Abstract
Malacostracan crustaceans have evolved a conserved stereotyped cell division pattern in the post-naupliar germ band. This cleavage pattern is unique in arthropods investigated so far, and allows a combined analysis of gene expression and cell lineage during segmentation and organ development at the level of individual cells. To investigate the cell lineage in the germ band of the isopod Porcellio scaber, we used a 4D-microscopy system, which enables us to analyse every cell event in the living embryo. The study was combined with the analysis of the expression of the gene engrailed (en) at different stages of germ band formation. Our findings confirm the results of earlier investigations of the cell division pattern in the posterior part of the isopod germ band. Furthermore, we can show that in the anterior region, in contrast to the posterior part, cleavage directions are variable and cell sorting takes place—similar to other arthropod germ bands. Additionally, the gene expression pattern of en in this region is not as regular as in the post-naupliar germ band, and only later becomes regulated into its characteristic stripe pattern. The comparison of the cell lineage of P. scaber with that of other malacostracan crustaceans shows an enhancement in the velocity of cell divisions relative to the arrangement of these cells in rows in the isopod germ band. The striking similarity of the formation of the genealogical units in the anterior part suggests a sister group relationship between the peracarid taxa Tanaidacea and Isopoda.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Arthropod development has been modified in different ways during evolution. Early developmental variations range from total cleavage to syncytial cleavage (e.g. Scholtz 1997). During germ band formation, variations include short vs long germ band development, the formation of different larval stages and hetero- and holometabolic development (e.g. Anderson 1973). The main sources of knowledge about arthropod germ band formation, segmentation, neurogenesis and leg development are the intensively investigated arthropods, the fruitfly Drosophila (Lawrence 1992), the beetle Tribolium (e.g. Maderspacher et al. 1998), the grasshopper Schistocerca (e.g. Broadus and Doe 1995; Dearden and Akam 2001), the spider Cupiennius (e.g. Damen et al. 2000) and the crustacean Artemia (e.g. Averof and Akam 1993). Their germ bands do not show any indication of a specific cell lineage. Instead, cell sorting and migration dominate during the formation and differentiation of their germ bands. In contrast to all other arthropods, the malacostracan crustaceans studied in this respect show a unique mode of germ band formation and differentiation in the post-naupliar region (posterior to the mandibular segment) of the germ band (Dohle and Scholtz 1988; Scholtz 1997; Dohle et al. 2004). Their post-naupliar germ band consists of cells, that divide in a specific stereotypic pattern that is highly conserved (Dohle and Scholtz 1988; Scholtz and Dohle 1996). In contrast to this, the naupliar segments are initially formed like the segments in other arthropods, i.e. by scattered cell divisions without any obvious stereotypic pattern. There is a zone of transition between the two modes of cell arrangement and differentiation during segmentation, which is situated in the segments of the maxillae and the mandibles. It is known from several malacostracans that the cell lineage of the maxillary region, although stereotypic, differs from that of the more posterior post-naupliar segments (Dohle 1972, 1976; Scholtz 1984, 1990). Furthermore, there is some evidence that the posterior margin of the mandibular segment shows some regular cell divisions, which indicate a pattern of cell lineage in this area as well (Scholtz 1990). However, with the methods and techniques applied so far, this transitional region could not be analysed in sufficient detail.
With the use of a 4D-microscopy system (Schnabel et al. 1997; Hejnol and Schnabel 2005), we were able to extend the investigation of cell divisions in the living embryos of an isopod crustacean, the wood louse Porcellio scaber, to the formation of the anterior part of the post-naupliar germ band. Tracing of the cells in the embryo showed an irregular cell division pattern in this part of the germ band, as well as cell sorting including the immigration of single cells. This is similar to the conditions in other arthropods. To see if this irregularity is also observed at the level of gene expression, we used the antibody 4D9 against the en protein (Patel et al. 1989b). It is interesting to note that the expression of en is first irregular, but later starts to form the expected stripes at the posterior end of each segment. In contrast to the formation of the post-naupliar germ band, which is formed in the malacostracans by ectoteloblasts (except in amphipods, see Dohle and Scholtz 1988; Scholtz 1990; Browne et al. 2005; Pavlopoulos and Averof 2005), the segments in the anterior part of the post-naupliar germ band are formed in different ways, i.e. the origin and division patterns of the genealogical rows, differ between the peracarid taxa studied so far (Cumacea: Dohle 1970, 1976; Tanaidacea: Dohle 1972; Mysidacea: Scholtz 1984; Amphipoda: Scholtz 1990). Our 4D-microscopic study of the development of this region enabled us to follow the formation of the genealogical rows in isopods, which could not be clarified by the methods used in earlier studies (Hahnenkamp 1974; Vehling 1994). The results show that the formation of the genealogical rows of the anterior part of the posterior germ band shows a high degree of similarity to the way these rows are formed in the tanaidaceans (Dohle 1972). This similarity supports a sister group relationship between the Isopoda and Tanaidacea.
