Abstract
To investigate the evolutionary history of mesoderm in the bilaterian lineage, we are studying mesoderm development in the polychaete annelid, Capitella sp. I, a representative lophotrochozoan. In this study, we focus on the Twist and Snail families as candidate mesodermal patterning genes and report the isolation and in situ expression patterns of two twist homologs (CapI-twt1 and CapI-twt2) and two snail homologs (CapI-sna1 and CapI-sna2) in Capitella sp. I. CapI-twt1 is expressed in a subset of mesoderm derivatives during larval development, while CapI-twt2 shows more general mesoderm expression at the same stages. Neither twist gene is detected before the completion of gastrulation. The two snail genes have very distinct expression patterns. At cleavage and early gastrula stages, CapI-sna1 is broadly expressed in precursors of all three germ layers and becomes restricted to cells around the closing blastopore during late gastrulation; CapI-sna2 expression is not detected at these stages. After gastrulation, both snail genes are expressed in the developing central nervous system (CNS) at stages when neural precursor cells are internalized, and CapI-sna1 is also expressed laterally within the segmental mesoderm. Based on the expression patterns in this study, we suggest a putative function for Capitella sp. I twist genes in mesoderm differentiation and for snail genes in regulating CNS development and general cell migration during gastrulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mesoderm evolved near the base of the bilaterian lineage and is one of the defining characteristics that unite bilaterians. It is often speculated that the invention of this third germ layer allowed for the elaboration and diversification of body plan morphologies in the Bilateria (Perez-Pomares and Munoz-Chapuli 2002). To reconstruct the ancestral origins of mesoderm and discover how the mesoderm developmental pathway was modified in the lineages leading to extant bilaterian phyla, it is important to analyze mesoderm development in diverse taxa representing all three of the major bilaterian groups: Ecdysozoa, Lophotrochozoa, and Deuterostomia. Molecular and genetic studies from model organisms (mainly Drosophila and various vertebrates) have identified genes that show a conserved role in mesoderm development between vertebrates and Drosophila. These genes have been used as candidates in broader investigations of mesoderm development in a variety of species. In this study, we focus on the transcription factors twist and snail, which are two of the best-characterized regulators of mesoderm development in the bilaterian lineage (Technau and Scholz 2003).
Twist encodes a basic helix–loop–helix (bHLH) transcription factor and was first identified as a zygotic gene required for gastrulation and mesoderm specification in the long germ-band insect, Drosophila (Simpson 1983; Thisse et al. 1988). Further studies demonstrated that twist plays a dual role in mesoderm development in Drosophila. First, Twist is expressed on the ventral side of the blastula where it specifies the presumptive mesoderm and activates a number of genes required for mesoderm development, including snail (Leptin 1991). Second, after gastrulation, Drosophila twist is expressed in a subset of mesoderm derivatives and is required for proper muscle development (Baylies and Bate 1996; Castanon and Baylies 2002; Cripps et al. 1998). Studies in the short germ-band insect, Tribolium castaneum, demonstrate that Tribolium twist regulates both mesoderm specification and mesoderm differentiation (Handel et al. 2005; Sommer and Tautz 1994). Homologs of twist have been studied in numerous other species representing diverse phyla including vertebrates (Hopwood et al. 1989; Tavares et al. 2001; Wang et al. 1997; Wolf et al. 1991), a lancelet (Yasui et al. 1998), a nematode (Harfe et al. 1998), and one lophotrochozoan, the limpet Patella vulgata (Nederbragt et al. 2002). In most species, twist genes are expressed in a subset of the mesoderm after this tissue is specified where they regulate mesoderm patterning and muscle differentiation (see references above). The prevailing theme that emerges from these studies is that the function of twist in mesoderm specification may be unique to insects, while a function in mesoderm differentiation represents an ancestral bilaterian trait.
The snail genes encode zinc-finger transcriptional repressors that regulate a variety of cell behaviors, including cell movement, cell adhesion, cell division, and cell survival (reviewed in Barrallo-Gimeno and Nieto 2005). The first Snail family member was identified as an embryonic patterning gene in Drosophila (Grau et al. 1984; Nusslein-Volhard et al. 1984) and was later shown to be essential for mesoderm development (Alberga et al. 1991; Leptin 1991). Snail family members have also been described as regulators of mesoderm development in a variety of other species, including Tribolium (Sommer and Tautz 1994), ascidians (Wada and Saiga 1999), amphioxus (Langeland et al. 1998), Xenopus (Mayor et al. 1993; Sargent and Bennett 1990), zebrafish (Thisse et al. 1995; Thisse et al. 1993), mouse (Nieto et al. 1992; Smith et al. 1992), and chick (Nieto et al. 1994). The snail genes are key regulators of cell motility, and careful molecular and genetic analyses demonstrate that the conserved function for snail genes in mesoderm development is in inducing the movement of mesodermal cells during gastrulation rather than in mesoderm specification (Barrallo-Gimeno and Nieto 2005; Nieto 2002). In many contexts, Snail proteins regulate cell movement by inducing epithelial to mesenchymal transitions (EMT) via direct repression of E-cadherin (Batlle et al. 2000; Cano et al. 2000). However, snail genes also regulate cell motility in EMT-independent processes where cells move while maintaining contact with their neighbors in processes such as mesoderm migration during gastrulation in Drosophila, Xenopus, and zebrafish (reviewed in Barrallo-Gimeno and Nieto 2005).
Expression patterns of snail genes (or their proteins) have been examined in two lophotrochozoans, the leech annelid Helobdella robusta (Goldstein et al. 2001) and the gastropod mollusk P. vulgata (Lespinet et al. 2002). In H. robusta, labeling with an anti-Snail antibody reveals broad expression in the early embryo including precursors of ectoderm and mesoderm (Goldstein et al. 2001). In P. vulgata, the expression of snail genes was examined by whole-mount in situ hybridization and transcripts were detected almost exclusively in ectodermal derivatives (Lespinet et al. 2002). Thus, in the two lophotrochozoan species studied to date, snail genes are not exclusively expressed in the mesoderm or presumptive mesoderm; based on these data, it was hypothesized that snail genes regulate cell motility but not mesoderm development in the lophotrochozoan lineage (Lespinet et al. 2002).
As described above, characterization of twist and snail genes in lophotrochozoans is limited to just a few published studies, and for the snail genes, there is variation in expression patterns between species; thus, it is difficult to draw conclusions about conserved functions for twist and snail genes in this lineage. To further investigate mesoderm development in the Lophotrochozoa, we are analyzing the Twist and Snail families in the polychaete annelid Capitella sp. I. In this paper, we report the isolation of two twist homologs (CapI-twt1 and CapI-twt2) and two snail homologs (CapI-sna1 and CapI-sna2) in Capitella sp. I, and characterize the in situ expression patterns for all four genes during embryonic and larval development. Our findings do not indicate a function for Capitella sp. I twist or snail genes in mesoderm specification. Instead, expression for CapI-twt1 and CapI-twt2 correlates with later stages of mesoderm differentiation while expression of the snail genes correlates with cell movements during gastrulation and/or central nervous system (CNS) development.
