Abstract
Cabut (cbt) is a Drosophila melanogaster gene involved in epidermal dorsal closure (DC). Its expression is dependent on the Jun N-terminal kinase (JNK) cascade, and it functions downstream of Jun regulating dpp expression in the leading edge cells. The Cbt protein contains three C2H2-type zinc fingers and a serine-rich domain, suggesting that it functions as a transcription factor. We have identified single cbt orthologs in other Drosophila species, as well as in other insects and invertebrate organisms like ascidians and echinoderms, but not in nematodes. Gene structure and protein sequence are highly conserved among Drosophilidae, but are more diverged in the other species of invertebrates analyzed. According to this, we demonstrate that cbt expression is detected in the embryonic lateral epidermis in several Drosophila species, as it occurs in D. melanogaster, thus suggesting that the cbt orthologs may have a conserved role in these species during DC. We have also analyzed the genomes of several vertebrate species, finding that the cbt orthologous genes in these organisms encode proteins that belong to the TIEG family of Sp1-like/Krüppel-like transcription factors. Phylogenetic analysis of the invertebrate and vertebrate proteins identified indicates that they mainly follow the expected phylogeny of the species, and that the cbt gene was duplicated during vertebrate evolution. Because we were not able to identify cbt orthologous genes neither in yeast nor in plants, our results suggest that this gene has been probably conserved throughout metazoans and that it may play a fundamental role in animal biology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cabut (cbt) is a gene involved in epidermal dorsal closure (DC) in Drosophila melanogaster (Muñoz-Descalzo et al. 2005). DC occurs when two lateral epidermal sheets move dorsally over the extra-embryonic amnioserosa and converge at the dorsal midline, sealing the dorsal side of the embryo. These movements are driven by multiple forces, including tissue-specific changes in the shape of individual cells, as well as the tension generated by a supracellular contractile actin–myosin cable that arises at the interface of the lateral epidermis and amnioserosa, which is known as the leading edge (Kiehart et al. 2000; Kaltschmidt et al. 2002; Franke et al. 2005). Genetic analyses have identified numerous genes required for DC. Among them, cbt encodes a putative transcription factor containing three C2H2 zinc finger motifs and a serine-rich region and is expressed in the yolk cell nuclei and embryonic epidermis. It functions downstream the Jun N-terminal kinase (JNK) cascade regulating the dpp expression in the leading edge cells (Muñoz-Descalzo et al. 2005).
The accessibility of genome sequence data from several organisms, together with the development of efficient computer-based search tools, has revolutionized modern biology, allowing in-depth comparative analysis of genomes. When applied to a certain gene, interspecies comparative analysis can be an especially interesting tool to obtain information about the sequence conservation, the expression regulation, and/or function of that gene. In this paper, we report the identification and analysis of cbt orthologous genes in invertebrates and vertebrates. To do that, we have performed searches in the completely and partially sequenced genomes of several Drosophila species, mosquito, silkworm, honeybee, red flour beetle, sea urchin, and sea squirt, identifying a single ortholog in all cases. However, we could not find a clear cbt ortholog in nematodes. Using this approach, we have annotated previously unknown genes that encode Cbt orthologous proteins in those invertebrate organisms. In addition, we have studied the expression pattern of the cbt genes in some Drosophila species, finding that it is very similar to the pattern found in D. melanogaster (Muñoz-Descalzo et al. 2005). We have also analyzed the genomes from vertebrate species used as model organisms, like chimpanzee, mouse, rat, zebrafish, and frog, as well as from humans, finding that the cbt gene probably underwent a duplication event during vertebrate evolution. Moreover, we have determined that the putative cbt orthologs in the species of vertebrates analyzed belong to the TIEG family of transcription factors. These proteins behave as cell growth repressors with antiproliferative and apoptosis-inducing functions in humans (Cook et al. 1999; Ellenrieder et al. 2002), and it seems that TIEG proteins also function as transcriptional repressors in mice (Yajima et al. 1997; Wang et al. 2004). Our results indicate that this gene has been conserved through animal evolution and that it may play a fundamental role in development.