Materials and methods
4D-microscopy
P. scaber was collected in Berlin/Kreuzberg and in the botanical garden in Braunschweig. For 4D-microscopic investigations, eggs in the stage of interest were collected from the breeding pouch of females and embedded on the slide with an agar pad in which a hole of the size of the egg was cut. The germ band was oriented so that the top of the germ band could be observed (ventral view). A cover slip (60×28 mm) was put on the drop of isopod saline (see Hejnol and Scholtz 2004) and was sealed with Vaseline to prevent evaporation. For the recording of the living embryos, an improved 4D-microscopy system (Schnabel et al. 1997) was used, which is described in detail in Hejnol and Schnabel (2005, 2006). Nomarski pictures of the embryos were taken every 3 min in Z-stacks composed of 25 levels with an increment of 1 μm. The stacks of pictures were analysed with the software SIMI°BioCell (SIMI, Unterschleiβheim, Germany). The recorded embryos are numbered 1, 2, 3..., respectively, and development can be seen in the movies in the supplementary material.
Immunostaining
For immunostaining of the germ bands, embryos were fixed in 4% formaldehyde in PBS for ∼15 min, then washed three times with PBS. Fixed embryos were incubated with the primary antibody (4D9 monoclonal anti-en, 1:3) overnight, followed by a few washes in PBS + BSA for 5 min each. The germ bands were incubated in secondary antibody (alkaline–phosphatase-conjugated anti-mouse IGg, 1:400) overnight at 4°C, followed by several washes. Detection of the immunoreaction was achieved with the alkaline phosphatase substrate BCIP/NBT. DAPI was used as counter-staining for better identification of the nuclei. For microscopy, the germ bands were embedded in 50% glycerol and sealed with Vaseline.
Nomenclature
The naming of the cells was done according to Dohle (1970) and Scholtz (1990) for the cells of the first maxillae. Cell rows of non-ectoteloblastic origin in this work are named regarding their future fate, e.g. row E(1) gives mainly rise to the segment of the first maxillae, E(2) to the second maxillae and E(3) mainly to the first thoracopod. Cell rows formed by the derivatives of the ectoteloblasts are named eI, eII, etc. The innermost cells of one genealogical unit are numbered from 1 with increasing numbers for the more lateral cells. The four cell rows are a, b, c and d from anterior to posterior. The nomenclature of the first maxillary segment differs due to the nature of its formation (see Scholtz 1990).
Results
Formation of the ectoteloblasts in P. scaber
After gastrulation (see Supplementary material S1) the row of ectoteloblasts is differentiated anterior to the prospective proctodaeum. The cells form a ring around the gastrulation centre and the cells grow in size (Fig. 1a). Anterior to the row of ectoteloblasts are smaller scattered cells which are not ectoteloblast derivatives. These cells form the naupliar and the anterior part of the post-naupliar germ band. During development, the posterior population of these cells is arranged in two rows, which are named row A and row B (Fig. 1b,c). Later row B gives rise to the daughter rows E(2) and E(3), and row A transforms to row E(1) which gives rise to most of the segment of the first maxilla. The cells anterior to row A form the naupliar part of the germ band (first + second antennae and the mandible) (Fig. 2). Every ectoteloblast gives off cells only to the anterior direction, thus successively forming the posterior part of the post-naupliar germ band. The median ectoteloblast is named ET0 and its derivatives will form the midline of the embryo. In the 4D-recording, it is shown that inner cells of the anterior rows divide before the ectoteloblasts begin to give off their derivatives (Fig. 2b).