Materials and methods
Animal husbandry
A Capitella sp. I colony was maintained in the laboratory using published culture methods (Grassle and Grassle 1976). Adults were kept at 19°C in bowls of 0.2-μM filtered seawater (FSW) with organically enriched ocean mud as a food source and brood tubes were recovered by filtering bowl contents through a fine sieve as previously described (Seaver et al. 2005). Embryos and larvae were dissected from brood tubes, rinsed in FSW to remove mud and other debris, and fixed for in situ hybridization or phallacidin staining (see methods below).
Gene isolation
Fragments of CapI-twt1 and CapI-sna1 were amplified by degenerate polymerase chain reaction (PCR) using genomic DNA as template. A ∼200-bp fragment of CapI-twt1 was isolated using a nested PCR strategy with degenerate primers designed to correspond to the highly conserved bHLH domain of Twist proteins. A 67-bp fragment of CapI-sna1 was isolated after a nested PCR using degenerate primers designed to correspond to highly conserved portions of the second and third zinc-finger domains. Degenerate PCR fragments were ligated into the pGEM-T-easy vector (Promega) and sequenced at Macrogen (South Korea).
The CapI-twt1 fragment was extended using a mixed stage complementary DNA (cDNA) library as template. Nested PCR was performed in the 5 and 3′ directions using vector primers in combination with primers specific to the CapI-twt1 fragment. The CapI-sna1 sequence was extended in the 5 and 3′ directions using the BD SMART rapid amplification of cDNA ends (RACE) cDNA amplification kit (BD Biosciences Clontech). RACE cDNA template was generated from mixed stage (5–9) Capitella sp. I larvae. RACE PCR was performed according to the BD SMART RACE protocol using primers specific to the CapI-sna1 fragment. RACE fragments were ligated into a pGEM T-easy vector (Promega) and sequenced (Macrogen). Sequences obtained from the original degenerate PCR products were conceptually spliced together with the 5 and 3′ RACE products; the resulting 968-bp composite transcript for CapI-twt1 and 2,171-bp composite transcript for CapI-sna1, which predict complete open reading frames for both genes, were submitted to GenBank (CapI-twt1 EF192048 and CapI-sna1 EF192044). An additional 3′ RACE fragment representing a unique snail sequence was also isolated; this gene was named CapI-sna2.
CapI-twt1 and CapI-sna1 sequences were used to search sequences of Capitella sp. I expressed sequence tag (EST) libraries. Searches identified one EST representing CapI-sna2 and three ESTs representing a second twist gene, which we named CapI-twt2. A pBluescript SK phagemid (Stratagene) containing the 1,195-bp CapI-twt2 insert was recovered from glycerol stocks and then sequenced for verification (Macrogen). Sequence of the CapI-sna2 EST was used to design primers to amplify additional sequence of CapI-sna2 from mixed stage cDNA template. The resulting 1,269-bp PCR fragment was ligated into pGEM T-easy vector (Promega) and sequenced at Macrogen. Capitella sp. I ESTs identified in our searches were sequenced by the Joint Genome Institute (Department of Energy, Walnut Creek, CA, USA) as part of the Capitella sp. I 8× genome sequencing project. EST sequences used in this study have been submitted to GenBank (CapI-twt2 EF192049 and CapI-sna2 EF192045). Degenerate and gene-specific primer sequences are available upon request (seaver@hawaii.edu).
Gene orthology analyses
Phylogenetic analyses of the Twist and Snail families were performed to determine gene orthology. Capitella sp. I genes were used as query sequences in BlastX searches of the GenBank database (NCBI) to identify related genes for orthology analyses. Alignments of amino acid sequences were made with the ClustalW program included in the MacVector package and corrected by hand for obvious alignment errors. MrBayes (Huelsenbeck and Ronquist 2001) was used to perform Bayesian phylogenetic analyses with the “wag” amino acid model and 1,000,000 generations sampled every 100 generations with four chains. For both gene families, a consensus tree was produced with PAUP*4.01b10 (Swofford 2000) using 9,500 trees representing 950,000 stable generations. A second independent method for determining gene orthology, maximum likelihood (ML), was performed with PhyML (Guindon and Gascuel 2003) using the model of evolution selected via ProtTest (JTT+G for Twist and Dayhoff I+G for Snail; Abascal et al. 2005) and 1,000 bootstrap replicates. The nexus alignment files are available upon request (seaver@hawaii.edu), and accession numbers for sequences used in the alignments are in the supplementary material ESM 1.
Whole mount in situ hybridization
Embryos and larvae were fixed and processed for whole mount in situ hybridization according to published protocols (Seaver and Kaneshige 2006; Seaver et al. 2001). Digoxigenin-labeled riboprobes were generated with the Megascript kit (Ambion), and each probe was diluted to a working concentration of 1 ng/μl. Riboprobes for CapI-twt2 and CapI-sna2 include the complete sequences for these genes as submitted to GenBank. The CapI-twt1 riboprobe is a 458-bp 3′ RACE fragment corresponding to bp 510–968 of the twt1 GenBank sequence, and the CapI-sna1 riboprobe is a 1,204-bp 5′ RACE fragment corresponding to bp 1–1,204 of the sna1 GenBank sequence.
Phallacidin-staining
Capitella sp. I larvae were stained with BODIPY FL-phallacidin (Molecular Probes) as described previously (Seaver et al. 2005).
Results
Molecular characterization of twist and snail orthologs from Capitella sp. I
We isolated two putative twist orthologs and two putative snail orthologs from Capitella sp. I (see “Materials and methods”). Each of the Capitella sp. I twist genes encode proteins that contain bHLH domains and WR motifs typical of the Twist family (Fig. S1a). To assess the orthology of CapI-twt1 and CapI-twt2, we performed phylogenetic analyses using an alignment of the bHLH domains from various Twist proteins as well as more distantly related bHLH proteins (Paraxis and Hairy). Results indicate that the Capitella sp. I Twist proteins group within the Twist family (Fig. S2).
CapI-sna1 and CapI-sna2 encode proteins that each contains five Snail-type zinc-fingers, and in addition, CapI-Sna1 contains a SNAG domain at the N terminus (Fig. S1b). The Snail superfamily is subdivided into two subfamilies, Snail and Scratch, which are thought to have originated from a duplication in an ancestral snail/scratch gene early in the metazoan lineage (reviewed in Nieto 2002). We also identified two scratch genes in Capitella sp. I, which to our knowledge represent the first report of the presence of scratch genes in lophotrochozoans (Fig. S3). To assign orthology to the Capitella sp. I snail and scratch genes, we performed phylogenetic analyses using an amino acid alignment of the zinc-finger regions of Snail and Scratch sequences from Capitella sp. I and a diverse representation of other metazoan species. Figure S3 shows a Bayesian inference consensus tree, which clearly indicates that CapI-sna1 and CapI-sna2 are members of the Snail family.
