Abstract
The basic helix-loop-helix transcription factor twist plays a key role during mesoderm development in Bilateria. In this study, we identified a twist ortholog in the polychaete annelid Platynereis dumerilii and analyze its expression during larval development, postlarval growth up to the adult stage, and caudal regeneration after amputation of posterior segments. At late larval stages, Pdu-twist is expressed in the mesodermal anlagen and in developing muscles. During adulthood and caudal regeneration, Pdu-twist is expressed in the posterior growth zone, in mesodermal cells within the newly forming segments and budding parapodia. Our results indicate that Pdu-twist is involved in mesoderm formation during larval development, posterior growth, and caudal regeneration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The mesoderm is a systematic characteristic of the Bilateria. It has been assumed that invention of the mesoderm facilitated the diversification of bilaterian body plan morphologies (Perez-Pomares and Munoz-Chapuli 2002). Several studies examining mesoderm development in species of the three major bilaterian groups, i.e., Deuterostomia, Ecdysozoa, and Lophotrochozoa, uncovered similar mechanisms of mesoderm formation (Technau and Scholz 2003). In the polychaete annelid Platynereis dumerilii, the mesoderm arises from cells that invaginate through the blastopore and accumulate between ectoderm and endoderm. During further development, these cells arrange into mesodermal packages in the three simultaneously formed anteriormost segments (Dorresteijn 1990). Starting from the larval nectochaete stage, segments are added continuously from a posterior subterminal proliferation zone throughout the entire life-span (Fischer and Dorresteijn 2004).
On the molecular level, a conserved regulator of mesoderm development is the gene twist (Technau and Scholz 2003). The basic helix-loop-helix (bHLH) transcription factor has been shown to be essential for mesoderm induction and gastrulation in Drosophila (Simpson 1983; Thisse et al. 1988; Leptin et al. 1992). Drosophila Twist is expressed on the ventral side of the blastoderm and activates genes that are required for mesoderm development (Leptin 1991; Baylies and Bate 1996; Castanon and Baylies 2002). In the Drosophila embryo, a subset of myoblasts, the so-called adult muscle progenitor cells (AMPs) continues to express Twist whereas differentiating larval muscles lose Twist expression (Currie and Bate 1991; Bate et al. 1991; Cripps and Olsen 1998; Castanon and Baylies 2002). During metamorphosis, the AMPs adopt their differentiation fate and contribute to the formation of adult muscles that lose Twist expression during further myogenesis (Fernandes et al. 1991).
A twist ortholog is also essential for the specification and invagination of the mesoderm in the short germ beetle Tribolium where it is additionally expressed in a posterior growth zone during the developing germ band (Sommer and Tautz 1994; Handel et al. 2005). In the nematode Caenorhabditis elegans, twist activity revealed by reporter fusions and antibody staining was not observed during early embryonic development. However, a twist reporter was highly active in the postembryonic single blast cell (M) and all undifferentiated descendants of M that lose twist activity upon differentiation into body wall and sex muscles (Harfe et al. 1998). Recently, the existence of two potential twist paralogs (CapI-twi1 and CapI-twi2) was reported in the polychaete Capitella sp. I. CapI-twi1 is a zygotic gene expressed during larval development within the mid-body mesoderm, in cells of the head mesoderm, in the stomodaeum, in regions of the developing foregut, as well as in the hindgut. CapI-twi2 shows a broad expression in the developing mesoderm where it partially overlaps with CapI-twi1 (Dill et al. 2007).
In contrast to invertebrate development, Twist is not involved in chick and mouse gastrulation (Wolf et al. 1991; Tavares et al. 2001; Technau and Scholz 2003). Moreover, while high Twist levels promote somatic myogenesis in the Drosophila embryo (Baylies and Bate 1996), Twist is a negative regulator of vertebrate myogenesis that blocks the activity of myogenic bHLH and MEF2 members and is therefore excluded from the developing myotome (Spicer et al. 1996; Technau and Scholz 2003). These results indicate significant differences in Twist function between vertebrates and insects (Technau and Scholz 2003). Controversially, Anant et al. (1998) reported that the persistent expression of twist inhibits the differentiation of indirect but not direct flight muscles in Drosophila. These results indicate that Twist functions may not only differ between taxa but also within a single organism, depending on developmental- and tissue-specific contexts. The identification of twist orthologs from further species and the analysis of their spatiotemporal expression during development could therefore elucidate our understanding of twist gene function.