Materials and methods
Sequence analysis
Database searches looking for cbt homologous sequences were performed using the basic local alignment search tool (BLAST) program (Altschul et al. 1997) at the Flybase (http://flybase.bio.indiana.edu/) or at the National Center for Biotechnology Information (NCBI). The GenScan program (Burge and Karlin 1997) was used for gene predictions (http://genes.mit.edu/GENSCAN.html). The accession numbers of the annotated proteins are shown in Table 1. The analysis of the primary structure of the putative protein sequences deduced from the DNA was performed using the Expasy–Prosite (http://us.expasy.org/prosite/) and MotifScan programs (http://myhits.isb-sib.ch/cgi-bin/motif_scan). To confirm that all the proteins identified are the real Cbt orthologs, we performed the symmetrical best hits (SymBets) approach (Koonin 2005). ClustalX was used for multiple sequence alignments (Thompson et al. 1997). We analyzed our data using ProtTest (Abascal et al. 2005); this program suggested the Jones, Taylor, and Thornton (JTT; Jones et al. 1992) correction method for the calculation of genetic distances. The phylogenetic trees were constructed with the neighbor-joining method (Saitou and Nei 1987), and the bootstrap tests were carried out with 500 iterations. These analyses were conducted using the Mega platform, version 3.1 (Kumar et al. 2004).
In situ hybridization
In situ hybridizations to whole-mount embryos were performed as described (Tautz and Pfeifle 1989), using 51°C as hybridization and washes temperature. Antisense RNA probes were generated with the DIG RNA labeling kit SP6/T7 (Roche). The cbt expression pattern was determined in D. melanogaster (OrR strain), D. affinis, D. azteca, and D. pseudoobscura 0- to 24-h embryos, using the SD06353 complementary DNA (cDNA) as template for riboprobe synthesis.
Results and discussion
Characterization of cabut genes in invertebrates
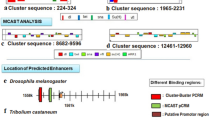
To identify putative cbt orthologs in invertebrates, the amino-acid sequence of the D. melanogaster Cbt protein (accession number AAF51489) was used to search the databases of sequences from several genome projects available through the NCBI. We searched the databases from several Drosophila species (D. simulans, D. sechellia, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. persimilis, D. willistoni, D. mojavensis, D. virilis, and D. grimshawi), from other insects like mosquito (Anopheles gambiae and Aedes aegypti), silkworm (Bombyx mori, p5OT strain), honeybee (Apis mellifera), and red flour beetle (Tribolium castaneum), as well as from other invertebrates like the echinoid sea urchin (Strongylocentrotus purpuratus), and the ascidia sea squirt (Ciona intestinalis; Table 1). For most of these organisms there is no protein annotation already made, therefore we used the TBLASTN algorithm (Altschul et al. 1997) to identify the cbt orthologs in the databases of genomic DNA sequences. However, in the case of D. pseudoobscura, A. gambiae, A. mellifera and S. purpuratus, the BLASTP algorithm (Altschul et al. 1997) was used to search the databases of annotated proteins. As a result of these searches we identified a single cbt ortholog in all species of invertebrates analyzed (see Materials and Methods). However, using the same strategy we could not find a clear cbt ortholog in the invertebrate C. elegans, and other model organisms like S. cerevisiae or A. thaliana. In D. pseudoobscura, the cbt gene has been annotated as GA-18176, in A. gambiae as agCG44033, in A. mellifera as GA-18176, and in S. purpuratus as TIEG (TGFβ inducible early growth factor response). To determine the structure of the cbt genes identified in the other species, we performed gene predictions by using the genomic sequences and coordinates obtained in the TBLASTN searches. In all cases, we identified the hypothetical full-length cbt-coding regions, except in B. mori and S. purpuratus. In B. mori, it is truncated at the 3′ end (Fig. 1c) because the genomic contig containing the cbt-coding region ended at that point and no overlapping contigs harboring additional sequences were found. In S. purpuratus, the TIEG protein annotated by the Sea Urchin Genome Project was incomplete. However, we analyzed in detail the genomic contig containing that sequence and were able to identify the complete cbt-coding region (Fig. 1c).