Capitella sp. I development
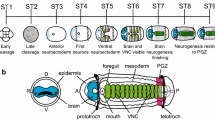
The development of Capitella sp. I has been described previously (Bhup and Marsden 1981; Eisig 1899; Seaver et al. 2005; Werbrock et al. 2001). Figure 1 provides an overview of embryonic and larval development (Fig. 1a), highlighting features relevant for interpreting the expression patterns in this study (Fig. 1b–h). Early embryogenesis follows a stereotypic pattern of unequal spiral cleavage. After cleavage and gastrulation, Capitella sp. I develops through a trochophore larval phase, which is characterized by anterior (prototroch) and posterior (telotroch) cilliary bands separated by the mid-body, where the segments and ventral nerve cord (VNC) form (Fig. 1b,g, and h). The larval phase is brief and nonfeeding, and metamorphosis involves only minor morphological modifications; the prototroch and telotroch are lost but most other structures are maintained in the juvenile and adult.
Summary of Capitella sp. I embryonic and larval development. Anterior is towards the left in each image or diagram. a Schematic of embryonic and larval stages. Each stage is approximately 24 h in length at 19°C; the transition from embryonic to larval life (hatching) occurs at stage 4 when the cilliary bands of the prototroch (p) and telotroch (t) penetrate the egg membrane. Segmentation begins at stage 5, and 13 segments form during larval development. b DIC image of a stage-6 larva in ventral view. The mid-body is shown with a bracket. c Magnification of the boxed region in b showing a clear morphological distinction between ectodermal (e) and mesodermal (m) tissue layers (red dashed line). d In post-gastrula embryos (late stage 4), the lateral growth zones, which appear as longitudinal bands of dividing cells located in ventral lateral positions, expand circumferentially around the larvae giving rise to ectodermal and mesodermal tissue of the first ten segments (Seaver et al. 2005). e During stage 6, the region of dividing cells transitions to a posterior position just anterior to the telotroch and represents a more classically described posterior growth zone, which gives rise to the posterior larval and juvenile segments (Seaver et al. 2005). f Brain development begins at stage 3 and is morphologically apparent as a thickening of the anterior neural ectoderm (Bhup and Marsden 1981; Frobius and Seaver 2006). g Two bilateral cerebral ganglia have formed by stage 5 (N. Meyer, personal communication). The VNC forms in the segmental region of the body; ventral ectodermal cells on either side of the midline give rise to the ganglia of the VNC (open circles in g; Eisig 1899; Seaver et al. 2005). h The brain is fully internalized by stage 9, and the VNC has fused at the ventral midline to form one ganglion in each segment (Eisig 1899 and N. Meyer, personal communication)
Mesoderm development in Capitella sp. I, including embryonic origins and larval development, was described morphologically more than 100 years ago (Eisig 1899). Mesoderm arises from three distinct blastomeres in the early cleavage stage embryo: third quartet micromeres, 3c and 3d, and fourth quartet micromere, 4d (Eisig 1899). After blastopore closure, two large mesodermal stem cells located posteriorly divide asymmetrically producing bilateral rows of mesoderm cells (mesodermal bands) that extend towards the anterior of the larva; these mesodermal bands proliferate and give rise to segmental mesoderm (Anderson 1973; Eisig 1899; E. Seaver, unpublished observations). An analysis of cell division patterns in Capitella sp. I indicates that the segmental tissue arises from two separate zones of cell proliferation (Seaver et al. 2005): lateral growth zones generate ectoderm and mesoderm of the first ten segments, and a posterior growth zone generates ectoderm and mesoderm of the posterior larval and juvenile segments (Fig. 1d,e). The mesodermal bands described by Eisig likely represent the mesodermal component of the lateral growth zones. Tissue originating from lateral growth zones expands circumferentially around the larvae (Seaver et al. 2005), and by stage 6, a morphological distinction between the ectoderm and mesoderm is apparent (Fig. 1b,c).
Expression of twist genes in Capitella sp. I
To determine the spatiotemporal expression of CapI-twt1, we performed whole mount in situ hybridization using a probe encompassing a 458-bp fragment of this transcript. No expression is detected in embryos during cleavage, blastula, or gastrula stages (data not shown). Soon after blastopore closure (late stage 3), CapI-twt1 transcript is detected in a small patch of cells at the anterior midline in the head and in two longitudinal bands in the mid-body (Fig. 2a). Expression in both of these domains persists and expands throughout larval development. Expression associated with the foregut appears at stage 5 (Fig. 2c,d), and expression around the hindgut appears at stage 7 (Fig. 2i,j).
The CapI-twt1 transcript is detected after gastrulation in a subset of mesodermal derivatives. Subpanels a, b, c, e, g, i, and m are ventral views, anterior to the left; d, h, j, and k are lateral views with ventral down and anterior to the left. Indicated in each subpanel: st. stage; asterisk stomodeum; t telotroch; br brain. a CapI-twt1 is expressed in a small patch of cells in the head (diagonal arrow) and in longitudinal bands in left and right ventro-lateral positions (horizontal arrows). b Expression is more prominent at stage 4 with a larger patch of CapI-twt1 positive cells in the head (diagonal arrow) and longitudinal bands spanning the entire mid-body region (horizontal arrows). c Longitudinal bands of CapI-twt1 expression are maintained at stage 5 (arrows). d A lateral view at stage 5 shows that the CapI-twt1 positive cells are off-set towards the dorsal side of the larva in the anterior ∼two thirds of the mid-body and are positioned closer to the ventral midline (solid line) in the posterior ∼third (compare horizontal arrows in d). A third domain of expression at stage 5 is associated with the developing foregut (arrowheads in c and d). e, f At stage 6, expression is detected on either side of the stomodeum in the region of the developing foregut (arrowheads) and in the mid-body mesoderm (horizontal arrows in e). There is also an arc of expression in the head (diagonal arrow), which co-localizes with muscle fibers (arrow in g). g Phallicidin stains F-actin filaments in the longitudinal (double-headed arrows) and circumferential (arrowheads) muscle fibers in the mid-body and the anterior extension of the longitudinal muscle fibers forming an arc in the head (arrow). The ventral nerve cord is also visible as longitudinal stripes on either side of the ventral midline. h At stage 6, CapI-twt1 positive cells in the anterior portion of the mid-body are closer to the ventral midline than at stage 5 (solid line, compare to d). i CapI-twt1 expression is confined to the mesoderm of the larval segments. j A deep focal plane showing expression associated with the foregut (staining on the anterior and posterior sides of the stomodeum), in the hindgut (arrowhead), and in the head muscle (arrow). k Close-up of j in a surface focal plane. CapI-twt1 expression is detected in three longitudinal stripes (arrowheads) in the mid-body. l Posterior view showing expression in a ring of cells around the posterior end of the hindgut. m At stage 9, expression is limited to the mesoderm of the posterior growth zone (arrowhead) and a small patch in the head (arrow)
The longitudinal bands of CapI-twt1 expression in the mid-body at stages 3 and 4 (Fig. 2a,b) correspond to the approximate position of dividing cells in the lateral growth zones at these stages (Seaver et al. 2005 and summarized in Fig. 1d). Analysis with differential interference contrast (DIC) microscopy reveals that these CapI-twt1 expressing cells are located between the surface ectodermal cells and the yolk rich endoderm; we therefore judge the expression to be restricted to the mesodermal portion of the lateral growth zones (mesodermal bands). At stage 5, when the lateral growth zone tissue has expanded dorsally and ventrally around the larva (Seaver et al. 2005), longitudinal bands of CapI-twt1 expression encompass only a subset of the segmental mesoderm (Fig. 2c,d). Each ‘band’ consists of two non-overlapping stripes of expression occupying different dorsal-ventral positions; a stripe in the anterior ∼two thirds of the mid-body is displaced towards the dorsal side of the larva while a stripe in the posterior ∼one third of the mid-body is positioned closer to the ventral midline (Fig. 2d). The difference in the position of CapI-twt1 positive cells in the anterior vs posterior of the mid-body is less dramatic at stage 6 when the anterior stripe, lying closer to the ventral midline, is only slightly dorsal of the posterior stripe (Fig. 2h). At stage 7, CapI-twt1 is expressed in segmentally iterated patches of cells arranged in three longitudinal rows on either side of the mid-body: a mid-lateral row spanning the entire mid-body region and dorsal-lateral and ventral-lateral rows extending only through the anterior-most ∼7–8 segments (Fig. 2k, arrowheads). The posterior boundary of the dorsal-lateral and ventral-lateral rows corresponds to the approximate position of the thoracic/abdominal boundary. In a ventral view at stage 7, the morphological distinction between mesoderm and ectoderm is clearly visible; CapI-twt1 transcript is detected in the mesodermal layer and is excluded from the ectoderm (Fig. 2i). The transition in the CapI-twt1 mid-body expression between stages 6 and 7 is abrupt, and there are no apparent morphological features that correlate with either of these patterns. Expression in the mid-body diminishes during stage 8, and at stage 9, CapI-twt1 transcript is detected in the mesoderm of the posterior-most segments and within the mesoderm in the posterior growth zone (Fig. 2m, arrowhead).