In this study, we report the identification of a twist ortholog (referred to as Pdu-twist) in the marine annelid Platynereis dumerilii and utilize Pdu-twist gene expression to follow mesoderm formation during larval development, growth up to adulthood, and regeneration of the posterior body segments. Our findings imply differences between larval mesoderm development and the formation of mesodermal tissues from a posterior growth zone during adulthood and regeneration revealing alternative segmentation modes during the animal’s life cycle.
Material and methods
Platynereis dumerilii culture
Standard Platynereis dumerilii culture methods were followed (Hauenschild and Fischer 1969). Regenerates were produced by amputation of posterior ends from atoke specimens (for a detailed protocol, see Pfeifer et al. 2012).
Cloning of partial and full-length Pdu-twist cDNAs
We isolated a fragment of Pdu-twist by semi-nested PCR after reverse transcription of mRNA derived from 48 h embryos with the Omniscript RT Kit (Qiagen). The following degenerate primers for Pdu-twist were used: forward GCC AAC GTC CGC GA(AG) (AC)G(AGCT) CA(AG) (AC)G, nested forward CCT TCG CCC CGC TG(AC) G(AGCT)A A(AG)A T(ACT)(AG) T, and reverse TCC ATG CGC CAC ACG (GC)(AT)(AG) AA(AGCT) GC(AG)TA. A PCR product of the expected size was gel purified using QIAquick Gel Extraction Kit (Qiagen) subcloned into pCRII-TOPO vector (Invitrogen) and sequenced (Seqlab).
Full-length cDNA clones were obtained by 5′ and 3′ rapid amplification of cDNA ends (RACE) on an embryonic 48-h cDNA library using sequence-specific primers for Pdu-twist (forward ACG TGC GCG AGC GAC AGA GTA CAC and reverse GCC ACG GTA ACT GCA GGA CGG GGG AA) and the primers supplied with the GeneRacer-Kit (Invitrogen). The resulting fragments were gel purified using QIAquick Gel Extraction Kit (Qiagen), subcloned into pCRII-TOPO vector (Invitrogen), and sequenced (Seqlab).
The complete cDNA sequence of Pdu-twist will be submitted to the NCBI (Acc. Nr. KF241717) nucleotide sequence database.
Gene orthology assessment
Protein sequences of a selected number of species were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov) and aligned using CLUSTALX (Thompson et al. 1997). This alignment spanning the conserved bHLH domain was used to calculate a 1,000-fold bootstrapped phylogenetic tree using the Neighbor-Joining method (Saitou and Nei 1987), excluding all positions with gaps in the alignment and correcting for multiple substitutions, using the program Tree-puzzle 5.2 (Schmidt et al. 2002).
Preparation and fixation of larvae and animal tissues
Larvae, regenerates, and posterior ends were fixed overnight in 3.7 % formaldehyde in phosphate-buffered saline (PBS, pH 9.5) at 4 °C. After several washes in PBS containing 0.1 % Tween-20 (PTw), specimens were rinsed twice with methanol and stored at −25 °C for later in situ hybridization. After rehydration in a descending series of 5-min methanol/PTw washes, regenerates and posterior ends were digested for 10 min with 10 μg/ml proteinase K in PTw, while larvae were digested 5 min. Digestion was stopped by washing the samples with 2 mg/ml glycine in PTw for 2 × 5 min.
In situ hybridization
In situ hybridization was performed according to Pfeifer et al. (2012). For probe synthesis, the 632-bp 5′ RACE fragment of Pdu-twist, encoding the entire bHLH-domain, was subcloned into the pCRII-TOPO vector (Invitrogen). Probes were generated using the Sp6- and T7-Megascript Kits (Ambion/Lifescience) and applied to the samples in a final working concentration of 2 ng/μl. Before mounting in Euparal (Carl Roth), samples were dehydrated in an increasing ethanol/water (v/v) series (30, 50, 70, 80, 90, 96, 98, and 100 %).
