Abstract
The formation of abdominal appendages in insects is suppressed by the Hox genes Ultrabithorax (Ubx) and abdominal-A (abd-A), but mechanisms of the suppression can differ among species. As the function of Ubx and abd-A has been described in only a few species, more data from various insects are necessary to elucidate the evolutionary transition of regulation on abdominal appendages. We examined the function of Ubx in the silkworm Bombyx mori (Bm-Ubx) by embryonic RNA interference (RNAi). This is the first case in which functional analysis for Ubx is performed in lepidopteran insects. Larvae treated with Bm-Ubx dsRNA displayed an additional pair of thoracic leg-like protuberances in A1, whereas the other abdominal segments had no transformation. Our results suggest that Bm-Ubx is a suppressor of leg development in A1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The homeotic complex (Hox) genes are required to assign segmental identity along the anterior-posterior body axis of an embryo. In insects, the abdominal appendages are suppressed by the Hox genes Ultrabithorax (Ubx) and abdominal-A (abd-A; reviewed by Hughes and Kaufman 2002), but mechanisms of the suppression could differ among species. In Drosophila melanogaster, both Ubx and abd-A act to suppress the development of abdominal appendages by repressing the expression of Dll (Vachon et al. 1992). Although Ubx/abd-A also suppresses appendage development in the butterfly Junonia coenia and the moth Manduca sexta, the downregulation of Ubx/abd-A expression as circular holes in abdominal segments A3–A6 allow Dll to be derepressed, and, thus, prolegs are developed in these abdominal segments (Suzuki and Palopoli 2001; Warren et al. 1994). In the sawfly, prolegs did not express Dll at any time, and expressed Ubx/abd-A throughout development, so it is suggested that larval prolegs have evolved independently in Lepidoptera and Hymenoptera (Suzuki and Palopoli 2001). In the A1 appendage (pleuropodia) of the beetle Tribolium castaneum and grasshopper Schistocerca americana, and in the A1 (ventral tube), A3 (retinaculum), and A4 (furca) appendages of the springtail Folsomia candida, Dll is expressed despite high levels of Ubx/abd-A (Palopoli and Patel 1998). It is suggested that Ubx/abd-A does not repress Dll expression in certain segments in these species. In the milkweed bug Oncopeltus fasciatus, Ubx and abd-A act independently and repress Dll expression in the abdominal segments, but are redundant in D. melanogaster (Angelini et al. 2005; Castelli-Gair and Akam 1995; Vachon et al. 1992). Ubx and abd-A have not yet been identified outside the onychophora/arthropod clade, and in the onychophora and the arthropod groups except for insects, the regulatory interaction between Ubx/abd-A and Dll has not been reported (e.g., Grenier et al. 1997). The Ubx proteins of the onychophoran Akanthokara kaputensis and the crustacean Artemia fransicana that were expressed in transgenic D. melanogaster were unable to repress Dll expression or prevent the initiation of appendage development (Galant and Carroll 2002; Ronshaugen et al. 2002; reviewed by Pavlopoulos and Averof 2002). These results indicate that the regulatory interaction between Ubx/abd-A and Dll has evolved and diverged among insects (e.g., Palopoli and Patel 1998; Suzuki and Palopoli 2001; reviewed by Angelini and Kaufman 2005).
In Bombyx mori, a number of homeotic mutants with extra crescent markings and extra legs in the abdominal segments have been reported, and these mutant genes belong to the E group (reviewed by Tazima 1964). The E loci are located at position 21.1 on the sixth-linkage group (Banno et al. 1997) and include homeotic genes specifying the identities of the larval abdominal segments, the Ubx, abd-A and, possibly, Abd-B genes (Ueno et al. 1992; Yasukochi et al. 2004). For instance, larvae homozygous for E N express thoracic-type legs in A1–A7 segments and intermediate thoracic/abdominal-type legs in A8 segment (Ueno et al. 1992). As a further example related to our work, E Cw heterozygous larvae show extra crescents in A1, and E Cw homozygous larvae show not only the extra crescents but also extra thoracic legs in A1 and rudimentary thoracic leg-like protuberances in A2 (Hirokawa 1998). However, the relationships between defects of each Hox gene and mutant phenotypes are still unclear. Intriguingly, unlike D. melanogaster, E mutants of B. mori have a different transformation between the dorsal and ventral sides (Itikawa 1943; Tazima 1964).