a Phylogeny of the analyzed Drosophilidae and insect species as suggested by the Flybase database. b Gene structure of the cabut orthologs in Drosophilidae. The last five amino acids of the first exon and the first five of the second are indicated. c Gene structure of the cabut orthologs in other species of invertebrates analyzed. The cabut-coding region of B. mori is incomplete (shown with a black triangle at the end of the gene, respectively). In b and c, coding regions are shown in black, non-coding regions in white, and introns are depicted as open triangles
Gene predictions showed that the exon–intron structure of the cbt genes is largely conserved in the Drosophila species, mainly consisting of two exons and one intron (Fig. 1b). In all these species, the coding region in the first exon is shorter (63–78 bp) than in the second (1,209–1,326 bp). Moreover, the position and size of the intron (1,208–1,518 bp) are conserved, as well as the amino acid sequence around the exon–intron boundary (Fig. 1b). However, in D. willistoni, neither the position/length of the intron (4,072 bp) nor the amino acid sequence around the exon–intron boundary is conserved. It is likely that this gene prediction is incorrect because the first 31 amino acids of the protein do not align with the other Drosophila Cbt proteins in a ClustalX analysis (data not shown). However, we were not able to identify any adjacent region that could encode an amino acid sequence similar to the N terminus of the D. melanogaster Cbt protein. Strikingly, cbt genes in D. pseudoobscura and D. persimilis, although similar to the others in size, contain three exons due to the existence of a small intron that splits the large second exon in two (Fig. 1b). In these species, the position and length of both introns are conserved, as well as the amino acid sequence around the exon–intron boundaries. Considering the phylogenetic relationships of the Drosophila species analyzed (Fig. 1a), this splitting would have occurred after the divergence between the melanogaster and obscura groups 25 Ma ago (Russo et al. 1995). In most of the other invertebrates analyzed, the cbt orthologs also contain two exons and one intron like in D. melanogaster, although the size and position of the intron is variable (Fig. 1c and data not shown). The exceptions are the cbt genes from A. mellifera and C. intestinalis that contain three exons/two introns, and from S. purpuratus with four exons/three introns. Despite these differences, we see that the coding region of all the cbt orthologs starts in the first exon, which may be relevant for the regulation of cbt expression.
We also tested whether the chromosomal location of the cbt orthologs was conserved in D. simulans, D. yakuba, and D. pseudoobscura for which the genome locations are available. The D. melanogaster gene is located on the left arm of chromosome 2, between the Arc105 and the u-shaped (ush) genes (Muñoz-Descalzo et al. 2005; data not shown). In D. simulans and D. yakuba, other species of the melanogaster subgroup, the cbt gene is positioned on the same chromosome (2L). Similarly, the D. pseudoobscura cbt gene is located on chromosome 4, which is ortholog to chromosome 2L of the melanogaster subgroup (Richards et al. 2005). Besides, we performed gene synteny analyses to determine whether there is a conservation of the orientation of the invertebrate cbt orthologs identified. These analyses were restricted to one of the neighboring genes, Arc105, which is located very close to the 3′ end of cbt and in opposite orientation. ush is located about 44 kb apart from cbt (Muñoz-Descalzo and Paricio, unpublished), making this analysis impossible in most of the invertebrate genomes available due to the absence of data. In all the Drosophila species analyzed, we confirmed the presence of an Arc105 orthologous gene adjacent to cbt. Gene orientations were also conserved in all cases (data not shown). In the other species of invertebrates analyzed (A. aegypti, A. gambiae, T. castaneum, A. mellifera, C. intestinalis, and S. purpuratus), the coding regions located at the 3′ end of the cbt orthologs are not related to the D. melanogaster Arc105 gene (data not shown).