Results
Cloning of a Platynereis twist ortholog
A Platynereis twist fragment of 243-bp length spanning the bHLH domain was isolated by RT-PCR with degenerated primers derived from conserved regions in the bHLH domain. The fragment was used to design sequence-specific primers for 5′ and 3′ RACE. Due to RLM-RACE PCR, we isolated a fragment of 632-bp length in the 5′ RACE reaction and a fragment of 1,292-bp length in the 3′ RACE reaction. With the sequence information obtained from the RACE, we built a contiguous nucleotide sequence of 1,895 bp representing the complete cDNA of Pdu-twist and, as proven by PCR on genomic DNA (data not shown), also the complete genomic sequence of Pdu-twist. Analysis of this sequence revealed a potential open reading frame of 666 bp, followed by a 1.1-kb-long 3′-UTR which is often found in genes encoding the bHLH domain (Spring et al. 2000). The open reading frame encodes a polypeptide of 222 amino acids, which shares a high similarity to other known members of the Twist family (Fig. 1). The deduced sequence of Platynereis Twist contains a basic DNA binding and dimerization motif common to bHLH proteins and a C-terminal motif of 14 amino acid residues (WR motif) characteristic for most members of the Twist subfamily (Fig. 1), but absent from all less related bHLH proteins (Spring et al. 2000).
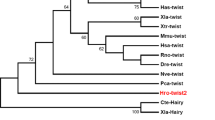
Phylogenetic analysis of Platynereis dumerilii Twist. a Proposed domain architecture of Pdu-Twist. b Sequence alignment of Platynereis Twist with Twist proteins from other species and Twist2-Hs (Dermo1-Hs) in the regions of the bHLH domain and the WR motif. Members of the Lophotrochozoa and vertebrates show the highest degree of conservation. c Phylogenetic tree of Twist-like bHLH and related transcription factors reconstructed with the Neighbor-Joining method. Sequences were taken from the NCBI database. Bootstrap values result from 1,000 replicates. All branches with a bootstrap value <50 % were collapsed. Pdu-Twist clusters with other known Twist homologs and this cluster is clearly separated from the clusters built by other bHLH domain containing proteins. Abbreviations: Pd, Platynereis dumerilii; Ec, Enchytraeus coronatus; Hr, Helobdella robusta; Tt, Transenella tantilla; Ha, Helix aspers; Pv, Patella vulgata; Xl, Xenopus laevis; Dm, Drosophila melanogaster; Mm, Mus musculus; Hs, Homo sapiens; Ce, Caenorhabditis elegans; Pc, Podocoryne carnea
Platynereis Twist (Pdu-Twist) clusters with other members of the Twist-like family of transcription factors
The analysis of the aligned sequences (Fig. 1b) revealed that the Pdu-Twist bHLH domain shows the highest score of similarity to the lophotrochozoan (90–94 %) and vertebrate (89–90 %) members of the Twist family. The Twist ortholog of the ecdysozoan Caenorhabditis elegans shares only a 62 % sequence identity which is just slightly higher than the score for mouse Scleraxis (59 %). However, inside, the Twist cluster Pdu-Twist groups together with Hr-Twist (Soto et al. 1997) from another unequal spiral cleaving annelid. The sequence of the WR motif (ERLSAYSVRMEG) is equivalent to the sequence found in almost all other Twist orthologs (Fig. 1b). Phylogenetic analysis was performed with amino acid sequences from the bHLH region of Pdu-Twist, known members of the Twist family and other bHLH factors using the Neighbor-Joining algorithm. In the resulting tree (Fig. 1c), Pdu-Twist forms a cluster with the proteins encoded by twist and its orthologs. Other proteins containing a bHLH domain formed distinct groups that are clearly separated from the Twist cluster. We therefore suggest that Platynereis Twist is a real member of the Twist family of bHLH transcription factors.
Pdu-twist expression during larval development
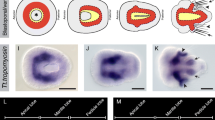
To examine the expression pattern of Pdu-twist, we performed in situ hybridization using a ribo-probe generated from a 632-bp 5′ RACE fragment of Pdu-twist. The expression of the Pdu-twist mRNA was examined from 24 h post-fertilization (hpf) up to 120 hpf (Fig. 2). Analysis with differential interference contrast microscopy reveals that Pdu-twist expressing cells are located directly between ectodermal and endodermal cells in the 24 hpf larva (Fig. 2 (a, a′, b)). At this time, expression is detectable in three bilateral domains along the anterior–posterior axis (Fig. 2 (a, a′, b)) while another expression domain can be observed in the position of the presumptive stomodaeum (Fig. 2 (a, b, arrowheads)). In addition, two small Pdu-twist expressing patches can be observed on both sides of the 24 hpf larva (Fig. 2 (a, a′, arrows)). According to their location in the episphere, it is most convincing that these cells refer to the transitory larval head kidneys (Hasse et al. 2010). During further development, Pdu-twist-positive cells of the three bilateral domains expand around the ventral and dorsal chaetal sacs and give rise to the trunk musculature (Fig. 2 (c–i)), while the stomodeal expression increases in intensity and labels the developing pharyngeal muscles (Fig. 2 (c, e, arrowheads, i, g)). During nectochaete stages, Pdu-twist expression of the trunk musculature is slightly decreased (Fig. 2 (i–j′)) compared to earlier stages whereas a robust expression can be observed within the pharyngeal musculature including the pharyngeal retractors (Fig. 2 (i, arrows)). Expression within the larval head kidneys is no longer detectable at this stage. We also observed Pdu-twist expression in two cells between the pygidium and the last larval segment 120 hpf (Fig. 2 (j′, red arrow)).
