagent (collectively designated TNB). Embryos were incubated overnight at 4 °C with peroxidase-conjugated anti-digoxygenin (Roche Applied Science) at a 1:1000 dilution in TNB. Subsequent washes (3 × 20 min) in TNT at room temperature were followed by a single 1 × 30-min rinse in NEN TSA Plus amplification solution. The color reaction was initiated by adding a 1:50 dilution of reconstituted cyanine-3 tyramide in NEN amplification solution. For fluorescence microscopy, some embryos were counterstained with 4,6-diamidino-2-phenylindole (DAPI, Sigma) to visualize cell nuclei. FWMISH was visualized using a Leica compound microscope and a spinning disk confocal microscope (Leica Microsystems).
Lineage tracing and cross sectioning
To determine the embryonic origin of the cells expressing specific Hau-twist, teloblasts or proteloblasts of embryos in stages 5 and 6 were injected with a fluorescently labeled, fixable dextran lineage tracer, fluorescein-conjugated dextran amine (FDA), as previously described (Gline et al. 2011). Injected embryos were cultured in Helobdella triserialis saline (HTR) at 23 °C to the desired embryonic stage, then fixed and processed by FWMISH as described above. After FWMISH, the embryos were dehydrated in ethanol and propylene oxide, followed by infiltration with plastic embedding medium (Poly/Bed 812; Polysciences). Then, embryos were imaged on a Leica spinning confocal microscope (Figs. 2 (I), 3 (B and D–E″), and 4 (C–C″)). Also, embryos were sectioned by using a glass knife on a microtome (MT-2B; Sorvall, Newtown, CT) or hand cut by razor blade into 0.1-mm sections. Sections were imaged on a Leica compound microscope to obtain combined fluorescence and brightfield images (Leica Microsystems).
Results and discussion
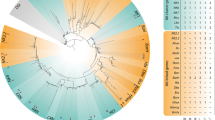
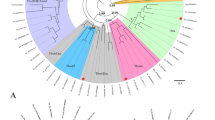
A phylogenetic analysis was conducted using predicted amino acid sequences of the basic helix-loop-helix domains of Hau-twist1, Hau-twist2, and several other twist orthologs using sequences from the hairy gene family as the outgroup. A maximum likelihood analysis grouped Hau-twist1 with other twist orthologs with good bootstrap support (Fig. 1a). In contrast, while Hau-twist2 did group with other twist genes, its position was not well supported. Sequences of the hairy gene family were grouped together outside the twist group at the base of the tree. The commonly accepted phyletic relationships of the species sampled were not recovered, presumably due to the small number of informative sites used in the analysis. Together, these results suggest that Hau-twist1 sequences belong to the twist class.
a Phylogenetic analysis of twist genes. Phylogenetic tree of two Helobdella twist homologs and related transcription factors reconstructed with the maximum likelihood method. Bootstrap value <50% was collapsed. Analysis of basic helix-loop-helix domain of Helobdella twist genes and of other twist family members, with hairy genes as outgroup. Hro Helobdella robusta (JGI Protein ID 155122 (twist1) and JGI Protein ID 176734 (twist2)), Pdu Platynereis dumerilii, Tta Transenella tantilla, Pvu Patella vulgata, Has Helix aspersa, Dme Drosophila melanogaster, Dvi Drosophila virilis, Nve Nematostella vectensis, Pca Podocoryne carnea, Hsa Homo sapiens, Mmu Mus musculus, Xla Xenopus laevis, Xtr Xenopus tropicalis, Rno Rattus norvegicus, Dre Danio rerio. b Temporal expression of Hau-twist1. Digital images of ethidium bromide-stained agarose gels showing replicated developmental semi-quantitative RT-PCR experiments for Hau-twist1 (30 and 35 cycles) and 18s rRNA (30 cycles). Semi-quantitative RT-PCR reveals the that the presence of Hau-twist1 at low levels demonstrates during cleavage (stages 1–7) and gastrulation (stages 7 and 8) with peak expression during organogenesis (stages 9–11)
As a first step in characterizing the expression of Hau-twist1, we carried out semi-quantitative RT-PCR using 18S ribosomal RNA as an internal control and starting with RNA samples from all stages of development (Weisblat and Huang 2001), to obtain the temporal expression profile for Hau-twist1 (Fig. 1b). Hau-twist1 was detected at low levels throughout cleavage stages 1 to 8, and then increased considerably from stage 9 onward, peaking at stage 10. The relatively high levels of Hau-twist1 transcripts during late stages, when segmentation and organogenesis are taking place, are consistent with the proposed role for Hau-twist1 in differentiation, consistent with observations from in other systems. On the other hand, the expression of Hau-twist1 seen at earlier stages is similar to that obtained by Soto et al. (1997), which is reminiscent of the early twist expression in Drosophila (Thisse et al. 1988). This observation raised the possibility that twist is involved in specification of mesodermal fate as well as mesoderm differentiation in Helobdella.