Taken together, our results suggest that the last common ancestor of bilaterians harbored a cbt gene, although we were not able to identify a clear cbt ortholog in nematodes. During evolution, the gene structure and location have been well conserved in Drosophilidae, but they are more diverged in the other insects and invertebrate organisms analyzed.
Analysis of cabut proteins in invertebrates
In D. melanogaster, the Cbt protein is 428-amino-acid long and contains three classical zinc finger domains of the C2H2 type located in tandem at the carboxy terminal region and a serine-rich region at the amino terminus (Fig. 2; Muñoz-Descalzo et al. 2005). In general, proteins with three C2H2 zinc fingers may bind DNA, RNA, or proteins through the fingers, although most of them are DNA-binding proteins that participate in transcriptional regulation of target genes (Iuchi 2001). Serine-rich regions are low-complexity regions, but it has been proposed that they could regulate protein activity through phosphorylation and might be necessary for protein–protein interaction and/or signal transduction (Okamura et al. 2004; Barrasa et al. 2005). Our results show that the size of the predicted Cbt proteins is similar among the Drosophilidae but it is shorter in the other species of the insects analyzed (Table 1). However, Cbt proteins from the invertebrates S. purpuratus and C. intestinalis are larger in size (Table 1). Functional domains were predicted in the Cbt orthologs by using the ScanProsite and MotifScan programs. All of them contain the three zinc finger domains of the C2H2 type at the carboxy terminal region (Fig. 2), suggesting that they have to be essential for Cbt function. However, the serine-rich region is not conserved in all the Cbt orthologs. In most Drosophila species, a serine-rich region of similar size is found at the amino terminal region of the Cbt proteins, however, in other species, is absent, and some of them contain additional serine, glutamine, or alanine-rich regions (Fig. 2). The analysis of the Cbt orthologous proteins in other invertebrates revealed that the serine-rich region is only present in A. gambiae protein (Fig. 2). In B. mori and C. intestinalis, Cbt proteins contain a proline-rich and a threonine-rich region at the carboxy terminus, respectively. In addition, the Cbt ortholog from A. aegypti has a glutamine-rich region at the carboxy terminus (Fig. 2). As suggested for the serine-rich regions, proline-, threonine-, and glutamine-rich regions are low-complexity regions that could also act as protein–protein interaction domains during signal transduction (Gill et al. 1994; Triezenberg 1995; Kay et al. 2000). Strikingly, we found no obvious low-complexity regions in the Cbt proteins from A. mellifera, T. castaneum, and S. purpuratus (Fig. 2). Maybe these proteins have developed new strategies for its regulation or to mediate interactions with other proteins, if they are essential for their function.
Structure of the Cabut proteins in invertebrates. The relative positions of the domains identified by the ScanProsite program in these proteins are indicated by colored boxes: Zn finger domains are shown as blue boxes, serine-rich regions are in red, glutamine-rich regions in green, alanine-rich regions in yellow, proline-rich regions in orange, and threonine-rich regions in purple
To further analyze the sequence conservation between the D. melanogaster Cbt protein and the invertebrate orthologs identified, we performed BLAST2 comparisons, both using the full length predicted proteins or only the zinc fingers domain (Table 1). As expected, the percentages of similarity are higher when the species compared are more closely related to D. melanogaster. Moreover, the percentages of similarity are also higher when the comparison is restricted to the zinc finger domains (Table 1).