In situ hybridization against Pdu-twist mRNA in different larval stages of Platynereis dumerilii. Asterisks indicate the stomodaeum. a, a′ Pdu-twist expression in different focal planes of a 24-hpf larva. Arrows mark Pdu-twist expression domains that flank the stomodaeum. Pdu-twist expression in the developing stomodaeum is highlighted by arrowheads. Anterior is up, posterior is down. b Posterior view of a 24-hpf larva. Pdu-twist expression in the developing stomodaeum is highlighted by arrowheads. Ventral is down, dorsal is up. c, c′ Pdu-twist expression at 36 hpf. Arrowheads point to Pdu-twist expression in two bilateral domains around the stomodaeum. Two different focal planes are shown, anterior is up and posterior is down. d Posterior view of a 36-hpf larva. Dorsal is up, ventral is down. e, e′ Pdu-twist expression in different focal planes of a 48-hpf larva. Pdu-twist expression ambilateral of the stomodaeum is marked by arrowheads. Anterior is up, posterior is down. f Lateral Pdu-twist expression around the chaetal sacks of a 48-hpf larva. g, g′ Pdu-twist expression at 58 hpf in two different focal planes. Anterior is up and posterior is down. h Posterior Pdu-twist expression at 58 hpf. i, i′ Pdu-twist expression in different focal planes of a 72-hpf larva. Arrows indicate Pdu-twist expression in the pharyngeal retractor muscles. Anterior is to the left, posterior is to the right. j, j′ Different focal planes of the Pdu-twist expression in a 120-hpf larva. The red arrow marks Pdu-twist expression in two cells within the pygidium. Anterior is to the left, posterior is to the right. Scale bars 50 μm
Pdu-twist expression during growth, i.e., posterior addition of segments
Our analysis revealed Pdu-twist mRNA-positive cells within the simultaneously forming segments of developing larvae. In contrast to larval segment formation, new segments are consecutively formed by a posterior growth zone up to adulthood (Fischer et al. 2010). Hence, we were interested in whether the mesodermal marker gene Pdu-twist is also expressed in the posterior segmentation zone of adult worms (Fig. 3). Pdu-twist mRNA expression is detected in the posterior growth zone but also in iterated stripes in the youngest body parts suggesting first signs of segmentation (Fig. 3b–d). Within the posterior growth zone, Pdu-twist expression is detectable in lateral domains and some dorsal extensions, which might indicate the presence of defined mesodermal anlagen from which cells contribute to the formation of the new segments. No expression is detected along the ventral side (Fig. 3a) but Pdu-twist is expressed in cells within the lateral forming parapodia (Fig. 3a, f). In older segments, Pdu-twist-positive cells are restricted to the parapodial base, whereas in younger segments, Pdu-twist expression extents dorsally (Fig. 3d, f). In fully differentiated segments, we detected only faint Pdu-twist expression in cells located at the anterior parapodial base (data not shown).
Pdu-twist expression in posterior ends of Platynereis dumerilii. Posterior is to the right in all panels. a–f In situ hybridization against Pdu-twist mRNA in posterior ends. a Pdu-twist expression in a posterior end from a ventral view. d Dorsal view of a posterior end showing Pdu-twist expression. b, c, e Close-up of a posterior end with Pdu-twist expression from a dorsal view. f Close up of a. Pdu-twist expression in the anterior part of the parapodia in differentiated segments of the posterior end. Scale bars 100 μm
Taken together, these results indicate that the mesodermal tissues in each newly formed segment arise from defined anlagen within the posterior growth zone that are characterized by Pdu-twist expression.