To gain further insight into the function of twist in leech development, we examined the spatial expression profiles of Hau-twist1 in H. austinensis embryos by ISH. Leech embryos fixed at stage 1 to stage 9 were processed for chemical ISH, while embryos fixed at stage 9 to stage 11 were processed for FWMISH. Results of ISH and FWMISH are shown in Figs. 2, 3 and 4. Hau-twist1 transcripts were associated with domains of specialized cytoplasm called teloplasm during cleavage stages 1 to 4 (Fig. 2 (A–C)). Teloplasm, a characteristic feature associated with unequal cleavage and D quadrant specification in clitellate annelids, consists of cytoplasm that is deficient in yolk but rich in messenger RNA (mRNA) and organelles (Weisblat et al. 1984). Teloplasm is segregated to the D macromere at the second cleavage and thence to macromere D′ at third cleavage (stage 3, not shown) (Weisblat and Kuo 2014). At fourth cleavage, macromere D′ divides to form ectodermal and mesodermal precursors, proteloblasts DNOPQ and DM, respectively (stage 4b). We observed that Hau-twist1 transcripts remain associated with teloplasm in both cells (Fig. 2 (C), asterisks). Hau-twist1 staining was also detected in micromeres and in pro-endodermal cell A′. Teloplasm segregation is involved in specifying the fate of stem cells that generate ectodermal and mesodermal lineages (Astrow et al. 1987; Nelson and Weisblat 1991, 1992). Thus, detection of Hau-twist1 mRNA in the teloplasm of both precursors instead of finding it in only the DM proteloblast argues against the possibility that Hau-twist1 mRNA localization functions in specifying the mesodermal fate specification, consistent with previous work (Farooq et al. 2012).
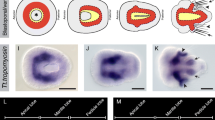
Expression patterns of Hau-twist1 during Helobdella development (from stage 1 to stage 9). Lateral view, animal pole up; during stage 1, Hau-twist1 is associated with the domains of teloplasm at the animal and vegetal poles (asterisks) (A). Roughly animal pole (prospective dorsal) views (B and C). In stage 2, Hau-twist1 remains associated with teloplasm primarily in cell CD, following the unequal first cleavage into cells AB and CD (B). By stage 4b, teloplasm (asterisks), still containing Hau-twist1, has been partitioned into the mesodermal (DM) and ectodermal (DNOPQ) proteloblasts (C). Vegetal view; in stage 5, Hau-twist1 is expressed in columns of mesodermal founder cells (arrows) produced by the bilateral mesodermal stem cells (ML and MR) derived from mesodermal precursor DM (D). Animal pole (prospective dorsal) view; in stage 7, Hau-twist1 is expressed more strongly in discontinuous sets of cells (black arrows) within the germinal bands (gb) (E). Ventral views, anterior up; Hau-twist1 is expressed in segmentally iterated sets of cells within the germinal bands (gb) and germinal plate (gp) (F–H). Fluorescence microscopy image of an early stage 8 embryo (oriented as in F) showing Hau-twist1 expression (red) and a unilaterally labeled M lineage (green); the embryo was then sectioned transversely near the junction of the germinal bands (yellow dashed arrow) (I). Combined fluorescence and brightfield images showing spots of Hau-twist1 expression pattern in germinal band (J, red) and the presence of M teloblast-derived cells injected with FDA lineage tracer (J′, green) (J–J″). The merge image (J″) demonstrates that Hau-twist1 is expressed within the M lineage (yellow spots). Hau-twist1 expression (blue) in segmental and non-segmental tissues during stage 9 (K–L). Lateral (K) and ventral (L) views of the germinal plate highlight the segmentally iterated pattern of Hau-twist1 expression. In addition, Hau-twist1 is expressed in non-segmental tissues at the anterior end (ventral view of the boxed section in K is enlarged in K′), in the region of the future proboscis. a anterior, p posterior. Scale bar 120 μm (A–I, K, L); 60 μm (J–J″, K′)