Expression of cabut in several Drosophila species
As mentioned above, cbt has a role in DC during D. melanogaster embryogenesis. Although it is expressed in the epidermis, yolk cell nuclei, and posterior gut (Fig. 3a, and data not shown), Cbt is only required in the epidermis for DC to be completed (Muñoz-Descalzo et al. 2005). To test whether the role of cbt during DC could be conserved in flies, we analyzed the expression pattern of the cbt orthologs in several Drosophila species. In situ hybridizations were performed using a D. melanogaster cbt riboprobe in 0- to 24-h embryos of D. affinis, D. azteca (from the obscura group, affinis subgroup), and D. pseudoobscura (from the obscura group, pseudoobscura subgroup). Our results show that the cbt genes are expressed in the lateral epidermis in the three species analyzed (Fig. 3). However, the expression in the yolk cell nuclei is only detected in D. affinis (Fig. 3b–d). cbt expression in the posterior gut was not detected even in D. melanogaster, probably due to the hybridization conditions used in this experiment. Considering that cbt expression in the epidermis is essential for the role of this gene during DC in D. melanogaster (Muñoz-Descalzo et al. 2005), our results suggest that the Cbt proteins in D. affinis, D. azteca, and D. pseudoobscura will probably have a similar role in this process.
Cabut expression in embryos of stage 13 from several Drosophila species analyzed by whole-mount in situ hybridization. a Wild-type D. melanogaster embryo, b D. affinis embryo, c D. azteca, and d D. pseudoobscura. Arrows show the expression of cbt in the epidermis, and arrowheads in the yolk cell. Anterior is to the left and dorsal is up in all cases
Analysis of cabut orthologs in vertebrates
To identify putative cbt orthologous genes in vertebrates, we analyzed several species used as model organisms like chimpanzee (Pan troglodytes), mouse (Mus musculus), rat (Rattus norvegicus), zebrafish (Danio rerio), and frog (Xenopus tropicalis), as well as humans (Homo sapiens). The sequence of the D. melanogaster Cbt protein was used to search the reference sequence (RefSeq) protein database of each organism available through the NCBI by using the BLASTP algorithm (Altschul et al. 1997). Comparative genomic analysis led to propose the one-to-four rule that says that one gene in invertebrates could have duplicated during vertebrate evolution in two or four (Ohno 1999). According to this, we identified two putative cbt orthologous genes in all the vertebrate species analyzed but in frog (Fig. 4, Table 2). In humans, two genes were previously reported as cbt orthologs by Suske et al. (2005), encoding the TIEG 1 and 2 transcription factors. These proteins belong to the subgroup III of the Sp1-like/Krüppel-like (Sp1-/KLF) family of transcription factors (Subramaniam et al. 1995; Cook et al. 1998; Kaczynski et al. 2003). In mice, three TIEG genes were initially described (TIEG1-3; Yajima et al. 1997; Scohy et al. 2000; Wang et al. 2004). However, recent studies have suggested that murine TIEG2 could represent the human TIEG2 sequence, and therefore, this gene was not included in this study (Suske et al. 2005; K. Krieglstein and C. Szpirer, personal communication). In each of the other species analyzed, we identified two orthologs that are also classified as TIEG transcription factors, except in X. tropicalis, in which we only identified an “unknown” protein as Cbt ortholog (Table 2). Because the genomic sequence of this organism is incomplete, it is possible that a second Cbt ortholog could be present in its genome. According to the structure of the gene, the protein identified could be the frog TIEG2 ortholog (see below).