Pdu-twist expression during different stages of caudal regeneration
A characteristic feature of Platynereis dumerilii is the ability to regenerate caudal segments after the posterior end has been amputated from the adult worm (Hauenschild and Fischer 1969). Since Pdu-twist mRNA is expressed in distinct domains within the posterior growth zone, we wondered how the cascade of events that leads to the establishment of mesodermal tissues within each segment in normogenesis is recovered after a removal of the growth zone. Therefore, we employed in situ hybridization against Pdu-twist as a marker for mesoderm development and followed its expression during the regeneration periods of 1, 2, 3, 4, 5, and 8 days post-amputation (dpa).
Already 1 day after wounding, Pdu-twist expression is detectable below an epithelial cell layer posterior to the amputation site (Fig. 4 (a, a′)). Two discrete expression domains are located laterally while one domain appears on the dorsal side of the regenerate (Fig. 4 (a, a′, arrowheads)). Two days post-amputation, the three expression domains have extended to a horseshoe-like structure leaving a ventral opening within the formed blastema (Fig. 4 (b, b′)). Compared to 1 dpa, the expression has increased in intensity. Three days post-amputation, the budding of the anal cirri can be observed at the posterior end of the regenerate (Fig. 4 (c, c′); de Rosa et al. 2005). Pdu-twist is expressed at the base of the anal cirri buds (Fig. 4 (c′-c′′′, arrowheads)) and around the anus opening, which probably refers to the prospective sphincter muscle cells (Fig. 4 (c, green arrowhead)). In addition to the horseshoe-like expression domain already observed 2 dpa, a ventral expression patch is now detectable (Fig. 4 (c′, c′′, arrow)). Four days post-amputation, the beginning of the segmentation process is revealed by stripes of Pdu-twist-positive cells in the center of the prospective segments while expression is still detectable around the anus opening (Fig. 4 (d)). At this time, single Pdu-twist-positive cells are also detectable at the base and within the anal cirri (Fig. 4 (d)). After one more day, a new pygidium has formed and the segmentation of the regenerate becomes apparent by the forming parapodial buds (Fig. 4 (e′, arrows)). The circular expression of Pdu-twist within the pygidium suggests the formation of the anus sphincter musculature (Fig. 4 (e′, green arrowhead)). Pdu-twist is also expressed in iterated stripes between the pygidium and the original segment (Fig. 4 (e and e′, arrows)) while the newly established posterior growth zone is still denoted by the horseshoe-like expression domain of Pdu-twist at this stage (Fig. 4 (e, red arrowhead)). Eight days post-amputation, up to seven segments have formed that vary in their differentiation status. While the oldest segments already bear parapodia, only lateral buds are present in the middle segments and the youngest segments show no morphological signs of parapodia formation. Interestingly, while lateral stripes of Pdu-twist-positive cells are detectable in the youngest segments (Fig. 4 (h)), Pdu-twist expression intensity decreases during parapodia development (Fig. 4 (f–g)). Pdu-twist is still expressed in the posterior subterminal proliferation zone in a horseshoe-like domain and within the sphincter musculature (Fig. 4 (h)). Taken together, our results indicate that Pdu-twist might be involved in the reestablishment of mesodermal tissues within the blastema and the subsequent formation of new segments after caudal amputation.