Expression pattern of Hau-twist1 during stage 9. Schematic of a stage 9 embryo highlighting the germinal plate (white) and the provisional integument (gray) (adapted from Cho et al. 2010) (A). Confocal image (maximum projections of stacks) of embryo processed for fluorescent in situ hybridization to detect Hau-twist1 transcripts (FWMISH, red) and in which the M teloblast on one side had been injected earlier with FDA lineage tracer (M-FDA, green) (B). Ventral view of four mid-body segments shows Hau-twist1 and M-FDA overlapping (yellow) within the mesoderm on the left side of the midline (dashed white line). Combined fluorescence and brightfield micrographs of a transverse section cut as indicated by the red plane in A, showing Hau-twist1 expression (C, red), M-FDA lineage tracer (C′, green) only, and merged vies (C″) (C–C″). Confocal images as in B of a similar embryo viewed laterally show Hau-twist1 (D, red), M-FDA (D′, green) and merged views (D″) expression in both the germinal plate (thick arrow) and in circumferential muscle fibers of the provisional integument (thin arrow) (D–D″). Corresponding views at higher magnification of the region indicated by the box in D″ (E–E″). Scale bar 120 μm (D–D″); 60 μm (B–C″); 40 μm (E–E″)
Expression of Hau-twist1 during stages 10 and 11. Lateral views at two different focal planes, showing a stage 10 embryo processed by FWMISH for Hau-twist1 (red), show transcripts in the everted proboscis region (arrow in A) and in the caudal segments (arrow in A′) (A–A′). Confocal image (maximum projections of stacks) of a sagittal section through the proboscis shows Hau-twist1 mRNA transcripts (red) staining strongly the cells of the forming outer ring proximal to the ventral side of the embryo, and DAPI-stained nuclei (blue) (B). Confocal stacks, ventral view showing Hau-twist1 transcripts only (C, red), lineage tracer M-FDA only (C′), and co-localization of Hau-twist1 and M-FDA (yellow in merged view C″) along the ventral midline (dotted line), in the mesoderm of three mid-body segments from a stage 10 embryo (C–C″). The labeling outlines the segmental ganglia. Lateral views at two different focal planes, showing a stage 11 embryo processed by fluorescent in situ hybridization for Hau-twist1 (red), show transcripts in the proboscis region (left arrow in D), in the caudal segments (right arrow in D), and in body wall of mid-body segments (arrow in D′) (D–D′). Combined fluorescence and brightfield of a cross section of the proboscis at the level corresponding to the dashed white line in D shows that Hau-twist1 is expressed in the ventral portion of the inner ring (E). Scale bar 120 μm (A–A′, D–D′); 60 μm (B); 20 μm (C–C″, E)
During stage 5, Hau-twist1 mRNA transcripts were no longer detected in the NOPQ proteloblasts formed from ectodermal precursor. In contrast, Hau-twist1 was uniformly expressed in the left and right m bandlets (Fig. 2 (D), arrows) arising; the bilateral pair of mesodermal stem cells (left and right M teloblasts) descended from the DM mesodermal precursor. Newly born m blast cells already show mesoderm-specific gene expression, such as Hau-pax3/7A (Woodruff et al. 2007). Thus, while we cannot affirm that Hau-twist1 is involved in mesoderm specification (due to the fact that Hau-twist1 transcript is not asymmetrically segregated between DNOPQ and DM division), the spatial expression pattern of Hau-twist1 at stage 5 suggests that this gene is involved in very early development of the mesodermal germ layer.