Expression of CapI-twt1 in the head is initiated at stage 3 in a small cluster of subepidermal cells at the anterior midline (Fig. 2a) and expands to additional cells at stage 4 (Fig. 2b). There is a transient downregulation of expression in the head at stage 5 (Fig. 2c,d). At stage 6, CapI-twt1 is detected prominently in a thin layer of cells forming an arc in the head (Fig. 2e,f, arrows), a pattern that corresponds to the shape and position of muscle fibers in the head at the same stage (Fig. 2g, arrow). This expression is maintained, and in lateral view, it is apparent that the CapI-twt1 expression domain is positioned beneath the surface ectoderm in an arc of cells along the anterior edge of the brain (Fig. 2j, arrow). Thus, in the head, CapI-twt1 is expressed in the mesoderm, likely in a developing muscle fiber, which lies between the brain and the anterior ectodermal epithelia. Head expression is downregulated during stage 8 and is only detected in a small domain at the anterior midline at stage 9 (Fig. 2m, arrow).
CapI-twt1 is also expressed in association with the developing foregut and hindgut. Initially, expression in the foregut is weak (stages 4 and 5; Fig. 2b–d, arrowheads) and becomes more pronounced at stage 6 (Fig. 2e,f, arrowheads). CapI-twt1 is maintained at lower levels in the anterior foregut at stage 7 (Fig. 4i,j) but is not detected in this tissue at subsequent larval stages. Beginning at stage 7, CapI-twt1 transcript is detected in a ring at the position of the developing hindgut (Fig. 2l and vertical arrowheads in i and j). Hindgut expression is downregulated during stage 8 (data not shown) and no longer detected at stage 9. The foregut and hindgut are likely derived from ectoderm, but are also associated with muscles used for feeding and excretion (Saulnier-Michel 1992). CapI-twt1 expression in the foregut is external to the epithelium in what appear to be muscles around the stomodeum (Fig. 2f,g).
In comparison to CapI-twt1, CapI-twt2 exhibits broader expression throughout most of the larval mesoderm. CapI-twt2 transcript is first detected at stage 4 (Fig. 3a) in two longitudinal bands located ventro-laterally in the mid-body, in the foregut (arrowheads), and in the brain. Expression is maintained through stage 9, and although it overlaps with CapI-twt1 expression in several domains, there are distinct characteristics to the CapI-twt2 pattern. We elaborate on each CapI-twt2 expression domain below.
CapI-twt2 is broadly expressed in the developing mesoderm and is most prominent in tissues that also express CapI-twt1. Subpanels a, b, d, e, g, h, and k are ventral views with anterior to the left; c, f, i, and j are lateral views with ventral down and anterior to the left. Indicated in each subpanel: st. stage; asterisk stomodeum; t telotroch; br brain. Red dashed lines (d, g, and k) mark the boundary between ectoderm (e) and mesoderm (m) and brackets (d, f, g, j, and k) mark the mid-body. a CapI-twt2 expression is first detected at stage 4 in longitudinal bands in the mid-body (arrows), in the region of the developing foregut (arrowheads), and in the region of the developing brain. b, c At stage 5, expression expands in the region of the foregut (arrowheads) and in the mid-body (arrows) while expression in the brain appears more faint. d–f At stage 6, CapI-twt2 is expressed throughout the entire mid-body mesoderm (see red dashed line in d); expression is also prominent in the developing foregut (arrowheads in e and f) and in a broad domain forming an arc in the head (arrows). g, h Expression in the mid-body mesoderm persists into stage 7, and transcript is still excluded from the ectodermal layer (see red dashed lines, h is a magnification of the boxed region in g). i Lateral view of a deep focal plane; expression in the developing foregut (arrowhead) is maintained at stage 7, while expression in the head (arrow) is now localized between the brain and surface ectoderm. j Same larva as i, surface focal plane showing dorsal-, ventral-, and mid-lateral stripes of CapI-twt2 expression (arrowheads). k At stage 9, CapI-twt2 is expressed at low levels throughout head and mid-body mesoderm
CapI-twt2 expression in the longitudinal bands (Fig. 3a) is similar to CapI-twt1 expression in this domain at stage 4; thus, it appears that CapI-twt2 may also be expressed in the mesodermal bands at this stage. CapI-twt2 expression in the lateral mid-body continues at stage 5 in a subset of the segmental mesoderm (Fig. 3c). The anterior portion of this expression pattern is not displaced dorsally, as CapI-twt1 is at this stage (Fig. 3c compare to Fig. 2d). At stages 6 and 7, CapI-twt2 expression expands to encompass all of the segmental mesoderm (Fig. 3d, f–j). A lateral view at stage 7 also shows broad expression of CapI-twt2 in the mid-body; however, more pronounced expression is apparent within dorsal-lateral, ventral-lateral, and mid-lateral rows (Fig. 3j, arrowheads). These rows are very similar in position to the three rows of CapI-twt1 expression at this stage (Fig. 2k). At stage 9, CapI-twt2 expression is diffuse throughout the segmental mesoderm and contrasts with the more restricted expression of CapI-twt1 at this stage (compare Fig. 3k to Fig. 2m).