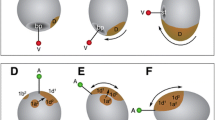
a Cells surrounding the gastrulation center (gz) in Porcellio scaber. Anterior to the top. Around the gastrulation centre are cells whose progeny enter the invagination site. The inner ring of cells (bordered by white lines) will later give off cells that will form the proctodaeum at the posterior end of the germ band. Surrounding these cells, the ring of ectoteloblasts is differentiating (ET). The arrow-marked ectoteloblasts divide unequally giving off cells in the anterior direction (beginning the formation of cell row eI). b Cell rows of non-ectoteloblastic origin anterior to the ectoteloblasts (red spheres in c). c SIMI°BioCell reconstruction of the arrangement of the cells inb. Cells of the row B are in violet. Single cells of the cell row anterior touch ectoteloblasts (green sphere touches red sphere in comparison with the nomarski image b). The posterior descendants of the green row will form the row A
First divisions of the ectoteloblasts and the divisions in the anterior rows [sequence out of movie 2 (S2), anterior to the top]. a Start of the recording. The row of ectoteloblasts is bordered through a dotted line. The gastrulation center is posterior (bottom) to the row of ectoteloblasts. b After 3 h, the median cells in front of the ectoteloblasts have divided. c After 2 h, the row of ectoteloblasts gives off their first derivatives (eI). d After 5 h, the cells of non-ectoteloblastic origin divide mainly in an anteriorposterior (arrowheads) direction but also mediolateral divisions are observed. e Arrangement of the cells after the cell division round. Upper right, reconstruction of with the software SIMI°BioCell. Cell of row A [E(1)] are represented as white spheres. The anterior cell row (red spheres) will form the mandibular segment. The anterior cells of row B are in cyan [row E(2)], the posterior sister cells are in green [row E(3)]. The violet spheres show undivided cells of row B. The median cells are in yellow, the derivatives of the ectoteloblasts are in orange. The bars in this and the following figures are connecting sister cells and show the division angle
Cell divisions in rows A and B and ectoteloblasts
Formation of the first ectoteloblast derivatives (row eI)
The recording 2 (Supplementary material S2; Fig. 2) shows that the ectoteloblasts do not give off derivatives in a regular fashion from the middle to the lateral side of the embryo, as they do later in development. In this embryo, ET2 divides before ET1 and ET5 before ET2 (Fig. 2c). The median ET0 divides later than the innermost ectoteloblasts. Within 5 h, all ectoteloblasts have given off their respective derivative cell eI, thus forming a row of small cells in front of the ectoteloblasts (Fig. 2d). In addition, no bilateral symmetry in the sequence of the ectoteloblast divisions is observed. During the progressive formation of the ectoteloblast-derived rows, the ectoteloblasts give off cells in a wave from the medial to the lateral direction, although sometimes irregularities can be observed. Also, the degree of the bilateral symmetry of the cell division waves is enhanced in successive divisions. The formation of a row of ectoteloblast derivatives starts always after the previous cell division round has been finished.
The formation of the rows of non-ectoteloblastic origin in P. scaber
The division of the rows A and B
In the embryo 2, after the formation of the row eI from the ectoteloblasts, the anterior cells divide (Fig. 2e). The cell divisions in row B and the formation of row A are part of a mitotic wave, which progresses from the anterior of the germ disc to the posterior (see Supplementary material S2). Thus, the cells of row B divide before the cells of row A undergo their first round of division. The divisions of all cells take the same amount of time as the formation of the row eI from the ectoteloblasts (300 min). The last cells formed are also the last cells that divide in the next round (Fig. 3). The descendants of the row A form most of the first maxillary segment. Row B gives rise to the cell rows E(2) and E(3) which generate the material for the posterior margin of the first maxillary segment up to the anterior part of the first thoracic segment.