By stage 7, discontinuous spots of more intense Hau-twist1 expression emerged, starting from the anterior ends of the germinal bands and progressing rearward (Fig. 2 (E)). During stage 8, as the left and right germinal bands coalesce along the ventral midline, forming the germinal plate, the spots of Hau-twist1 expression appeared to intensify and enlarge, but remained discontinuous (Fig. 2 (F–H)); by late stage 8, the pattern was clearly suggestive of a segmentally iterated sets of cells (Fig. 2 (H)). During stage 9, the segmentally iterated pattern of Hau-twist1 expression persisted in the germinal plate with subtle refinements (Fig. 2 (K and L)). In addition, Hau-twist1 expression was observed in a ring around the presumptive stomodeum and posterior to that in the forming proboscis (Fig. 2 (K′), arrow).
To test the assumption that Hau-twist1 expression in the germinal bands and germinal plate is associated with the mesodermal derivatives, we carried out FWMISH analyses on embryos in which the mesodermal lineage had been labeled by injecting a M teloblast at earlier stages with fluorescent lineage tracer (details described in “Materials and methods”). The expression of Hau-twist1 RNA in germinal bands of a mid-stage 8 embryo, in which the right M proteloblast had been injected with FDA lineage tracer at stage 5, is shown in Fig. 2 (I). The examination of such embryos in section confirmed that Hau-twist1 was expressed exclusively in subsets of mesodermal derivatives in the germinal band during stage 8 (Fig. 2 (J–J″)).
To further characterize the expression of Hau-twist1 at the level of a readily identified mesodermal cell type, we examined a set of early differentiating circumferential muscle fibers underlying squamous epithelium within the provisional integument that transiently covers the surface of the embryo, dorsal to the edges of the germinal plate during stages 8–10 (Fig. 3 (A), gray area). These muscle fibers arise by the outmigration of cells from the m blast cell clones in the germinal bands (Weisblat et al. 1984; Weisblat and Shankland 1985). They provide for peristaltic movements of the provisional integument starting at late stage 8, and it has been speculated that they assist in the expansion and dorsal closure of the germinal plate by providing traction to the leading edges of the germinal plate as it expands toward the dorsal midline during stages 9–10. The provisional integument is eventually replaced by body wall epidermis and musculature when dorsal closure is completed.
To determine if the circumferential muscle fibers of the provisional integument express Hau-twist1 as part of their differentiation, we labeled them by injecting mesodermal precursor DM or newborn M teloblast with FDA lineage tracer. In the example shown, Hau-twist1 expression overlapped with the FDA linage tracer on the left side of the ventral midline of the embryo (Fig. 3 (B)). Examination of sectioned and FWMISH-stained stage 9 embryos in which newborn M teloblast had been injected with lineage tracer revealed doubly labeled cells within the germinal plate, consistent with the results from the germinal bands at earlier stages of development (Fig. 3 (C–C″)). In addition to the expression of Hau-twist1 within germinal plate mesoderm, lateral views of embryo showed that the Hau-twist1 signal appeared to overlap with the FDA lineage tracer in circumferential muscles of the provisional integument, visible as elongated, circumferentially oriented cells outside the germinal plate (Fig. 3 (D–D″)). Higher magnification images confirm the presence of Hau-twist1 transcripts primarily in perinuclear cytoplasm of these cells (Fig. 3 (E–E″). Thus, FWMISH combined with FDA lineage tracer analysis confirms that Hau-twist1 is expressed in the differentiating circumferential muscles of the provisional integument.