CapI-twt2 is initially expressed more broadly in the head than CapI-twt1 and is detected throughout the developing brain at stages 4 and 5 (Fig. 3a,c). Expression is reduced in the brain at stage 6 and is instead concentrated in tissue anterior to the brain (Fig. 3d–f, arrows). At stage 7, CapI-twt2 is expressed in head mesoderm in a thin arc of cells located between the brain and the anterior ectoderm (Fig. 3i, arrow). Thus, at early larval stages, CapI-twt2 head expression correlates with brain development and is quite distinct from the CapI-twt1 head expression. However, at stage 7, both transcripts are expressed in association with a muscle in the head.
CapI-twt2 is also expressed in the developing foregut from stage 4 to 7 (Fig. 3a,b,c,e,f, and i, arrowheads). In contrast to the CapI-twt1 foregut expression, which appears to be in foregut-associated muscles, CapI-twt2 is expressed within the ectodermally derived foregut epithelium.
Expression of snail genes in Capitella sp. I
Expression of CapI-sna1 transcript was examined from early cleavage stages through the end of larval development (stage 9). CapI-sna1 is detected in all four blastomeres in four-cell stage embryos (Fig. 4a). At later cleavage stages, transcript is detected in a subset of cells, and although we did not determine the identity of each CapI-sna1 positive cell, it is clear from the overall pattern that the transcript is not restricted to precursors of a particular germ layer or embryonic quadrant (data not shown). Towards the end of gastrulation, CapI-sna1 expression is downregulated in most of the embryo, and transcript is only detected in surface cells around the closing blastopore (Fig. 4b). Expression in this region continues through the end of gastrulation. After blastopore closure, CapI-sna1 positive cells are located deep within the embryo and coincide with the position of endodermal cells (Fig. 4c).
CapI-sna1 has a dynamic expression pattern through cleavage, embryonic, and larval stages. Subpanel a is an animal view, b–g, k, l, and m are ventral views with anterior to the left; h, i, and j are lateral views with ventral down and anterior to the left. Indicated in each subpanel: st. stage; asterisk stomodeum; t telotroch; br brain. Solid lines (f, h, and i) mark the ventral midline, and red dashed lines (k, l, and m) mark the boundary between the ectoderm (e) and mesoderm (m). Images d and e show the same larva at surface (d) and deep (e) focal planes; this same scheme follows for f/g and h/i. a The CapI-sna1 transcript is detected in all four blastomeres at the four-cell stage. During stage 3, CapI-sna1 is expressed along the blastopore (b, arrow) and after gastrulation is complete (c), in the developing brain and deep within the center of the embryo (arrow). d, e At stage 4, CapI-sna1 transcript is detected in the developing foregut (arrowheads), laterally in stripes that extend along the entire mid-body (d, arrows), and in the brain (e). f–i At stage 6, CapI-sna1 expression persists in the brain (g–i), foregut (arrowheads in h and i), and lateral mesodermal stripes (arrows in f, g, and h). Additionally, expression in the ventral nerve cord is detected flanking the ventral midline (arrowheads in f, h, and i). j At stage 8, CapI-sna1 exhibits general mesoderm expression and is also expressed in the ganglia of the VNC (arrows point to three of the ganglia), the position of which shifts ventrally at the thoracic/abdominal boundary (dashed line). CapI-sna1 is expressed in the posterior growth zone (arrowhead) at stage 8 (k) and stage 9 (l). Subpanel m is a close-up view of the posterior end of the larva shown in l illustrating expression in both mesoderm and ectoderm of the posterior growth zone (arrows)
After gastrulation at stage 4, CapI-sna1 is detected in bilateral stripes that extend through the mid-body and correlate with the position of the lateral growth zones (Fig. 4d, arrows); these stripes are located below the surface ectoderm and likely represent segmental mesoderm. CapI-sna1 expression persists in the lateral mesoderm through the end of stage 6 (Fig. 4f–i, arrows) and is downregulated in this domain during stage 7. Expression of CapI-sna1 in the lateral mesoderm (stages 4–6) partially overlaps with the longitudinal bands of CapI-twt1 and CapI-twt2, but the patterns are not identical. At stages 8 and 9, CapI-sna1 is expressed in both the mesoderm and ectoderm of the posterior growth zone and at lower levels in the mid-body and head mesoderm (Fig. 4j–m).
After gastrulation, the most prominent expression of CapI-sna1 is in the developing CNS. CNS development in Capitella sp. I has been described previously and is referenced in Fig. 1. The brain develops as paired cerebral ganglia in the head and is derived from neural precursors in the anterior ectoderm (Fig. 1f–h). The VNC develops in the segmental mid-body; ganglia of the VNC are derived from neural precursors located on either side of the ventral midline (Fig. 1f–h). CapI-sna1 transcript is detected in the brain very early in its development, before significant thickening of the neuroectoderm (Fig. 4c). Over time, the maturation of the pattern in the anterior neural ectoderm mimics the progression of brain development (Fig. 4e,g,h,i). At stage 4, there is increased thickening of the anterior neural ectoderm, and CapI-sna1 expression in this region likewise expands (Fig. 4e). At stage 6, internalization of neural precursor cells is complete, and the brain is now segregated from the non-neural ectoderm (see Fig. 3h). CapI-sna1 brain expression persists at stage 6 and is notably absent from the overlying non-neural ectoderm (Fig. 4g–i). Also at stage 6, CapI-sna1 expression is detected on either side of the midline in the VNC (Fig. 4f,h, and i, arrowheads). At stage 8, expression in the VNC has matured into discrete patches within each segment that correlate with the position of the VNC ganglia (Fig. 4j).
CapI-sna1 also has an expression domain in the foregut. Expression in the foregut is first detected at stage 4, lateral to the stomodeum (Fig. 4d,e, arrowheads) and persists through stage 6 (Fig. 4h,i, arrowheads). The CapI-sna1 foregut expression is present within the foregut epithelium.
CapI-sna2 expression was also examined and is detected almost exclusively in the CNS; the only exception is a brief period of expression in the foregut epithelium at stages 6 and 7 (Fig. 5f,g, arrowheads). In contrast to CapI-sna1, the CapI-sna2 transcript is not detected before larval stages. Expression in the brain is initially detected at stage 4 (Fig. 5b). At stage 5, brain expression diminishes (Fig. 5c,d); a second transient wave of CapI-sna2 expression in the brain is detected at stage 6 (Fig. 5e,f). Brain expression is again downregulated by the beginning of stage 7 (Fig. 5g).