Gene structure of the cabut orthologs in vertebrates. Coding regions are shown in black, non-coding regions in white, and introns are depicted as open triangles. Sequence accession numbers: human TIEG1, AF050110; human TIEG2, NM_003597; chimpanzee TIEG1, XM_528205; chimpanzee TIEG2, XM_515296; mouse TIEG1, NM_013692; mouse TIEG3, NM_178357; rat TIEG1, NM_031135; rat TIEG2, NM_001037354; zebrafish TIEG2, genomic contig BX248136; zebrafish TIEG3, XM_682384; frog sequence, BC121242
The structure of the genes identified in the searches was determined using the messenger RNA (mRNA)/genomic sequences available trough the NCBI database (Fig. 4). The overall genomic organization of these genes is similar in most of the species analyzed and more complex than in invertebrates, consisting of four exons and three introns. However, the putative cbt orthologous genes contain six exons and five introns in P. troglodytes. In all cases, the first exon is small in size and contains the translation start codon, as occurs in invertebrates, again suggesting that this could be relevant for the regulation of cbt expression. The size of the vertebrate Cbt orthologous proteins is variable (458–734 amino acids, see Table 2), but all of them contain three classical zinc finger domains of the C2H2 type at the carboxy terminal region (data not shown), as in D. melanogaster and other members of the Sp1-like/KLF family (Kaczynski et al. 2003; Muñoz-Descalzo et al. 2005). However, the serine-rich region found at the amino terminus of the D. melanogaster Cbt protein was not identified in any of them. Conversely, they contain a proline-rich region (Subramaniam et al. 1995; Yajima et al. 1997; Cook et al. 1998; Wang et al. 2004; data not shown), which may associate with the SH3 domains of src tyrosine kinases and be involved in signal transduction processes (Subramaniam et al. 1995; Yajima et al. 1997; Ellenrieder et al. 2002). Pair-wise comparisons with the D. melanogaster Cbt protein revealed that the percentages of similarity are higher when only the zinc finger domains are compared (Table 2), as seen in invertebrates, suggesting that this region may play an important role in Cbt function. Several studies have shown that TIEG transcription factors in humans and mice are involved in cellular growth control and pancreatic cancer, and biochemical characterization have demonstrated that TIEGs are transcriptional repressors (Cook et al. 1998, 1999; Cook and Urrutia 2000). Moreover, recent studies on TIEG1 knock-out mice have shown that this gene is involved in flexor tendon healing, cardiac hypertrophy, and skeletal development and maintenance (Bensamoun et al. 2006; Tsubone et al. 2006; Rajamannan et al. 2007). Further analyses will be required to demonstrate whether the proteins identified in this study could play a similar role than TIEGs in humans or mice.
Phylogenetic analysis of the cabut proteins
To confirm whether the TIEG proteins identified in this study are the vertebrate orthologs of the D. melanogaster Cbt protein, we performed a phylogenetic analysis of them but also including other D. melanogaster proteins containing C2H2 zinc finger domains. As can be seen in Fig. 5a, Cbt and the TIEG proteins appear clearly clustered and separated from the other D. melanogaster proteins analyzed (Fig. 5a). This result confirms our previous assumption that they are real orthologs. Subsequently, a phylogenetic analysis of all the invertebrate and vertebrate proteins described in this study was performed. It shows that they fall into two clusters (Fig. 5b). One of these clusters contains sequences from insects, including all the Drosophila species. Within Drosophila, Cbt proteins mainly follow the accepted phylogeny of the species (compare with Fig. 1a). However, Cbt proteins from A. gambiae and A. aegypti should be forming a clade with the Drosophila sequences because they are also dipterans, and that is not the case. An explanation for these inconsistencies could be that the proteins in insects have been predicted in silico from genomic sequences, and in most cases, there are no expressed sequence tags (ESTs) available that could support these predictions. Regarding the second cluster of Cbt-like sequences, it includes not only the vertebrate TIEG proteins but also the sequence from the tunicate sea squirt, which is also a chordate. However, our results also show that the Cbt ortholog from the echinoderm S. purpuratus is more closely related to vertebrates than the one from tunicates. Regarding this, we also find that the structure of the cbt gene from sea urchin is very similar to TIEG1, one of the vertebrate cbt orthologs (compare Fig. 1c with Fig. 4). In vertebrates, we find that the TIEG protein clade is monophyletic. In humans, chimpanzees, mice, and rats, two TIEG proteins (TIEG1 and TIEG2/3) have evolved independently after a gene duplication that occurred in their common ancestor. Besides, the only protein identified in X. tropicalis is grouped with the mammalian TIEG2/3 sequences, thus, indicating that it is the TIEG2 ortholog in frogs. Further sequencing of the genome of this species will be required to identify the gene encoding the TIEG1 protein. In zebrafish, the two TIEG proteins identified are clustered and grouped with the TIEG2/3 orthologs, thus, suggesting that the TIEG ancestor was probably more similar to TIEG2/3 than to TIEG1.