Helobdella and other glossiphoniid leech species feed via an eversible piercing and sucking organ called the proboscis. The proboscis is a muscular tubule that develops in an everted position during stage 9 through early stage 10, and then is withdrawn through the oral opening during late stage 10 onward (Weisblat and Huang 2001). The Helobdella proboscis is organized into three concentric rings of cells, namely (i) the inner ring, a layer of columnar epithelial cells surrounding the triangular lumen; (ii) the middle ring, a thin layer comprised of presumptive circumferential muscle fibers situated roughly midway along the radius of the proboscis; and (iii) the outer ring, which includes multiple layers of cells such as presumptive radial and longitudinal muscle cells, nerves, and secretory ductules (Kang et al. 2003; Gline et al. 2011). At stage 10, Hau-twist1 was expressed in the developing proboscis (Fig. 4 (A)), specifically in the ventral portion of the inner ring (Fig. 4 (B)). Expression at this stage was also strong in the posterior mesoderm including the developing rear suckers (Fig. 4 (A′)), and persisted at somewhat lower levels in mid-body segments (Fig. 4 (C–C″)). Hau-twist1 expression continued within the inverted proboscis during early stage 11 (Fig. 4 (D)). Transverse sections of stage 11 embryos suggest that Hau-twist1 was expressed in columnar epithelial cells from the inner ring of the proboscis (Fig. 4 (E)). Finally, during stage 11, Hau-twist1 expression was maintained in the segmental mesoderm flanking the midline (Fig. 4 (D′), arrow) and was particularly prominent in segments forming the rear sucker (Fig. 4 (D)).
The results obtained in the present study have expanded previous characterizations of twist expression in Helodella (Soto et al. 1997; Farooq et al. 2012), by adding the characterization of its expression throughout stages 9 to 11. These results are similar to those reported for embryos of the mollusca Patella vulgata and the polychaete annelid Capitella teleta in that twist homologs are expressed in subsets of mesodermal derivatives throughout differentiation and organogenesis (Dill et al. 2007; Nederbragt et al. 2002). In contrast to these other lophotrochozoan systems, we detected low levels of Hau-twist1 expression (by semi-quantitative RT-PCR and in situ hybridization) throughout cleavage stages. Since Hau-twist1 RNA appeared in both mesoderm and ectoderm precursors, however, this early expression does not support a role for this gene in mesoderm specification, unless one also postulates differential expression of the early transcripts. In any event, the expression pattern of Hau-twist1 in late stages does seem to indicate a role in mesoderm differentiation. Future functional studies are recommended to understand the exact role of Hau-twist1 in proboscis formation.
References
Astrow S, Holton B, Weisblat D (1987) Centrifugation redistributes factors determining cleavage patterns in leech embryos. Dev Biol 120(1):270–283
Baylies MK, Bate M (1996) twist: a myogenic switch in Drosophila. Science 272:1481
Cho SJ, Vallès Y, Giani VC Jr, Seaver EC, Weisblat DA (2010) Evolutionary dynamics of the wnt gene family: a lophotrochozoan perspective. Mol Biol Evol 27(7):1645–1658
Dill KK, Thamm K, Seaver EC (2007) Characterization of twist and snail gene expression during mesoderm and nervous system development in the polychaete annelid Capitella sp. I. Dev Genes Evol 217:435–447
Farooq M, Choi J, Seoane AI, Lleras RA, Tran HV, Mandal SA, Nelson CL, Soto JG (2012) Identification of 3'UTR sequence elements and a teloplasm localization motif sufficient for the localization of Hro-twist mRNA to the zygotic animal and vegetal poles. Develop Growth Differ 54(4):519–534
Gline SE, Nakamoto A, Cho SJ, Chi C, Weisblat DA (2011) Lineage analysis of micromere 4d, a super-phylotypic cell for Lophotrochozoa, in the leech Helobdella and the sludgeworm Tubifex. Dev Biol 353:120–133