Recently, it was reported that abd-A expression is required for proleg development in B. mori by RNAi experiments (Tomita and Kikuchi 2009; Pan et al. 2009). Although Ubx expression has been examined in various insect species (reviewed by Hughes and Kaufman 2002), functional analysis of Ubx by RNA interference (RNAi) has been limited to Tribolium (Lewis et al. 2000), Acheta (Mahfooz et al. 2007), and Oncopeltus (Angelini et al. 2005; Mahfooz et al. 2007). For evolutionary comparisons of regulation in abdominal appendages, more data from various insects groups are necessary. Here, we applied Ubx RNAi to B. mori in order to analyze the function of Ubx in a lepidopteran insect.
In the present study, we cloned full-length cDNA for the Ubx homolog from B. mori (Bm-Ubx) and analyzed the expression pattern in embryos of B. mori by whole-mount in situ hybridization. The expression of Bm-Ubx in the embryonic stages is very similar to that in the moth M. sexta (Zheng et al. 1999) and butterfly J. coenia (Warren et al. 1994). We also analyzed the function of Bm-Ubx with an embryonic RNAi experiment. Consequently, the larvae treated with Bm-Ubx dsRNA displayed an additional pair of thoracic leg-like protuberances in A1. This phenotype is very similar to that of E Cw homozygous larvae (Hirokawa 1998). Our results suggest that Bm-Ubx is a suppressor of leg development in A1. Furthermore, we show that the embryonic RNAi method is efficient in B. mori.
Materials and methods
Insects
Strain No. 459 (sex-linked black eggs) and a polyvoltine strain (N4) of the silkworm, B. mori, were used. Eggs were incubated at 25°C and the embryos were staged according to morphological markers as described by Ohtsuki (1979) and Morita et al. (2003).
Cloning
Total RNA was extracted from 48-h-old female B. mori embryos (No. 459) with TRIzol (Gibco BRL) according to the manufacturer’s instructions. The first-stranded cDNA was synthesized with a SMART polymerase chain reaction (PCR) cDNA Amplification kit (Clontech) using 1 μg total RNA. Bm-Ubx cDNA fragments were amplified by PCR with the following pair of degenerate primers corresponding to the highly conserved amino acid sequences found in the sequences of Ubx from several arthropods and an onychophora.
The degenerate primer set for Ultrabithorax (Ubx):
-
Ubx-1: 5′-CARACITAYACIMGITAYCARAC-3′ (23 mer)
-
Ubx-3: 5′-TGIGCYTGYTTYTCYTGYTCRTT-3′ (23 mer)
-
(R = A + G, M = A + C, Y = T + C, I = inosine)
PCRs were performed using 2.5 μl of the tenfold diluted first-stranded cDNA, the above pair of primers, and AmpliTaq Gold (Perkin Elmer).
To obtain full-length cDNA, 5′ RACE and 3′ RACE were performed with the following gene-specific primers and the SMART PCR cDNA Amplification kit (Clontech) according to the manufacturer’s instructions.
Gene-specific primers for 5′ RACE:
-
Bm-Ubx-3: 5′-GCGTCTCCTTCGCGTAAGGTAGTGGTTC-3′ (28 mer)
-
Bm-Ubx-4: 5′-TGATTTGCCTCTCCGTGAGGCACAACGC-3′ (28 mer)
Gene-specific primers for 3′ RACE
-
Bm-Ubx-1: 5′-GAACCACTACCTTACGCGAAGGAGACGC-3′ (28 mer)
-
Bm-Ubx-2: 5′-GCGTTGTGCCTCACGGAGAGGCAAATCA-3′ (28 mer)
The 5′ RACE and 3′ RACE were performed using 2.5 μl of the tenfold diluted first-stranded cDNA from 32-h-old male embryos, 10× Universal Primer Mix, Bm-Ubx-4 for 5′ RACE or Bm-Ubx-1 for 3′ RACE, and Advantage 2 Polymerase Mix. The nested PCRs for 5′ RACE and 3′ RACE were performed using 0.2 μl of the primary PCR product, Nested Universal Primer, Bm-Ubx-3 for 5′ RACE or Bm-Ubx-2 for 3′ RACE, and an Advantage 2 Polymerase Mix.