Cell proliferation in the area anterior to the ectoteloblasts. Continuation sequence of movie 2 (S2) from Fig. 2, anterior to the left, except for i where anterior is to the top. In the left column are pictures of the recording, in the right column are the corresponding stages as SIMI°BioCell reconstructions. The lines in the figures are bordering the cell rows. Time stamps show the time after the beginning of the recording in hours and minutes. a and b Stage before the start of the divisions in cell row E(1). The cells of row E(1) (white spheres) are bordered by the continuous line (posterior), the dotted lines separates it from the anterior sister row (red spheres). The cells of row B are posterior (right). Cells of row E(2) are bright blue spheres, cells from E(3) are represented as green spheres. The first row of cells of ectoteloblastic origin is in orange. Median cells are yellow. c and d After 5.5 h, the rows started with the divisions. The anterior daughter cells of the red cells are colored in green, the posterior ones in blue. Anterior cells are in bright yellow and the posterior ones in orange. The arrow points to a cell, which is positioned anteriorly to other cells of the row of the same origin due to a mediolateral division of a neighbour cell. e and f Nearly all cells of the rows have divided either in the anterioposterior or mediolateral direction. g and h After 33 h, the start of the recording all cells of the rows have divided. The SIMI°BioCell reconstruction clearly shows the rearrangements of the cells in contrast to their former position and origin (arrowheads). i Clonal composition of the descendants of row E(1) and their anterior sister row in the recorded embryo. The cell division angles are not in the AP direction; cells of anterior rows can become members of posterior rows
After formation of the second row of ectoteloblast descendants (eII) and before the divisions in the rows E(2) and E(3), the descendants of row A [corresponding to E(1)] and more anterior cells, start to divide. In embryo 2, these cell divisions do not follow a regular pattern (Supplementary material S2; Fig. 3). Lateral cells divide before median cells and no bilateral symmetry in the sequence is observed. The cell divisions follow the same sequence as that one cell cycle earlier (compare Figs. 2 and 3). The cells of row A are not organised bilaterally, as is found in rows E(2) and E(3). In the median region, not all cells divide in an anteroposterior direction, mediolaterally oriented cell divisions also take place (Fig. 3c,d). In embryo 2, single cells change their position from one row to the other, which demonstrates the absence of a positional determination by their lineage (Fig. 3e–h). This observation explains the impossibility of the re-identification of single cells in germ band preparations.
The first divisions of E(2) and E(3)
The development and the cell divisions of the rows E(2) and E(3) were analysed from the recording of embryo 3 (Supplementary material S3; Figs. 4 and 5). During formation of the row eIII by the ectoteloblasts and before the cells of row eIV are given off, row E(2) divides into the row E(2)ab and E(2)cd. At the same time E(3)ab and E(3)cd are formed from cells of row E(3). The cells of row E(2) and E(3) divide in an anterioposterior direction as the cells in the other rows of the post-naupliar germ band do during later development with the result, that all daughter cells lie behind each other. A mitotic wave, which progresses in one direction as in the posterior rows of ectoteloblastic origin, is not present.
Formation of the rows E(2) and E(3) in Porcellio scaber. Sequence of movie 3 (S3), anterior to the left. In the left column are pictures of the recording; in the right column are the corresponding stages as SIMI°BioCell reconstructions. Ectoteloblasts are in red; ectoteloblast derivatives are in orange. Cells of row E(2) are represented as magenta spheres, cells of row E(3) are in cyan. Median cells in yellow. Timestamp is in hours and minutes. a and b The ectoteloblasts give off the third row of their derivatives in the anterior direction (left) beginning from the midline and proceeding laterally. c and d The cells of the rows E(2) and E(3) start dividing after all ectoteloblasts have given off their third derivatives. e and f After 2.5 h, the cells of row E(2) and E(3) have nearly completed their divisions. The right half of the recorded embryo is slightly more progressed. g and h After 4.5 h, the cells of row E(2) and E(3) have divided before the ectoteloblasts will start to give off the next derivatives (row eIV)
Immigration of cells of the midline area into the embryo. Movie 4 (S4) shows immigrating cells of the ectoderm row which forms the mandible. A sequence of this movie is shown in this study together with the reconstruction with SIMI°BioCell, a–e anterior to the left, e view from posterior. a Asterisks mark the two cells which will immigrate into the embryo. Arrowheads point on the cells which will move to anterior thus closing the immigration point. b Asterisks mark the spheres of the immigrating cells in the SIMI°BioCell reconstruction. c and d Immigration and closure of the immigration point. e Position of the ectodermal cells after immigration of the two cells. f Same stage as e. The SIMI°BioCell reconstruction was rotated to show the position of the immigrated cells (arrows)
The first division of the row eI and the divisions of other ectoteloblast derivatives
In embryo 3 (Figs. 4 and 5; Supplementary material S3), the first division of a row of ectoteloblast descendants starts after the rows E(2) and E(3) have divided and before the row eIV has been formed. The divisions of the cells of row eI also do not progress in a wave from the midline laterally (see Supplementary material S3). The cell divisions of the ectoteloblast derivatives always start after two rows of ectoteloblast derivatives are formed by the ectoteloblasts. The cell cycle of the smaller ectoteloblast derivatives is exactly three times longer than the cell cycle of the large ectoteloblasts, which is in average 6.5 h. This is maintained throughout the development of the post-naupliar germ band. With the establishment of the mitotic waves from the middle to the lateral regions of the germ band, the mitotic waves of the ectoteloblast derivatives become more regular on both sides of the bilateral germ band. The ectoteloblast derivatives form the two rows ab and cd (not shown) in each genealogical unit.