Pdu-twist expression during various stages of caudal regeneration in Platynereis dumerilii. Anterior is to the left and posterior is to the right in all panels. The red line marks the side of amputation. The regenerate is on the right side of the red line, the original segment is on the left side. a Arrowheads point to three Pdu-twist expression domains 1 dpa. Dorsal view. a′ Close-up of a. b Pdu-twist expression 2 dpa. Ventral view. b′ Close-up of b. c Dorsal Pdu-twist expression 3 dpa. Green arrowhead points to the circular expression domain around the anus. c′ Ventral Pdu-twist expression 3 dpa. Arrow marks Pdu-twist mRNA-positive cells in the region of the new established posterior growth zone. Arrowhead points to Pdu-twist expression on the base of the anal cirri buds. c′′ and c′′′ Close-up of c′. d Pdu-twist expression 4 dpa. e Five days post-amputation, the Pdu-twist expression is detected in a segmental iterated stripe pattern along the anterior posterior axis of the regenerate. Expression within the posterior growth zone is marked by a red arrowhead. e′ Pdu-twist expression in stripes is highlighted by arrows. Green arrowhead points to Pdu-twist expression around the anus. f, f′ Pdu-twist expression 8 dpa in different focal planes. g Pdu-twist is expressed within the newly developed parapodia. h Regenerate 8 dpa from a dorsal view. Red arrowhead marks Pdu-twist expression in the posterior growth zone. The green arrowhead points to the Pdu-twist expression around the newly formed anus. Scale bars 100 μm
Discussion
A twist ortholog is expressed during Platynereis dumerilii mesoderm development
Previous studies among a variety of bilaterian species revealed that the bHLH transcription factor Twist is essential for mesoderm development (Technau and Scholz 2003). In accordance, expression of twist within cells of mesodermal origin has been described in several lophotrochozoans (Nederbragt et al. 2002; Dill et al. 2007). In this work, we identified a potential twist ortholog in the annelid Platynereis dumerilii. Sequence analysis of Platynereis dumerilii twist (Pdu-twist) unveils the presence of the highly conserved bHLH domain and the C-terminal WR motif known from other orthologs like Drosophila Twist, the founding member of the Twist-like subfamily. A phylogenetic analysis based on amino acid identity in the bHLH domain sequences of the Twist homologs revealed between 59 and 85 % conserved residues in comparison to Drosophila Twist, whereas the bHLH domains show a higher degree of conservation within the vertebrates (Castanon and Baylies 2002). In the Neighbor-Joining tree derived from the phylogenetic analysis of the bHLH domains, the family of the Twist-like transcription factors shows a distinct systematic separation to the other families of bHLH domain containing transcription factors. The phylogenetic branch of the Twist homologs exhibits a pronounced polytomy clustering of the chordate members, the Lophotrochozoa, Ecdysozoa, and the cnidarian phyla. Only the Twist homolog of Caenorhabditis elegans displays an atypical position at the root of the phylogenetic tree, probably representing a long-branch attraction artefact of the Neighbor-Joining analysis. Platynereis twist can be found in the “Lophotrochozoan cluster” and in systematic neighborhood to the Twist homologs from Tubifex tubifex and Helobdella robusta, demonstrating that Pdu-twist represents a real member of the Twist-like family of transcription factors.
Former studies on Helobdella robusta and Patella vulgata (Soto et al. 1997; Nederbragt et al. 2002) reported the identification of only one twist ortholog, which would be in accordance to our findings in Platynereis. However, in the polychaete Capitella sp. I, the presence of two twist paralogs has been reported indicating differential numbers of twist genes within these closely related taxa (Dill et al. 2007). Since variations in the number of twist paralogs is common, even among vertebrate species (Germanguz et al. 2007), the number of twist genes in the genome might not be a valid criterion for understanding phylogenetic relationships. There are several known functions of twist in Bilateria. In the Drosophila embryo, Twist is required for the normal development of the larval musculature, whereas in vertebrates, Twist is supposed to inhibit myogenesis (Hebrok et al. 1994; Cripps and Olsen 1998; Technau and Scholz 2003). Whole mount in situ hybridization of Pdu-twist demonstrates that twist mRNA is specifically expressed in mesodermal cell packages during early larval development and within the musculature of larvae and young worms, indicating a similar function as presumed for other invertebrate embryos and larvae (Michelson 1996; Technau and Scholz 2003; Dill et al. 2007; Martín-Durán et al. 2010).
Pdu-twist expression in posterior ends during adulthood and caudal regeneration
Adult worms continuously add new segments from a subterminal posterior growth zone. Within this particular region, Pdu-twist is expressed in lateral domains with dorsal extensions, revealing a defined source for the mesoderm during posterior growth. Interestingly, while Pdu-twist mRNA is highly present in the youngest segments of adult worms, its expression decreases during further development and is absent within mature muscles and fully differentiated segments.
We also analyzed that the establishment of mesodermal tissues is recovered after removal of this posterior growth zone. According to the results presented here, not all cells joining in blastema formation are Pdu-twist mRNA positive although twist expression is already detectable in defined domains at 1 dpa. One explanation for such an early patterning might be that, in consequence of a trauma, de-differentiating cells descend from different tissues along the amputation site and contribute to blastema formation. Thus, the assumption that the blastema in Platynereis dumerilii consists of cells derived from already differentiated tissues that hold only restricted potentials for re-differentiation (Tanaka and Reddien 2011) would be more convincing than a de novo formation by adult stem cells or “neoblasts.” During the regeneration process, the blastema forms a pygidium and a posterior subterminal proliferation zone. Subsequently, the formation of new segments begins. A similar process has been described in the polychaete species Perinereis nuntia (Niwa et al. 2013). As reported by Niwa et al. (2013), the segment regeneration mode seems to be an identical but accelerated mode of the posterior segment formation process (Niwa et al. 2013). During adult segment formation in Platynereis, Pdu-twist expressing cell rows can be detected in the dorsolateral regions of the posterior ending. Comparing segment formation during the regeneration process, we detected less Pdu-twist expressing cells suggesting that the regeneration of segments in Platynereis might be an accelerated but identical mode of the normal segmentation process.