Sequencing and sequence analysis
The PCR product was subcloned into the EcoR V site of the pBluescript KS vector (Stratagene). Nucleotide sequence determination was performed by the dideoxy chain-termination method using an automatic DNA sequencer CEQ 2000XL (Beckman Coulter). Sequence analysis was performed using the DNASIS system (Hitachi Software Engineering). Deduced amino acid sequences were aligned with ClustalW to determine amino acid sequence identities. The DDBJ/EMBL/GenBank accession number for Bm-Ubx is AB505052.
Whole-mount in situ hybridization
Embryos (N4) between stage 16 (neural groove formation) and stage 22 (complete embryonic reversal) were dissected from their eggs in saline solution (0.75% NaCl) and fixed overnight with a chilled PLP fixative (4% paraformaldehyde, 30 mM NaPO4, 10 mM NaIO4, and 75 mM lysine, pH 6.8). After fixation, embryos were washed several times in 100% methanol and stored in 100% methanol at −20°C until use.
The in situ hybridizations were performed using standard procedures (Tomoyasu et al. 2005). Embryos were exposed to Proteinase K (10 μg/ml) for 20 min. Digoxigenin-labeled RNA probes were synthesized based on the Bm-Ubx ORF sequence (765 bp) using a DIG RNA Labeling kit (Boehringer). Embryos were subjected to hybridization with the probes in hybridization buffer at 55°C for 18 h, washed at 55°C for several hours, and treated with blocking buffer (Boehringer blocking reagent, Triton, and NGS) at room temperature for 1 h. Embryos were then stained with the alkaline phosphatase-conjugated anti-digoxigenin antibody at 4°C overnight.
Preparation of dsRNA
Two regions of Bm-Ubx cDNA were subcloned into the EcoR V site of the pBluescript KS vector, a 765 bp fragment (the ORF of Bm-Ubx cDNA), obtained using Bm-Ubx-5 (5′-ATGAACTCTTACTTCGAGCA-3′) and Bm-Ubx-8 (5′-TTAATGTTCGGGGTGTCCCT-3′), and a 445-bp fragment (encoding the N-terminus sequences to the conserved YPWM motif) was obtained using Bm-Ubx-5 and Bm-Ubx-6 (5′-ATGATTAGTAGGCTGTTGGT-3′) as primers (Fig. S1; Electronic Supplementary Material). For the production of a template for in vitro transcription, PCR was performed using plasmids containing either Bm-Ubx-ORF (the 765 bp PCR product) or Bm-Ubx-HDless (the 445-bp PCR product, which does not contain homeobox; Fig. S1; Electronic Supplementary Material), two universal primers (T7-KS, 5′-TAATACGACTCACTATAGGGAGACCACTCGAGGTCGACGGTATC-3′; T7-SK, 5′-TAATACGACTCACTATAGGGAGACCACCGCTCTAGAACTAGTGGATC-3′) containing the T7 polymerase promoter sequence at their 5′ ends, and AmpliTaq Gold (Perkin Elmer). As a negative control, DsRed ORF (678 bp) was amplified using two primers with a T7 promoter sequence at the 5′ end (T7-DsRed5, 5′-TAATACGACTCACTATAGGGAGACCACATGGTGCGCTCCTCCAAG-3′; T7-DsRed3, 5′-TAATACGACTCACTATAGGGAGACCACCTACAGGAACAGGTGGTG-3′). Sense and antisense transcripts were simultaneously synthesized using 1 μg PCR product and a MEGAscript T7 kit (Ambion) according to the manufacturer’s instructions. After DNase I treatment, RNA precipitated with LiCl was dissolved in RNase-free water. The RNA solution was heated at 65°C for 30 min and cooled slowly to room temperature for annealing of the dsRNA. The concentrations of dsRNA for Bm-Ubx-ORF, Bm-Ubx-HDless, and DsRed were 1.2 μg/µl. The quality of the dsRNA was examined by agarose gel electrophoresis and small aliquots of the dsRNA were stored at −80°C until use.
Injection of dsRNA into B. mori embryos
Injections were performed under a dissection microscope (Stemi 2000, Carl Zeiss). B. mori embryos (N4) were collected within 5 h of oviposition to perform dsRNA injection during the syncytial blastoderm stage. The dsRNA was injected ventrally into an egg using a micromanipulator (Narishige) and FemtoJet (Eppendorf). The injection into an embryo was performed with a special glass needle (uMPm-02, Natsume Optical Corporation).