The divisions of the descendants of row E(2) and E(3), ab and cd
Later cell divisions in E(2) and E(3) start in the row E(2)ab with a division oriented in direction of the a/p axis (Fig. 6). During this cell division, the ectoteloblast descendants eVII are formed by the ectoteloblasts (Fig. 6c). After that, the cells of row E(3)ab and E(2)cd start with cell divisions in the same orientation. Finally, the cells of the posterior row E(3)cd divide (Fig. 6c,d). All preparations of embryos of this stage show mitoses in each of these cell rows—but not in a reproducible sequence. The two halves of the germ band show no correlation in the sequence of the divisions but if one half is in progress in relation to the other, all rows of this germ band show an advanced development. This indicates an independence of the left from the right body half (Fig. 6c,d). The next row that divides is the row eIcd, followed by eIab, thus forming the rows a, b, c and d.
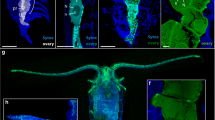
Different germ bands showing the variable pattern of en expression. a In this germ band anterior cells in the mandibular region express en still forming ‘bridges’ between adjacent en stripes. One cell is en positive although not connected to the other en-positive cells. b and c This germ band shows a small region of en expression in the mandibular segment. d The four rows of cells in the genealogical unit E(2) are formed. The cells of the anteriormost row a are expressing en in a stripe like pattern
Further divisions and expression of the gene en in E(1)
The divisions in the genealogical unit formed by row E(1) could not be reconstructed in detail. The en expression during these stages was analysed with the antibody 4D9 and could be compared between these preparations (Figs. 6 and 7). The cells of row E(2)ab and E(3)ab show a regular en expression pattern, so it is possible to identify them in every germ band preparation. Every embryo later forms rows composed of different precursor cells and no determined cell-lineage is observed in the mandibular and first maxillary part of the embryo at this early stage (Figs. 2 and 3). The lateral cells form a doubled row of cells. Because of the previously described sorting process in this region of the germ band and the later formation of the cell rows, a definition of row E(1) is required. The en expression in row E(2)ab can be used as a marker for the posterior border of E(1). E(1) divides into two rows of cells in which the anterior row first also expresses en, which is lost later in an irregular manner (Fig. 7a,b). Also, the midline cell of the genealogical unit of E(1) seems not to be determined early because it can be recruited from both sides (Fig. 3g,h; Supplementary material S2). In embryo 3, two of the median cells migrate into the embryo after row E(2) and E(3) have divided once (Fig. 5). The later fate of these cells remains unclear. The germ band preparations show at this stage three cells, and at later stages only one cell, which can be designated as the median cell. Two midline cells express en (Fig. 7a,b), which gets lost during the differential cell divisions in this area (Fig. 7e,f).