CapI-sna2 is expressed in the CNS and in the foregut. a–c, e, and h are ventral views with anterior to the left; d, f, g, and i are lateral views with ventral down and anterior to the left. Indicated in each subpanel: st. stage; asterisk stomodeum; t telotroch; br brain. CapI-sna2 transcript is first detected at stage 4 in two to three bilateral patches flanking the ventral midline (a) and at the anterior edge of the larva in the developing brain (b). Subpanel b is a deeper focal plane of the same larva shown in a. c, d At stage 5, CapI-sna2 is expressed in the developing VNC in segmentally iterated patches of cells that flank the ventral midline and extend ∼three quarters of the way to the telotroch. e, f At stage 6, expression in the VNC extends posteriorly to the telotroch and is diminished at the anterior end (arrows). Expression is also detected in the brain and the developing foregut (f, arrowhead). Similar domains of expression are detected at stage 7 (g); however, the ventral nerve cord expression is limited to a small patch just anterior to the telotroch (g and h, arrows). i At stage 9, CapI-sna2 expression is maintained only at the posterior end of the ventral nerve cord (arrows)
CapI-sna2 is also expressed in the developing VNC. At stage 4, CapI-sna2 is expressed in several patches of cells arranged in two rows flanking the ventral midline in the anterior mid-body (Fig. 5a, arrows). Based on the position and timing of this expression, we interpret it to be in precursors of the VNC (see Fig. 1f–h for details and references). During stage 5, expression in the VNC expands posteriorly ∼three fourths of the way to the telotroch (Fig. 5c,d, arrows). Expression in the VNC extends all the way to the telotroch at stage 6 and is downregulated in the anterior segments (Fig. 5e,f, arrows). By stage 7, when the axon tracts of the VNC are well formed, CapI-sna2 is only detected in the posterior VNC (Fig. 5g,h, arrows). CapI-sna2 expression is reduced after stage 7, and by the end of larval development (stage 9), transcript is only detected in the VNC ganglia of the posterior most ∼4–5 segments (Fig. 5i).
Discussion
Twist genes and mesoderm development in Capitella sp. I
In accordance with previously described ancestral functions for twist genes in the Bilateria (Technau and Scholz 2003), our data point towards a role for the Capitella sp. I twist genes in regulating mesoderm differentiation. Within the segmental region, CapI-twt1 is expressed in a discrete subset of mesoderm cells: bilateral bands of cells at stages 3–6 and three lateral-longitudinal rows of cells along each side of the larva at stage 7 (Fig. 2). CapI-twt2 is expressed broadly throughout the segmental mesoderm, and highest levels of expression are detected in the same subset of cells that also express CapI-twt1 (Fig. 3). We suggest that CapI-twt2 may have a specific function in these high-expressing cells in addition to a general function throughout the segmental mesoderm. There are no obvious morphological features within the segments that correspond to these twist expression domains, and we predict that the expression patterns mark precursors to a specific subset of mesoderm derivatives. Major derivatives of polychaete segmental mesoderm include muscle, coelomic mesoderm, nephridia, and gonads (Ruppert and Barnes 1994). In Capitella sp. I, nephridia and gonads are limited to a subset of mid-trunk segments (Eckelbarger et al. 1984; and E. Seaver, unpublished observations). Because the CapI-twt1 and CapI-twt2 expression patterns are segmentally iterated along the entire A/P-axis, these genes are unlikely to regulate nephridia or gonad development. Thus, we postulate that CapI-twt1 and CapI-twt2 may influence the development of the coelomic mesoderm or late-forming larval muscle (Seaver et al. 2005) or both. Expression domains for CapI-twt1 and CapI-twt2 in the head implicate these genes in the development of the muscle anterior to the brain.
CapI-twt1 and CapI-twt2 are not expressed during cleavage stages when mesoderm precursors are specified, indicating that these genes do not regulate the initial specification of mesoderm cell lineages. This feature distinguishes mesoderm development in Capitella sp. I from the mesoderm development in Drosophila and Tribolium where, in addition to regulating mesoderm differentiation after gastrulation, twist genes are required for specifying the mesoderm precursor territory during blastula stages (Handel et al. 2005; Leptin 1991; Simpson 1983; Sommer and Tautz 1994; Thisse et al. 1988). This molecular difference may be associated with the different cellular mechanisms of early development between Capitella sp. I and insects. In insects, mesoderm is defined by positional cues in the syncytial blastula before cellularization. In contrast, Capitella sp. I (Eisig 1899), the amphipod Parhyale hawaiensis (Gerberding et al. 2002), and many other crustaceans (Dohle et al. 2004) develop via stereotypic holoblastic cell cleavage programs whereby mesoderm precursors can be identified based on stereotyped cell lineages. Recently, a study in Parhyale found ph-twist expression in a subset of segmental mesoderm during mesoderm differentiation but not during early cleavage stages when mesoderm cell lineages first arise (Price and Patel 2007). The authors relate the difference in the timing of twist expression in Parhyale vs Drosophila to the distinct mechanisms of mesoderm development in these two groups and suggest that conserved mesoderm regulatory genes may be deployed with different timing in animals with different mechanisms of mesoderm cell fate specification (Price and Patel 2007). In this regard, we might find more similarities between the genetic regulation of mesoderm development in Capitella sp. I and animals that develop with stereotyped cell lineages than between Capitella sp. I and Drosophila.
Mesoderm development and the expression of Capitella sp. I snail genes
In the context of mesoderm development in bilaterians, the function of snail genes has been attributed to regulating morphogenetic movements of mesoderm precursors during gastrulation (reviewed in Nieto 2002). However, there are currently no data indicating that snail genes have a comparable function specific to the mesoderm during gastrulation in lophotrochozoans. In the mollusk, P. vulgata, snail genes are expressed almost exclusively in ectoderm (Lespinet et al. 2002). Among annelids, snail genes are expressed before and during gastrulation, but expression is not limited to mesodermal lineages. Snail proteins in H. robusta (Goldstein et al. 2001) are detected in the precursors of multiple tissue layers before gastrulation, and in this study, we show CapI-sna1 expression in multiple tissue layers before gastrulation and in the cells around the closing blastopore at late gastrula stages (Fig. 4). Thus, among the lophotrochozoans studied to date, it appears that snail genes might regulate cell movements during gastrulation, but this function is not likely exclusive to the mesoderm. After gastrulation, H. robusta Snail protein and CapI-sna1 messenger RNA transcripts are detected in a subset of segmental mesoderm, indicating a putative function for annelid snail genes in mesoderm differentiation. However, this putative late mesodermal function for annelid snail genes contrasts to the function for snail genes in the mesoderm during gastrulation in ecdysozoans and deuterostomes.
Expression of Capitella sp. I snail genes in the developing CNS
Both of the Capitella sp. I snail genes are expressed prominently in the CNS during early stages of neurogenesis at stages when neural precursors are internalized from the epithelial ectoderm (Figs. 4 and 5). Although CNS development in Capitella sp. I is not yet described in terms of individual cell behaviors, any process whereby cells leave an epithelium and become internalized must involve cell migration, changes in cell adhesive properties, and/or a regulated asymmetric cell division (e.g., delamination), all of which are cell behaviors known to be regulated by snail genes (reviewed in Barrallo-Gimeno and Nieto 2005). Among arthropods, snail genes are expressed in and required for the internalization of neuroblasts in Drosophila (Ashraf et al. 1999; Ashraf and Ip 2001; Cai et al. 2001), and expression patterns for Snail family genes in Glomeris marginata (myriapod; Pioro and Stollewerk 2006) and Cupiennius salei (chelicerate; Weller and Tautz 2003) indicate that this function is conserved among diverse arthropod groups. Our data suggest a similar function for snail genes in a lophotrochozoan, but it is not clear whether this represents an evolutionarily conserved trait between these groups. In P. vulgata, Pv-sna2 is expressed in a sensory structure associated with the apical tuft implying involvement in the neural differentiation of this structure (Lespinet et al. 2002). However, Snail protein expression in H. robusta does not support a role for Hro-snail in leech neurogenesis (Goldstein et al. 2001). Studies in additional representative lophotrochozoans are needed to determine whether or not the function of snail genes in CNS development is a shared trait between arthropods and lophotrochozoans.