Scanning electron microscopy
Dissected embryos were fixed with FAA solution (37% formaldehyde: acetic acid anhydride: 50% ethanol = 1:1:18), dehydrated in a graded ethyl alcohol series and then transferred to acetone. The embryos were dried in a critical-point dryer, coated with platinum and observed under a scanning electron microscope (S-3000N, Hitachi).
Results and discussion
Cloning of the B. mori Ubx homolog
Partial Bm-Ubx cDNA was PCR-amplified using the first-strand cDNA prepared from B. mori embryos with a pair of degenerate primers based on the highly conserved amino acid sequence. The size of the PCR product for Bm-Ubx was 160 bp. Following this, full-length cDNA for Bm-Ubx was cloned by 5′ and 3′ RACE and consisted of 1,458 bp (Fig. S1; Electronic Supplementary Material). The ORF for Bm-Ubx encoded 254 amino acid residues (Fig. S1; Electronic Supplementary Material). As shown in Fig. 1, the entire Ubx sequences of B. mori showed the highest in identity to the butterfly J. coenia Ubx (98%) when compared with those of J. coenia, the red flour beetle T. castaneum, and D. melanogaster. Bm-Ubx shares 61% identity with T. castaneum Ubx and 53% identity with D. melanogaster Ubx. Conservation was highest in homeodomain and the Ubd-A peptide, in which all of the amino acid residues are identical among Ubx sequences of these insects. A transcriptional repression domain in the C-terminal region is also highly conserved (Galant and Carroll 2002; Ronshaugen et al. 2002). Thus, sequence comparison demonstrates that the cloned B. mori cDNA is a homolog of the known Ubx.
Comparison of amino acid sequence of B. mori Ubx with those of Ubx orthologs from J. coenia, D. melanogaster, and T. castaneum. Asterisks indicate identical amino acids among the four species. The amino acids with conservative substitutions and semi-conservative substitutions based on the physiochemical criteria are marked with double dots (:) and single dots (.), respectively. Dm D. melanogaster (P83949), Jc J. coenia (AY074760), Tc T. castaneum (AY074761). Accession numbers are in parentheses
Expression of B. mori Ubx during embryogenesis
We examined the expression of Bm-Ubx transcript during embryogenesis. For embryos in stage 16 (60 h after oviposition), Bm-Ubx transcripts were detected in T3–A9 (Fig. 2a). The highest levels of staining were detected in A1. The expression in T3 and A9 were weak. No expression of Bm-Ubx was detected in the lateral regions of T3−A9. For embryos in stage 18 (80 h after oviposition), Bm-Ubx expression was detected in from the posterior half of T2 to A9 (Fig. 2b). The signal in anterior A1 was the highest level with diminishing levels in the more posterior segments. Signals in A3−A9 were lower than that in stage 16. Unlike embryos in stage 16, the signals were detected in the lateral region. For embryos in stage 20 (96 h after oviposition), the expression pattern of Bm-Ubx was the same as in stage 18 (Fig. 2c). What has to be noticed is that the signal was undetectable in the proleg primordia of A3−A6 (Fig. 2c). For embryos in stage 22 (120 h after oviposition), Bm-Ubx expression could not be detected due to nonspecific background that was probably due to cuticle deposition. No signal was observed with sense strand probes in stages 16−20 (Fig. 2d–f). Therefore, the abovementioned signals obtained by an antisense probe were specific for Bm-Ubx.
Expression pattern of Bm-Ubx transcripts in the B. mori embryo. Embryos were stained with a riboprobe (blue). Anterior is left. a Embryo in stage 16 (60 h after oviposition). Ventral view. Bm-Ubx expression is detected in T3–A9. The highest levels of staining are detected in A1. The expression in T3 and A9 is weak. No expression of Bm-Ubx is detected in the lateral regions of T3–A9. b Embryo in stage 18 (80 h after oviposition). Ventral view. Bm-Ubx expression is detected from the posterior half in T2–A9. The highest levels of staining are detected in A1. Arrow indicates the anterior limit of Bm-Ubx expression. c Embryo in stage 20 (96 h after oviposition). Lateral view. The expression pattern is similar to that in stage 18. Arrowheads indicate proleg primordia where the signal was undetectable. d–f Embryos with the sense strand probe in the same stage of a–c, respectively. No signal is detected. A1–9 abdominal segments, T1–3 thoracic segment. Bar = 500 μm
The expression of Bm-Ubx in the embryonic stages was almost the same as that in the moth M. sexta (Zheng et al. 1999) and the butterfly J. coenia (Warren et al. 1994). It indicates that Ubx expression pattern has been highly conserved in lepidopteran insects.