en expression during formation of the rows of non-ectoteloblastic origin in Porcellio scaber. DAPI counter-staining of the en distribution. a After the division of the cells posterior to the mandibular segment, the field of en expressing cells expands to posterior and to the lateral sides. This pattern differs between the individual germ bands. The anterior cells of the genealogical unit E(2) start with the en expression from the midline (ml) to the latral sides. b During the divisions of the cells of the genealogical unit E(2) into AP direction, the cells of the first maxillary segment start with their differential divisions. At this stage, the en expression is lost in the cells of the genealogical unit and only anterior cells are en positive. c Differential cell divisions of cells in the first maxillary segment. d Other germ band in a similar stage as in c. In this germ band, cells of first maxillary segment are still expressing en (arrowhead). e The cells of the genealogical units continue with the differential cell divisions and arrange into the grid like pattern. In the anterior part of the midline are two cells found in this germ band. f Germ band in a later stage after the first differential cell divisions in the segment of the first maxillae
Later in development, the cells show a stereotyped cell division pattern, in which the orientations of the cell divisions are bilaterally symmetrical (Fig. 7c–f). Four rows of cells result from these divisions, which are called a, b, c and d, as in the more posterior rows. Although en expression starts always in the midline area anterior to row E(1) (mandibular segment), the detailed pattern of the expression differs within each individual germ band (Figs. 6 and 7). Posterior cells gain en expression first and the field widens laterally. Later, midline cells lose their en expression. Subsequently to this development, more posterior cells establish another field of en expression, which will expand laterally to form the stripe of the next posterior segment anlage (first maxillary segment) (Fig. 6a). The two en stripes are separated by one cell row of which only a few cells express en (Fig. 6a). This is not observed in the more posterior rows of the P. scaber germ band or other malacostracan germ bands (Scholtz et al. 1993, 1994; Patel 1994;). The en expression in the intermediate cell row is lost later in development and the cells of this row begin the stereotyped cleavage pattern (Fig. 6a,b).
The field of the en expressing cells of the mandibular segment anterior to row E(1) has different dimensions in each investigated germ band. In some germ bands, the en expression domain is concentrated in an area of cells close to the midline (Fig. 6a); in others, it expands laterally up to seven cells wide (Fig. 7d). In the more posterior genealogical units of the germ band, the cells form four more or less regular rows (a, b, c and d) in which the anterior row a expresses en. The cells of the anterior row ab are also the first that divide with an anteroposteriorly oriented spindle (Fig. 6b). After this division, the posterior cells (descendant row b) lose the en expression resulting in the pattern of one anterior en positive row followed by three en negative rows of cells, a pattern which is also observed in other malacostracans (Patel et al. 1989a; Scholtz et al. 1993, 1994; Scholtz and Dohle 1996).
The differential cleavages
The differential cleavages are the first divisions after which the cells in every segment show distinct differences. At this stage, every cell in the segment divides in a taxon-specific stereotyped and conserved pattern (see Dohle et al. 2004). Neuroblasts, segment border cells and leg cells start to behave according to their future fate. The 4D-recording (supplementary material S4) of one segment (T5) of the post-naupliar germ band shows the timing, sequence and angles of the cell divisions and the morphogenetic events such as segment formation and limb bud formation during the process of development.
Discussion
The relationships within the Peracarida—phylogenetic implications from cell division pattern in the anterior part of the germ band
The phylogenetic relationships among the main taxa of the peracarids (Isopoda, Tanaidacea, Amphipoda, Mysidacea, Cumacea, Mictacea, Spelaeogriphacea) are controversial (Siewing 1951; Schram 1984, 1986; Richter and Scholtz 2001; Schram and Koenemann 2001). These relationships are crucially important, however, for the understanding of the evolution of complex characters e.g. the uniramous leg of amphipods and isopods (Hejnol and Scholtz 2004). Our study of the formation of the anterior post-naupliar segments of isopods fills a gap in the cell lineage studies previously performed in the different peracarid taxa. The first detailed description of the cell lineage was done by Dohle (1970, 1976) in the Cumacean Diastylis, followed by descriptions in the Tanaidacean Leptochelia (Dohle 1972), Mysidacea (Scholtz 1984) and Amphipoda (Scholtz 1990). These studies describe a field of cells in the anterior part of the germ band not formed by ectoteloblasts, which form the posterior segments of the post-naupliar germ band (Fig. 8). The slight modifications of the cell division patterns between the taxa, but with enough conserved features to allow homologisation, makes them useful