References
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105
Alberga A, Boulay JL, Kempe E, Dennefeld C, Haenlin M (1991) The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development 111:983–992
Anderson DT (1973) Embryology and phylogeny in Annelids and Arthropods. Pergamon, Oxford
Ashraf SI, Ip YT (2001) The Snail protein family regulates neuroblast expression of inscuteable and string, genes involved in asymmetry and cell division in Drosophila. Development 128:4757–4767
Ashraf SI, Hu X, Roote J, Ip YT (1999) The mesoderm determinant snail collaborates with related zinc-finger proteins to control Drosophila neurogenesis. EMBO J 18:6426–6438
Barrallo-Gimeno A, Nieto MA (2005) The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132:3151–3161
Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A (2000) The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2:84–89
Baylies MK, Bate M (1996) twist: a myogenic switch in Drosophila. Science 272:1481–1484
Bhup R, Marsden JR (1981) The development of the central nervous system in Capitella capitata (Polychaeta, Annelida). Can J Zool 60:2284–2295
Cai Y, Chia W, Yang X (2001) A family of snail-related zinc finger proteins regulates two distinct and parallel mechanisms that mediate Drosophila neuroblast asymmetric divisions. EMBO J 20:1704–1714
Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA (2000) The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2:76–83
Castanon I, Baylies MK (2002) A Twist in fate: evolutionary comparison of Twist structure and function. Gene 287:11–22
Cripps RM, Black BL, Zhao B, Lien CL, Schulz RA, Olson EN (1998) The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev 12:422–434
Dohle W, Gerberding M, Hejnol A, Scholtz G (2004) Cell lineage, segment differentiation, and gene expression in crustaceans. In: Scholtz G (ed) Evolutionary developmental biology of Crustacea. Sweets and Zeitlinger, Lisse, The Netherlands, pp 95–133
Eckelbarger KJ, Linley PA, Grassle JP (1984) Role of ovarian follicle cells in vitellogenesis and oocyte resorption in Capitella sp. I (Polychaeta). Mar Biol 79:133–144
Eisig H (1899) Zur Entwicklungsgeschichte der Capitelliden. Mittheilungen Aus der Zoologischen Station Zu Neapel 13:5–39
Frobius AC, Seaver EC (2006) Capitella sp. I homeobrain-like, the first lophotrochozoan member of a novel paired-like homeobox gene family. Gene Expr Patterns 6:985–991
Gerberding M, Browne WE, Patel NH (2002) Cell lineage analysis of the amphipod crustacean Parhyale hawaiensis reveals an early restriction of cell fates. Development 129:5789–5801
Goldstein B, Leviten MW, Weisblat DA (2001) Dorsal and snail homologs in leech development. Dev Genes Evol 211:329–337
Grassle JP, Grassle JF (1976) Sibling species in the marine polution indicator Capitella (Polychaeta). Science 192:567–569
Grau Y, Carteret C, Simpson P (1984) Mutations and chromosomal rearrangements affecting the expression of snail, a gene involved in embryonic patternin in Drosophila melanogaster. Genetics 108:347–360
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Handel K, Basal A, Fan X, Roth S (2005) Tribolium castaneum twist: gastrulation and mesoderm formation in a short-germ beetle. Dev Genes Evol 215:13–31
Harfe BD, Vaz Gomes A, Kenyon C, Liu J, Krause M, Fire A (1998) Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev 12:2623–2635
Hopwood ND, Pluck A, Gurdon JB (1989) A Xenopus mRNA related to Drosophila twist is expressed in response to induction in the mesoderm and the neural crest. Cell 59:893–903
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Langeland JA, Tomsa JM, Jackman WR Jr, Kimmel CB (1998) An amphioxus snail gene: expression in paraxial mesoderm and neural plate suggests a conserved role in patterning the chordate embryo. Dev Genes Evol 208:569–577
Leptin M (1991) twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev 5:1568–1576
Lespinet O, Nederbragt AJ, Cassan M, Dictus WJ, van Loon AE, Adoutte A (2002) Characterisation of two snail genes in the gastropod mollusc Patella vulgata. Implications for understanding the ancestral function of the snail-related genes in Bilateria. Dev Genes Evol 212:186–195
Mayor R, Essex LJ, Bennett MF, Sargent MG (1993) Distinct elements of the xsna promoter are required for mesodermal and ectodermal expression. Development 119:661–671
Nederbragt AJ, Lespinet O, van Wageningen S, van Loon AE, Adoutte A, Dictus WJ (2002) A lophotrochozoan twist gene is expressed in the ectomesoderm of the gastropod mollusk Patella vulgata. Evol Dev 4:334–343
Nieto MA (2002) The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol 3:155–166
Nieto MA, Bennett MF, Sargent MG, Wilkinson DG (1992) Cloning and developmental expression of Sna, a murine homologue of the Drosophila snail gene. Development 116:227–237
Nieto MA, Sargent MG, Wilkinson DG, Cooke J (1994) Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science 264:835–839
Nusslein-Volhard C, Weischaus E, Kluding H (1984) Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. I. Zygotic loci on the second chromosome. Wilheim Roux Arch Dev Biol 193:267–282
Perez-Pomares JM, Munoz-Chapuli R (2002) Epithelial-mesenchymal transitions: a mesodermal cell strategy for evolutive innovation in Metazoans. Anat Rec 268:343–351
Pioro HL, Stollewerk A (2006) The expression pattern of genes involved in early neurogenesis suggests distinct and conserved functions in the diplopod Glomeris marginata. Dev Genes Evol 216:417–430
Price AL, Patel NH (2007) Investigating divergent mechanisms of mesoderm development in arthropods: the expression of Ph-twist and Ph-mef2 in Parhyale hawaiensis. J Exp Zoolog B Mol Dev Evol (in press)
Ruppert E, Barnes R (1994) Invertebrate zoology. Thomson learning, Belmont
Sargent MG, Bennett MF (1990) Identification in Xenopus of a structural homologue of the Drosophila gene snail. Development 109:967–973
Saulnier-Michel C (1992) Polychaeta: Digestive system. Wiley-Liss, New York
Seaver EC, Kaneshige LM (2006) Expression of ‘segmentation’ genes during larval and juvenile development in the polychaetes Capitella sp. I and H. elegans. Dev Biol 289:179–194
Seaver EC, Paulson DA, Irvine SQ, Martindale MQ (2001) The spatial and temporal expression of Ch-en, the engrailed gene in the polychaete Chaetopterus, does not support a role in body axis segmentation. Dev Biol 236:195–209
Seaver EC, Thamm K, Hill SD (2005) Growth patterns during segmentation in the two polychaete annelids, Capitella sp. I and Hydroides elegans: comparisons at distinct life history stages. Evol Dev 7:312–326
Simpson P (1983) Maternal-zygotic gene interactions during formation of the dorsoventral pattern in Drosophila embryos. Genetics 105:615–632