Phenotypes of Bm-Ubx RNAi
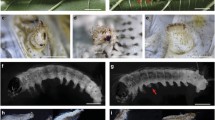
The B. mori larva has three pairs of thoracic legs from the first to the third thoracic segments (T1–T3), four pairs of abdominal legs in the A3–A6 segments (prolegs), and a pair of caudal legs in A10. There are no leg-like structures in A1 and A2 (Fig. 3a, b).
RNAi of Bm-Ubx. a A lateroventral view of a DsRed dsRNA injected embryo. Bar = 1 mm. b Enlargement of the same embryo in Fig. A. Bar = 300 μm. c A lateroventral view of a Bm-Ubx RNAi embryo. Bar = 1 mm. d Enlargement of the same embryo in c. Bar = 300 μm. e Thoracic leg of DsRed dsRNA injected embryo. Bar = 100 µm. f Abdominal leg (proleg) of DsRed dsRNA injected embryo. Bar = 100 μm. g Enlargement of the same embryo in d. Additional pair of legs in A1. Bar = 100 μm. h Lateral view of a DsRed dsRNA injected embryo. Spiracles in T1 and A1–A8 (A7 and A8 not shown). Bar = 500 μm. i Lateroventral view of a Bm-Ubx RNAi embryo. Spiracle remains in A1. Bar = 300 μm. A1–9 abdominal segments, apr anal proleg, cr crochets, pl planta, se setae, pr proleg, pta pretarsus (claws), sp spiracle, T1–3 thoracic segment, arrows additional pair of leg, arrowhead claw-like structure
We injected Bm-Ubx dsRNA into eggs within 5 h of oviposition. We dissected embryos in the last developmental stage (just before hatching), because many embryos could not hatch. In Bm-Ubx RNAi embryos, the additional pair of leg-like protuberances in A1 was observed (Fig. 3c, d). These protuberances had setae and shallow grooves, suggesting segments, and corresponded in position to the thoracic leg (Fig. 3g). Furthermore, this protuberance had a claw-like structure on the tip so that it was more similar to a thoracic-type leg than an abdominal-type leg (proleg; Fig. 3e−g). This result is the same as in O. fasciafus (Mahfooz et al. 2007) and T. castaneum (Lewis et al. 2000). In Ubx RNAi O. fascifus nymphs, however, A1 bears ectopic T3-like dorsal pigmentation (Angelini et al. 2005; Mahfooz et al. 2007). In T. castaneum, Utx (Ubx ortholog in T. castaneum) mutant larvae lack the A1 spiracle (as wild type T. castaneum larvae have spiracles in T2 and all abdominal segments, but lack them in T1 and T3). It was concluded that these larvae had transformations of the A1 segment toward the T3 segment (Angelini et al. 2005; Lewis 2000). In Bm-Ubx RNAi embryos, however, spiracles in A1 remained (Fig. 3i), as wild-type larvae have spiracles in T1 and in A1–A8, but are lacking in T2 and T3 (Fig. 3h). In E Cw homozygous larvae, ectopic A2-like dorsal pigmentation and a degenerated spiracle formed in the A1 segment (Hirokawa 1998). Therefore, in Bombyx, the Bm-Ubx RNAi phenotype might not simply be interpreted as "entire transformations of A1 segment toward thoracic identity" as in T. castaneum and O. fasciafus. On the contrary, the regulation mechanism of the A1 identity by Bm-Ubx may differ between the dorsal and ventral parts.