Hamamori Y, Wu HY, Sartorelli V, Kedes L (1997) The basic domain of myogenic basic helix-loop-helix (bHLH) proteins is the novel target for direct inhibition by another bHLH protein, twist. Mol Cell Biol 17:6563–6573
Kang Y, Massagué J (2004) Epithelial-mesenchymal transitions: twist in development and metastasis. Cell 118(3):277–279
Kang D, Huang F, Li D, Shankland M, Gaffield W, Weisblat DA (2003) A hedgehog homolog regulates gut formation in leech (Helobdella). Development 130:1645–1657
Kuo DH, Weisblat DA (2011) Intermediate filament genes as differentiation markers in the leech Helobdella. Dev Genes Evol 221(4):225–240
Kutschera U, Langguth H, Kuo DH, Weisblat DA, Shankland M (2013) Description of a new leech species from North America, Helobdella austinensis n. sp. (Hirudinea: Glossiphoniidae), with observations on its feeding behaviour. Zoosyst Evol 89:239–246
Leptin M (1991) twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev 5:1568–1576
Nederbragt AJ, Lespinet O, Van Wageningen S, Van Loon AE, Adoutte A, Dictus WJ (2002) A lophotrochozoan twist gene is expressed in the ectomesoderm of the gastropod mollusk Patella vulgata. Evol Dev 4:334–343
Nelson BH, Weisblat DA (1991) Conversion of ectoderm to mesoderm by cytoplasmic extrusion in leech embryos. Science 253(5018):435–438
Nelson BH, Weisblat DA (1992) Cytoplasmic and cortical determinants interact to specify ectoderm and mesoderm in the leech embryo. Development 115(1):103–115
O’Rourke MP, Soo K, Behringer RR, Hui CC, Tam PP (2002) Twist plays an essential role in FGF and SHH signal transduction during mouse limb development. Dev Biol 248(1):143–156
Price AL, Patel NH (2008) Investigating divergent mechanisms of mesoderm development in arthropods: the expression of Ph-twist and Ph-mef2 in Parhyale hawaiensis. J Exp Zool B Mol Dev Evol 310:24–40
Sablitzky F (2005) Proteins motifs: the helix motif. eLS
Simpson P (1983) Maternal-zygotic gene interactions during formation of the dorsoventral pattern in Drosophila embryos. Genetics 105:615–632
Soto JG, Nelson BH, Weisblat DA (1997) A leech homolog of twist: evidence for its inheritance as maternal mRNA. Gene 199(1–2):31–37
Thisse B, Stoetzel C, Gorostiza-Thisse C, Perrin-Schmitt F (1988) Sequence of the twist gene and nuclear localization of its protein in endomesodermal cells of early Drosophila embryos. EMBO J 7(7):2175–2183
Weisblat DA, Huang FZ (2001) An overview of glossiphoniid leech development. Can J Zool 79:218–232
Weisblat DA, Kuo DH (2014) Developmental biology of the leech Helobdella. J Dev Biol 58:429–443
Weisblat DA, Shankland M (1985) Cell lineage and segmentation in the leech. Philos Trans R Soc Lond Ser B Biol Sci 312:39–56
Weisblat DA, Kim SY, Stent GS (1984) Embryonic origins of cells in the leech Helobdella triserialis. Dev Biol 104:65–85
Woodruff JB, Mitchell BJ, Shankland M (2007) Hau-Pax3/7A is an early marker of leech mesoderm involved in segmental morphogenesis, nephridial development, and body cavity formation. Dev Biol 306(2):824–837
Acknowledgements
The authors would like to thank members of the Weisblat Lab for advice and support. This work was supported by the National Institute of Health [NIH R01 grant GM 074619 to DAW], “Cooperative Research Program for Agriculture Science and Technology Development [Project No. PJ011661]” Rural Development Administration, and by a grant from the Collaborative Genome Program (20140428) funded by the Ministry of Oceans and Fisheries, Republic of Korea. This work was supported by the research grant of the Chungbuk National University in 2014.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Mark Q. Martindale
Rights and permissions
About this article
Cite this article
Kim, JS., Jiménez, B.I.M., Kwak, HJ. et al. Spatiotemporal expression of a twist homolog in the leech Helobdella austinensis . Dev Genes Evol 227, 245–252 (2017). https://doi.org/10.1007/s00427-017-0585-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-017-0585-4