Taken together, these results reveal that we isolated a Platynereis twist ortholog, called Pdu-twist. Its expression within the developing mesoderm and forming muscles suggests a conserved function among invertebrate species.
Conclusions
In this study, we report the identification of a twist ortholog in the polychaete annelid Platynereis dumerilii and employ Pdu-twist expression to follow mesoderm formation during normal development and regeneration. The analysis of Pdu-twist expression in mesodermal cells and developing muscles at different larval stages points towards a conserved function during mesoderm development and myogenesis as postulated for other invertebrates. The continuous proliferation of segments from a posterior subterminal proliferation zone is one of the characteristic features of the Platynereis dumerilii life cycle. Our examination on mesoderm specification during this posterior segmentation process indicates the presence of defined Pdu-twist expressing anlagen within this posterior growth zone that could serve as a proliferative reservoir for the mesoderm during adulthood. Employing Pdu-twist expression as a marker, we could follow the re-establishment of the posterior subterminal proliferation zone after caudal amputation for the first time in Platynereis. Our results reveal that a distinct Pdu-twist expression pattern is already detectable within the early forming blastema. This would favor a model in which the blastema in Platynereis is composed of dedifferentiated cells that descend from tissues along the amputation site. Furthermore, segment regeneration could be accelerated but otherwise identical with the segment formation process observed during normal posterior growth. In contrast to simultaneous segment formation within the larva, the segment appearance in adults (independent of normal posterior growth or regeneration) follows a successive program.
References
Anant S, Roy S, VijayRaghavan K (1998) Twist and Notch negatively regulate adult muscle differentiation in Drosophila. Development 125:1361–1369
Bate M, Rushton E, Currie DA (1991) Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development 113:79–89
Baylies MK, Bate M (1996) twist: a myogenic switch in Drosophila. Science 272:1481–1484
Castanon I, Baylies MK (2002) A Twist in fate: evolutionary comparison of Twist structure and function. Gene 287:11–22
Cripps RM, Olson EN (1998) Twist is required for muscle template splitting during adult Drosophila myogenesis. Dev Biol 203:106–115
Currie DA, Bate M (1991) The development of adult abdominal muscles in Drosophila: myoblasts express twist and are associated with nerves. Development 113:91–102
de Rosa R, Prud'homme B, Balavoine G (2005) Caudal and even-skipped in the annelid Platynereis dumerilii and the ancestry of posterior growth. Evol Dev 7:574–587
Dill KK, Thamm K, Seaver EC (2007) Characterization of twist and snail gene expression during mesoderm and nervous system development in the polychaete annelid Capitella sp. I. Dev Genes Evol 217:435–447
Dorresteijn AWC (1990) Quantitative analysis of cellular differentiation during early embryogenesis of Platynereis dumerilii. Roux's Arch Dev Biol 199:14–30
Fernandes J, Bate M, Vijayraghavan K (1991) Development of the indirect flight muscles of Drosophila. Development 113(1):67–77
Fischer A, Dorresteijn A (2004) The polychaete Platynereis dumerilii (Annelida): a laboratory animal with spiralian cleavage, lifelong segment proliferation and a mixed benthic/pelagic life cycle. Bioessays 26:314–325
Fischer AH, Henrich T, Arendt D (2010) The normal development of Platynereis dumerilii (Nereididae, Annelida). Front Zool 7:31
Germanguz I, Lev D, Waisman T, Kim CH, Gitelman I (2007) Four twist genes in zebrafish, four expression patterns. Dev Dyn 236:2615–2626
Handel K, Basal A, Fan X, Roth S (2005) Tribolium castaneum twist: gastrulation and mesoderm formation in a short-germ beetle. Dev Genes Evol 215:13–31
Harfe BD, Vaz Gomes A, Kenyon C, Liu J, Krause M, Fire A (1998) Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev 12:2623–2635