for systematic analyses (Dohle 1989). Additional studies of the expression of the gene en in some of these groups (Amphipoda, Mysidacea) enhance the number of characters which can be used in comparisons (Scholtz et al. 1993; Scholtz and Dohle 1996). In all taxa of the peracarid malacostracans that have been investigated [Isopoda: (Hahnenkamp 1974; Vehling 1994; Hejnol and Scholtz 2004; this study) Tanaidacea: (Dohle 1972), Cumacea: (Dohle 1970, 1976) and Mysidacea: (Scholtz 1984)], rows of cells anterior to the ectoteloblasts are present that form the post-naupliar segments of the maxillae and part of the first thoracic segment but which are not of ectoteloblastic origin (Fig. 8). In the amphipods, no segments of the post-naupliar germ band are generated by ectoteloblasts due to the secondary loss of ectoteloblasts in amphipod evolution (Scholtz 1990; Gerberding et al. 2002; Scholtz and Wolff 2002). Outside the Peracarida, anterior post-naupliar rows of cells of non-ectoteloblastic origin also exist in the decapod Cherax destructor (Scholtz 1992). Later in development, these rows of cells form the rows E(2), E(3) and E(1) and in some taxa an additional cell row, which takes part in the formation of the first maxillae is already present [E(0)] (Fig. 8b). In the Cumacea, Mysidacea and Amphipoda, the rows of cells arrange out of scattered cells. In the Tanaidacea, the rows E(2) and E(3) are formed from a single row B through oriented cell divisions in the A/P direction (Fig. 8). The rows E(0) and E(1) are similarly formed from row A. Earlier results for isopods are conflicting: Hahnenkamp (1974) and Vehling (1994) could not clarify the formation of these rows. Our direct observation by using the 4D-microscopic system resolved the problems which a reconstruction by using series of fixed stages could not solve. We observed cells in front of the ectoteloblasts, which are not their derivatives. After formation of the first row of ectoteloblast derivatives, the cells in front of the ectoteloblast start with a division into the AP direction. During this process, the rows of cells become more and more regular. Four rows of cells are formed in front of the ectoteloblast derivatives. The first two rows have the same position and react similar to the rows Dohle (1972) describes as A and B in a tanaidacean (Fig. 8). In the tanaidacean and in P. scaber the row B divides before A, thus forming the genealogical cell rows E(2) and E(3). Row A forms the largest part of the segment of the first maxillae. In no other investigated peracarid do the rows in front of the ectoteloblasts derive from two single rows of cells. This similarity in the row formation via precursor rows A and B indicate a sister group relationship between tanaidaceans and isopods (see Richter and Scholtz 2001) because a comparable pattern is absent in the other malacostracans studied in this respect.
Comparison of the cell division pattern, en expression and the row formation between malacostracan taxa. a Modified drawings of the originals (Dohle 1972, 1976; Scholtz 1984, 1990; Scholtz et al. 1993). en expressing cells so far investigated are in blue (Scholtz et al. 1993, 1994). In all malacostracan crustaceans, the segment of the first maxillae is formed by four cell rows, which are en negative in the investigated species. b Schematic drawing of the non-ectoteloblastic row formation in the different malacostracan taxa. In the Amphipoda, Cumacea and Mysidacea cells are arranged into a grid like pattern. In the Tanaidacea and Isopoda at least the rows E(2) and E(3) are formed out of the cell row B. The dotted lines indicate the future segmental borders
Cell sorting vs cell-lineage in the malacostracan germ band
At first sight, the conserved stereotyped cleavage pattern in the post-naupliar germ band of malacostracan crustaceans suggests a role in lineage-dependent cell fate determination. However, recent investigations on the expression of the genes en and Distal-less in the malacostracan germ band at the cellular level dispute this view and suggest that gene expression patterns are independent of lineage (Scholtz et al. 1993; Scholtz and Dohle 1996; Hejnol and Scholtz 2004). Our study of the anterior part of the post-naupliar germ band shows that a regular cell division pattern is absent in this area and that cell sorting is present similar to that of other arthropod germ bands. The previously described development of other peracarid taxa shows a regular cell division pattern in this part of the germ band (Dohle 1970, 1972, 1976; Scholtz 1984, 1990, 1992). Furthermore, the species in which en expression was studied show a regular expression pattern from the beginning (Scholtz et al. 1994). Dohle (1970, 1972, 1976) and Scholtz (1984, 1990, 1992) were able to identify and name the cells present in the anterior part and to reconstruct the cell lineage from fixed germ bands of different stages. In our study on the isopod P. scaber, only the use of the 4D-microscopy system enabled us to study the cell lineage of this area and to detect the cell sorting. It is interesting to note that these cells that originate from an irregular cell division later show a stereotyped cell division pattern, which makes it possible to homologise single cells with that of some other peracarids even though they have a different lineage.