There was no presentation of a protuberance in A2 of Ubx RNAi embryos (Fig. 3d). This phenotype indicates that Dll in A2 is not repressed by Ubx singularly in B. mori. It might be that Ubx and abd-A act redundantly to inhibit appendage development in A2 in Bombyx such as in Drosophila (Castelli-Gair and Akam 1995; Vachon et al. 1992). Recently, however, it has been reported that abd-A RNAi does not cause extra appendage formation in A2 in Bombyx (Pan et al. 2009; Tomita and Kikuchi 2009). So it is highly possible that neither Ubx nor abd-A is important for appendage suppression in A2. Also in A3–A9, where Bm-Ubx expressed, there was no notable alteration in morphology, and the prolegs in A3–A6 remained the same as in the wild type. These morphologies are probably regulated by abd-A, which is essential for proleg development (Pan et al. 2009; Tomita and Kikuchi 2009). In addition, the phenotype observed in our study closely resembled E Cw homozygous larvae. Thus, E Cw may be associated with the loss-of-function of Bm-Ubx.
In our present work, it was found that Bm-Ubx suppresses leg development in A1. In two other lepidopterans, M. sexta and J. coenia (Zheng et al. 1999; Warren et al. 1994), Ubx expression pattern is almost the same as in B. mori (in particular, expression was highest in A1), and so these results indicate that Ubx in lepidopterans functions as a suppressor of leg development in A1. We support the idea that the strong repressive interaction between Ubx and Dll evolved in the dipteran/lepidopteran lineage (Palopoli and Patel 1998), since Ubx acts as a modifier rather than a suppressor of the abdominal appendage in T. castaneum (Lewis et al. 2000). Because Ubx and abd-A, and Ubx and Abd-B, overlap in their expression (Tomita and Kikuchi 2009), simultaneous RNAi analysis is necessary to elucidate the exact regulation of abdominal appendages.
Embryonic RNAi efficiency in B. mori
In this study, two dsRNA constructs were used for Bm-Ubx: a 765-bp fragment including the homeobox (Bm-Ubx-ORF) and a 445-bp fragment excluding the homeobox (Bm-Ubx-HDless; Fig. S1; Electronic Supplementary Material). It is possible that Bm-Ubx-ORF dsRNA could cross-react with Bm-abd-A transcripts because the amino acid sequence in homeodomain of Ubx and abd-A are resembled in many insects (the homology of them was 89% in B. mori). Thus, in order to elucidate the specific function for Bm-Ubx, we used the two constructs, mentioned above, for RNAi. The phenotype observed in these studies was indistinguishable. Additionally, no detectable effect was observed in DsRed RNAi larvae as the control (Fig. 3a, b). Therefore, phenotypes induced by Bm-Ubx RNAi must be specific for Bm-Ubx. This specific phenotype was ectopic A1 appendages, which were observed in 16.7% and 34.3% of developed embryos that were injected with two different constructs (Fig. 3c, d, g, Table 1). This result indicates that Bm-Ubx is a suppressor of leg development in A1. In this experiment, it is likely that B. mori embryos died during early embryogenesis due to physical damage from injection of dsRNA. Although improvement with respect to survival rate is required, we conclude that the embryonic RNAi method is effective in B. mori.
References
Angelini DR, Kaufman TC (2005) Insect appendages and comparative ontogenetics. Dev Biol 286:57–77
Angelini DR, Liu PZ, Hughes CL, Kaufman TC (2005) Hox gene function and interaction in the milkweed bug Oncopeltus fasciatus (Hemiptera). Dev Biol 287:440–455
Banno Y, Sakaida K, Nakamura T, Tsuchida K, Kawaguchi Y, Koga K, Doira H (1997) Reassessment of mapping of the E homeotic gene complex of Bombyx mori by linkage analysis and in situ hybridization with an Antennapedia clone as a probe. J Seric Sci Jpn 66:151–155
Castelli-Gair J, Akam M (1995) How the hox gene Utrabithorax specifies two different segments: the significance of spatial and temporal regulation within metameres. Development 121:2973–2982
Galant R, Carroll SB (2002) Evolution of a transcriptional repression domain in an insect Hox protein. Nature 415:910–913