Hasse C, Rebscher N, Reiher W, Sobjinski K, Moerschel E, Beck L, Tessmar-Raible K, Arendt D, Hassel M (2010) Three consecutive generations of nephridia occur during development of Platynereis dumerilii (Annelida, Polychaeta). Dev Dyn 239:1967–1976
Hauenschild C, Fischer A (1969) Platynereis dumerilii. Grosses Zoologisches Praktikum 10 b
Hebrok M, Wertz K, Fuchtbauer EM (1994) M-twist is an inhibitor of muscle differentiation. Dev Biol 165:537–544
Leptin M (1991) twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev 5:1568–1576
Leptin M, Casal J, Grunewald B, Reuter R (1992) Mechanisms of early Drosophila mesoderm formation. Dev Suppl:23–31
Martín-Durán JM, Amaya E, Romero R (2010) Germ layer specification and axial patterning in the embryonic development of the freshwater planarian Schmidtea polychroa. Dev Biol 340:145–158
Michelson AM (1996) A new turn (or two) for twist. Science 272:1449–1450
Nederbragt AJ, Lespinet O, van Wageningen S, van Loon AE, Adoutte A, Dictus WJ (2002) A lophotrochozoan twist gene is expressed in the ectomesoderm of the gastropod mollusk Patella vulgata. Evol Dev 4:334–343
Niwa N, Akimoto-Kato A, Sakuma M, Kuraku S, Hayashi S (2013) Homeogenetic inductive mechanism of segmentation in polychaete tail regeneration. Dev Biol. doi:10.1016/j.ydbio.2013.04.010
Perez-Pomares JM, Munoz-Chapuli R (2002) Epithelial-mesenchymal transitions: a mesodermal cell strategy for evolutive innovation in Metazoans. Anat Rec 268:343–351
Pfeifer K, Dorresteijn AW, Frobius AC (2012) Activation of Hox genes during caudal regeneration of the polychaete annelid Platynereis dumerilii. Dev Genes Evol 222:165–179
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schmidt HA, Strimmer K, Vingron M, von Haeseler A (2002) TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502–504
Simpson P (1983) Maternal-zygotic gene interactions during formation of the dorso-ventral pattern in Drosophila embryos. Genetics 105:615–632
Sommer RJ, Tautz D (1994) Expression patterns of twist and snail in Tribolium (Coleoptera) suggest a homologous formation of mesoderm in long and short germ band insects. Dev Genet 15:32–37
Soto JG, Nelson BH, Weisblat DA (1997) A leech homolog of twist: evidence for its inheritance as a maternal mRNA. Gene 199:31–37
Spicer DB, Rhee J, Cheung WL, Lassar AB (1996) Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science 272:1476–1480
Spring J, Yanze N, Middel AM, Stierwald M, Groger H, Schmid V (2000) The mesoderm specification factor twist in the life cycle of jellyfish. Dev Biol 228:363–375
Tanaka EM, Reddien PW (2011) The cellular basis for animal regeneration. Dev Cell 21:172–185
Tavares AT, Izpisuja-Belmonte JC, Rodriguez-Leon J (2001) Developmental expression of chick twist and its regulation during limb patterning. Int J Dev Biol 45:707–713
Technau U, Scholz CB (2003) Origin and evolution of endoderm and mesoderm. Int J Dev Biol 47:531–539
Thisse B, Stoetzel C, Gorostiza-Thisse C, Perrin-Schmitt F (1988) Sequence of the twist gene and nuclear localization of its protein in endomesodermal cells of early Drosophila embryos. EMBO J 7:2175–2183
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wolf C, Thisse C, Stoetzel C, Thisse B, Gerlinger P, Perrin-Schmitt F (1991) The M-twist gene of Mus is expressed in subsets of mesodermal cells and is closely related to the Xenopus X-twi and the Drosophila twist genes. Dev Biol 143:363–373
Acknowledgments
The authors would like to thank Brigitte Fronk, Renate Plaß, and Susanne Vasoldt-Kröckel for maintenance of the Platynereis dumerilii culture.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by David A Weisblat
Kathrin Pfeifer and Christoph Schaub contributed equally to this work.
Rights and permissions
About this article
Cite this article
Pfeifer, K., Schaub, C., Wolfstetter, G. et al. Identification and characterization of a twist ortholog in the polychaete annelid Platynereis dumerilii reveals mesodermal expression of Pdu-twist . Dev Genes Evol 223, 319–328 (2013). https://doi.org/10.1007/s00427-013-0448-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-013-0448-6