Phylogenetic analysis of the cabut proteins. a Unrooted tree constructed with several D. melanogaster proteins containing C2H2 zinc finger domains and vertebrate TIEGs. The fly proteins are Sp1-PA (NP_572579.2), Sp1-PB (NP_727360.1), luna-PB (NP_995811.1), buttonhead (NP_511100.1), Bteb2 (NP_572185.1), huckebein-PA (NP_524221.1), odd-paired PA (NP_524228.2), poils au dos-PA (NP_650534.1), and chorion factor 2-PA (NP_523474.1). b Unrooted tree constructed with all the cabut proteins described in this study. The D. melanogaster Sp1-PA, Sp1-PB, and CG5669 (NP_651232.1) Sp1/KLF proteins were used as outgroups, as they also contain C2H2 zinc fingers and show the highest score to Cbt in a BLASTP comparison (data not shown). Trees were constructed with the neighbor-joining method and a bootstrap test with 500 iterations (bootstrap values are indicated in bold at each branching position). Branch lengths are also indicated
In summary, we show that the Cbt proteins are present in invertebrate and vertebrate organisms and, with several exceptions, they seem to follow the accepted phylogeny of the species analyzed, thus suggesting that the cbt gene was present in the early ancestor of these species. However, we were not able to identify a clear cbt ortholog in C. elegans. In vertebrates, a specific duplication event led to the presence of two cbt orthologs that encode TIEG proteins that belong to the Sp1/KLF transcription factor family. Taken together, our results suggest that the cbt gene has been probably conserved throughout metazoans and that it may play a fundamental role in animal biology. However, whether their molecular function has been conserved through evolution is unclear. Further functional analysis will be required to clarify this issue.
References
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Barrasa MI, Harel NY, Alwine JC (2005) The phosphorylation status of the serine-rich region of the human cytomegalovirus 86-kilodalton major immediate–early protein IE2/IEP86 affects temporal viral gene expression. J Virol 79:1428–1437
Bensamoun S, Tsubone T, Subramaniam M, Hawse JR, Boumediene E, Spelsberg TC, An KN, Amadio PC (2006) Age-dependent changes in the mechanical properties of tail tendons in TGF{beta} inducible early gene-1 (TIEG) knockout mice. J Appl Phys 101:1419–1424
Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94
Cook T, Gebelein B, Mesa K, Mladek A, Urrutia R (1998) Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J Biol Chem 273:25929–25936
Cook T, Gebelein B, Belal M, Mesa K, Urrutia R (1999) Three conserved transcriptional repressor domains are a defining feature of the TIEG subfamily of Sp1-like zinc finger proteins. J Biol Chem 274:29500–29504
Cook T, Urrutia R (2000) TIEG proteins join the Smads as TGF-beta-regulated transcription factors that control pancreatic cell growth. Am J Physiol Gastrointest Liver Physiol 278:G513–G521
Ellenrieder V, Zhang JS, Kaczynski J, Urrutia R (2002) Signaling disrupts mSin3A binding to the Mad1-like Sin3-interacting domain of TIEG2, an Sp1-like repressor. EMBO J 21:2451–2460
Franke JD, Montague RA, Kiehart DP (2005) Nonmuscle myosin II generates forces that transmit tension and drive contraction in multiple tissues during dorsal closure. Curr Biol 15:2208–2221
Gill G, Pascal E, Tseng ZH, Tjian R (1994) A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA 91:192–196
Iuchi S (2001) Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci 58:625–635
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Kaczynski J, Cook T, Urrutia R (2003) Sp1- and Kruppel-like transcription factors. Genome Biol 4:206
Kaltschmidt JA, Lawrence N, Morel V, Balayo T, Fernandez BG, Pelissier A, Jacinto A, Martinez Arias A (2002) Planar polarity and actin dynamics in the epidermis of Drosophila. Nat Cell Biol 4:937–944
Kay BK, Williamson MP, Sudol M (2000) The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J 14:231–241
Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA (2000) Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J Cell Biol 149:471–490
Koonin EV (2005). Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39:309–338
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Muñoz-Descalzo S, Terol J, Paricio N (2005) Cabut, a C2H2 zinc finger transcription factor, is required during Drosophila dorsal closure downstream of JNK signaling. Dev Biol 287:168–179
Ohno S (1999) Gene duplication and the uniqueness of vertebrate genomes circa 1970–1999. Semin Cell Dev Biol 10:517–522
Okamura H, Garcia-Rodriguez C, Martinson H, Qin J, Virshup DM, Rao A (2004) A conserved docking motif for CK1 binding controls the nuclear localization of NFAT1. Mol Cell Biol 24:4184–4195
Rajamannan NM, Subramaniam M, Abraham TP, Vasile VC, Ackerman MJ, Monroe DG, Chew TL, Spelsberg TC (2007) TGFbeta inducible early gene-1 (TIEG1) and cardiac hypertrophy: discovery and characterization of a novel signaling pathway. J Cell Biochem 100:315–325
Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, Couronne O, Hua S, Smith MA, Zhang P, Liu J, Bussemaker HJ, van Batenburg MF, Howells SL, Scherer SE, Sodergren E, Matthews BB, Crosby MA, Schroeder AJ, Ortiz-Barrientos D, Rives CM, Metzker ML, Muzny DM, Scott G, Steffen D, Wheeler DA, Worley KC, Havlak P, Durbin KJ, Egan A, Gill R, Hume J, Morgan MB, Miner G, Hamilton C, Huang Y, Waldron L, Verduzco D, Clerc-Blankenburg KP, Dubchak I, Noor MA, Anderson W, White KP, Clark AG, Schaeffer SW, Gelbart W, Weinstock GM, Gibbs RA (2005) Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res 15:1–18
Russo CA, Takezaki N, Nei M (1995) Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol 12:391–404
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Scohy S, Gabant P, Van Reeth T, Hertveldt V, Dreze PL, Van Vooren P, Riviere M, Szpirer J, Szpirer C (2000) Identification of KLF13 and KLF14 (SP6), novel members of the SP/XKLF transcription factor family. Genomics 70:93–101
Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC (1995) Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res 23:4907–4912
Suske G, Bruford E, Philipsen S (2005) Mammalian SP/KLF transcription factors: bring in the family. Genomics 85:551–556
Tautz D, Pfeifle C (1989) A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98:81–85
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Triezenberg SJ (1995) Structure and function of transcriptional activation domains. Curr Opin Genet Dev 5:190–196
Tsubone T, Moran SL, Subramaniam M, Amadio PC, Spelsberg TC, An KN (2006) Effect of TGF-beta inducible early gene deficiency on flexor tendon healing. J Orthop Res 24:569–575
Wang Z, Peters B, Klussmann S, Bender H, Herb A, Krieglstein K (2004) Gene structure and evolution of Tieg3, a new member of the Tieg family of proteins. Gene 325:25–34
Yajima S, Lammers CH, Lee SH, Hara Y, Mizuno K, Mouradian MM (1997) Cloning and characterization of murine glial cell-derived neurotrophic factor inducible transcription factor (MGIF). J Neurosci 17:8657–8666
Acknowledgment
We are grateful to Rosa de Frutos for fly stocks. S. M.-D. and Y.B. were supported by fellowships from Consellería de Cultura, Educació i Ciència and from Comunidad Autónoma de La Rioja, respectively. This work was supported by grants from Ministerio de Educación y Ciencia (BFU2004-00498/BMC) and from Consellería de Cultura, Educació i Ciència (ACOMP06/116) to N.P.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Simpson
Rights and permissions
About this article
Cite this article
Muñoz-Descalzo, S., Belacortu, Y. & Paricio, N. Identification and analysis of cabut orthologs in invertebrates and vertebrates. Dev Genes Evol 217, 289–298 (2007). https://doi.org/10.1007/s00427-007-0144-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-007-0144-5