Smith DE, Franco del Amo F, Gridley T (1992) Isolation of Sna, a mouse gene homologous to the Drosophila genes snail and escargot: its expression pattern suggests multiple roles during postimplantation development. Development 116:1033–1039
Sommer RJ, Tautz D (1994) Expression patterns of twist and snail in Tribolium (Coleoptera) suggest a homologous formation of mesoderm in long and short germ band insects. Dev Genet 15:32–37
Swofford DL (2000) PAUP*: Phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland, MA
Tavares AT, Izpisuja-Belmonte JC, Rodriguez-Leon J (2001) Developmental expression of chick twist and its regulation during limb patterning. Int J Dev Biol 45:707–713
Technau U, Scholz CB (2003) Origin and evolution of endoderm and mesoderm. Int J Dev Biol 47:531–539
Thisse B, Stoetzel C, Gorostiza-Thisse C, Perrin-Schmitt F (1988) Sequence of the twist gene and nuclear localization of its protein in endomesodermal cells of early Drosophila embryos. EMBO J 7:2175–2183
Thisse C, Thisse B, Schilling TF, Postlethwait JH (1993) Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119:1203–1215
Thisse C, Thisse B, Postlethwait JH (1995) Expression of snail2, a second member of the zebrafish snail family, in cephalic mesendoderm and presumptive neural crest of wild-type and spadetail mutant embryos. Dev Biol 172:86–99
Wada S, Saiga H (1999) Cloning and embryonic expression of Hrsna, a snail family gene of the ascidian Halocynthia roretzi: implication in the origins of mechanisms for mesoderm specification and body axis formation in chordates. Dev Growth Differ 41:9–18
Wang SM, Coljee VW, Pignolo RJ, Rotenberg MO, Cristofalo VJ, Sierra F (1997) Cloning of the human twist gene: its expression is retained in adult mesodermally derived tissues. Gene 187:83–92
Weller M, Tautz D (2003) Prospero and Snail expression during spider neurogenesis. Dev Genes Evol 213:554–566
Werbrock AH, Meiklejohn DA, Sainz A, Iwasa JH, Savage RM (2001) A polychaete hunchback ortholog. Dev Biol 235:476–488
Wolf C, Thisse C, Stoetzel C, Thisse B, Gerlinger P, Perrin-Schmitt F (1991) The M-twist gene of Mus is expressed in subsets of mesodermal cells and is closely related to the Xenopus X-twi and the Drosophila twist genes. Dev Biol 143:363–373
Yasui K, Zhang SC, Uemura M, Aizawa S, Ueki T (1998) Expression of a twist-related gene, Bbtwist, during the development of a lancelet species and its relation to cephalochordate anterior structures. Dev Biol 195:49–59
Acknowledgements
The Capitella sp. I genomic and EST sequence data were produced by the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/). We thank lab members Néva Meyer and Michael Boyle for sharing unpublished data and commenting on this manuscript, and Lori Kaneshige for assistance with preliminary in situ experiments. We also thank David Matus for assistance with gene orthology analyses and two anonymous reviewers for insightful comments. This work was supported by NSF grants IOB05-44869 and EF0531558.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D.A. Weisblat
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
(A) Predicted amino acid sequences of the basic helix-loop-helix domains and WR motifs of CapI-Twt1 and CapI-Twt2 aligned with the corresponding domains of Twist homologs from other species. Consensus residues (as defined in Castanon and Baylies, 2002) for the ‘twist type’ basic helix-loop-helix domain and for the WR motif are shown in the bottom line of the alignment. (B) Predicted amino acid sequences of the 5 zinc-finger domains of CapI-Sna1 and CapI-Sna2 and the SNAG domain of CapI-Sna1 aligned with the corresponding domains of Snail homologs from other species. The first zinc-finger domain is not present in the human protein (Hs-sna) and the SNAG domain is not present in CapI-Sna2. Snail consensus residues in the zinc-finger sequences are shown in the bottom line of the alignment, shaded residues in CapI-Sna1 and CapI-Sna2 sequences indicate residues that differ from the conserved sequence. In each alignment, amino acid identities are indicated as dashes. Species abbreviations are as follows: CapI, Capitella sp. I; Pv, Patella vulgata; Dm, Drosophila melanogaster; Xl, Xenopus laevis; Hs, Homo sapiens (GIF 14 kb)
Fig. S2
Maximum likelihood consensus tree of the basic-helix-loop-helix domains of Twist family proteins from Capitella sp. I and other species. Also included are sequences of related basic-helix-loop-helix proteins, Paraxis and Hairy. Support values calculated from the maximum likelihood analysis are above the branches; values below 50 are not included. Species abbreviations are as follows: CapI, Capitella sp. I; Xl, Xenopus laevis; Ec, Enchytraeus coronatus (oligochaete) ; Am, Apis Mellifera (honey bee); Dm, Drosophila melanogaster; Io, Ilynassa obsoleta (snail); HRO, Helobdella robusta (leech); Tt, Transennella tantilla (bivalve); Dr, Danio rerio; Pv, Patella vulgata (limpet); Gg, Gallus gallus; Hs, Homo sapiens; Mm, Mus musculus; Tc, Tribolium castaneum (beetle); Nv, Nematostella vectensis (sea anemone); Pc, Podocoryne carnea, (jellyfish) (GIF 16 kb)
Fig. S3
Bayesian inference consensus tree of Snail and Slug family members from Capitella sp. I and other representative species. Sequences of Kruppel, a related zinc-finger protein, are included as an outgroup. Posterior probabilities from Bayesian analysis are reported above the branches and support values calculated from Maximum Likelihood analysis are below the branches; values below 50 are not included. Capitella sp. I genes are indicated with shaded boxes. Among the Snail family, lophotrochozoan genes are in purple and vertebrate genes are in green; the Scratch family is in blue. The two Capitella sp. I scratch genes included in the orthology analyses were isolated by degenerate PCR and RACE during our initial attempts at isolating snail fragments (Thamm and Seaver, unpublished). Species abbreviations are as follows: Hs, Homo sapiens; Mm, Mus musculus; Dr, Danio rerio; Gg, Gallus gallus; Xl, Xenopus laevis; CapI, Capitella sp. I; Pv, Patella vulgata (limpet); Nv, Nematostella vectensis (sea anemone); Am, Acropora millepora (coral); Pc, Podocoryne carnea, (jellyfish); HRO, Helobdella robusta (leech); Bf, Branchiostoma floridae (amphioxus); At, Achaearanea tepidariorum (spider); Dm, Drosophila melanogaster; Tc, Tribolium castaneum (beetle); Lf, Lithobius forficatus (centipede); Io, Ilynassa obsoleta (snail) (GIF 15 kb)
Rights and permissions
About this article
Cite this article
Dill, K.K., Thamm, K. & Seaver, E.C. Characterization of twist and snail gene expression during mesoderm and nervous system development in the polychaete annelid Capitella sp. I. Dev Genes Evol 217, 435–447 (2007). https://doi.org/10.1007/s00427-007-0153-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-007-0153-4