Grenier JK, Garber TL, Warren R, Whitington PM, Carroll S (1997) Evolution of the entire arthropod Hox gene set predated the origin and radiation of the onychophoran/arthropod clade. Curr Biol 7:547–553
Hirokawa M (1998) Genetic analysis of a new mutant expressing extra-legs and extra-wings at the adult stage in the silkworm, Bombyx mori. J Seric Sci Jpn 67:1–7
Hughes CL, Kaufman TC (2002) Hox genes and the evolution of the arthropod body plan. Evol Dev 4:459–499
Itikawa N (1943) Genetical and embryological studies of a dominant mutant, ‘new additional crescent’, of the silkworm. Jap J Genet 19:182–188
Lewis DL, DeCamillis M, Bennett RL (2000) Distinct role of the hoeotic genes Ubx and abd-A in beetle embryonic abdomina appendage development. Proc Natl Acad Sci USA 97:4504–4509
Mahfooz N, Turchyn N, Mihajlovic M, Hrycaj S, Popadic A (2007) Ubx regulates differential enlargement and diversification of insect hind legs. PLoS ONE 2:e866. doi:10.1371/journal.pne.0000866
Morita A, Niimi T, Yamashita O (2003) Physiological differentiation of DH-PBAN producing neurosecretory cells in the silkworm embryo. J Insect Physiol 268:1093–1102
Ohtsuki Y (1979) Silkworm eggs. Japanese Society of Sericultural Science (ed) A general textbook of sericulture (in Japanese). Nihon Sansei Shinbun-Sha, Tokyo, pp 156–173
Palopoli MF, Patel NH (1998) Evolution of the interaction between Hox genes and a downstream target. Curr Biol 18:587–590
Pan MH, Wang XY, Chai CL, Zhang CD, Lu C, Xiang ZH (2009) Identification and function of Abdominal-A in the silkworm, Bombyx mori. Insect Mol Biol 18:155–160
Pavlopoulos A, Averof M (2002) Developmental evolution: Hox proteins ring the changes. Curr Biol 12:R291–R293
Ronshaugen M, McGinnis N, McGinnis W (2002) Hox protein mutation and macroevolution of the insect body plan. Nature 415:914–917
Suzuki Y, Palopoli MF (2001) Evolution of insect abdominal appendages: are prolegs homologous or convergent traits? Dev Genes Evol 211:486–492
Tazima Y (1964) E-group as a tool of developmental genetics. Tazima Y (ed) The genetics of the silkworm. Logos Press, London, pp 60–75
Tomita S, Kikuchi A (2009) Abd-B suppresses lepidopteran proleg development in posterior abdomen. Dev Biol 328:403–409
Tomoyasu Y, Wheeler SR, Denell RE (2005) Ultrabithorax is required for membranous wing identity in the beetle Tribolium castaneum. Nature 433:643–647
Ueno K, Hui C, Fukuta M, Suzuki Y (1992) Molecular analysis of the deletion mutants in the E homeotic complex of the silkworm Bombyx mori. Development 114:555–563
Vachon G, Cohen B, Pfeifle C, McGuffin ME, Botas J, Cohen SM (1992) Homeotic genes of the Bithorax complex suppress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell 71:437–450
Warren RW, Nagy L, Selegue J, Gates J, Carroll S (1994) Evolution of homeotic gene regulation and function in flies and butterflies. Nature 372:458–461
Yasukochi Y, Ashakumary LA, Wu C, Yoshido A, Nohata J, Mita K, Sahara K (2004) Organization of the Hox gene cluster of the silkworm, Bombyx mori: a split of the Hox cluster in a non-Drosophila insect. Dev Genes Evol 214:606–614
Zheng Z, Khoo A, Fambrough D, Garza L, Booker R (1999) Homeotic gene expression in the wild-type and a homeotic mutant of the moth Manduca Sexta. Dev Genes Evol 209:460–472
Acknowledgment
We thank Dr. M. Kobayashi and Dr. M. Ikeda for productive discussions and express our gratitude to the National Institute of Agrobiological Science for providing silkworm strain No. 459. This study was supported in part by Grants-in-Aid from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Roth
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Fig. S1
The nucleotide sequence (top) and deduced amino acid sequence (bottom) of Bm-Ubx cDNA from B. mori. Arrows indicate primer sequences used in RACE and RT-PCR (See "Materials and methods"). Double underline indicates homeobox. Boxed nucleotides represent a putative polyadenylation signal (AATAAA; GIF 138 kb)
Rights and permissions
About this article
Cite this article
Masumoto, M., Yaginuma, T. & Niimi, T. Functional analysis of Ultrabithorax in the silkworm, Bombyx mori, using RNAi. Dev Genes Evol 219, 437–444 (2009). https://doi.org/10.1007/s00427-009-0305-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-009-0305-9