Gene expression, cell lineage, cell identity and the question of homology
A comparison of post-naupliar germ band development in malacostracans shows the conserved grid-like arrangement of the ectodermal cells, a similar gene expression pattern of the gene en in stripes and a similar cell division pattern. First, the grid-like pattern is established, either by the formation of the rows by ectoteloblasts or the arrangement of scattered cells, as in amphipods. Then, en is expressed in a regular sequence from the midline to the lateral sides. After the establishment of the en stripes, the cells start to cleave in a stereotypic fashion—the differential cleavages (see Dohle and Scholtz 1988). However, in the isopod germ band described in this study, this sequence differs. The irregularly arranged cells of the anterior post-naupliar region begin to express en in an irregular pattern. Later, with the arrangement of the cells into a grid-like pattern, the expression of en is lost in some cells and gained in others, thus forming the stripes typical for the other investigated groups. The subsequent sequence of stereotypic cell divisions is then performed. These processes are clearly heterochronically shifted relative to one another in P. scaber.
This separation of sub-processes raises questions about the homology of the developmental stages among the malacostracan groups. The homology of single cells can be detected by their ‘identity’ later in development even though these cells are of a different origin in each individual, due to their position in the grid-like pattern of cells and the expression of developmental genes. However, what we observe in P. scaber is not only an interspecific variation, but also an intraspecific variability between each individual embryo. But what is ‘sufficient’ to homologise single cells in the germ band? How can we homologise cells at later stages when it is not possible earlier? The still-debated question of whether gene expression patterns are sufficient to homologise whole organs (e.g. Dickinson 1995; Bolker and Raff 1996; Nielsen and Martinez 2003; Sanetra et al. 2005; Scholtz 2005) is shifted to the level of individual cells in our comparative study. Through our examination of en expression in combination with cell lineage data, it becomes obvious that gene expression of the gene en alone is not sufficient to claim homology of a single cell, and—due to the process of cell sorting—the cells which express en in the later grid-like pattern are not of the same origin from embryo to embryo. Such an uncoupling of en expression from cell genealogy and cell sorting has been described also for Drosophila (Vincent and O’Farrell 1992). The inclusion of the isopod species in our comparative study shows that cell origin, gene expression and cell fate can be separated at different stages of development yet still converge later to produce one homologous stage. In this stage, the homology of individual cells in the germ band seems to be clear, although we can show that these cells are not of the same origin. In the case of the segmental grid-like pattern, the future fate of the descendants of some cells contributes to an endopodite or exopodite, regarding the formation of a uniramous or biramous leg in the corresponding taxon (Hejnol and Scholtz 2004; Wolff 2004). All these structures can be homologised between taxa though also formed by cells of different origin. Our results support the notion of Dohle (1989) and Scholtz (2005) that developmental stages can be modified during evolution without affecting later stages, and that homologous developmental stages are not necessarily based on homologous earlier stages nor necessarily followed by homologous later stages. How this is achieved during development and how the information required for the specification of evolutionarily conserved later stages is transformed during variable ways of development still remains a mystery and is one of the biggest challenges of ‘evo–devo’ research.
References
Anderson DT (1973) Embryology and phylogeny in annelids and arthropods. Pergamon, Oxford
Averof M, Akam M (1993) HOM/Hox genes of Artemia: implications for the origin of insect and crustacean body plans. Curr Biol 3:73–78
Bolker JA, Raff RA (1996) Developmental genetics and traditional homology. BioEssays 18:489–494
Broadus J, Doe CQ (1995) Evolution of neuroblast identity: seven-up and prospero expression reveal homologous and divergent neuroblast fates in Drosophila and Schistocerca. Development 121:3989–3996
Browne WE, Price AL, Gerberding M, Patel NH (2005) Stages of embryonic development in the amphipod crustacean, Parhyale hawaiensis. Genesis 42:124–149
Damen WG, Weller M, Tautz D (2000) Expression patterns of hairy, even-skipped, and runt in the spider Cupiennius salei imply that these genes were segmentation genes in a basal arthropod. Proc Natl Acad Sci USA 97:4515–4519
Dearden PK, Akam M (2001) Early embryo patterning in the grasshopper, Schistocerca gregaria: wingless, decapentaplegic and caudal expression. Development 128:3435–3444
Dickinson WJ (1995) Molecules and morphology: where’s the homology? Trends Genet 11:119–121
Dohle W (1970) Die Bildung und Differenzierung des postnauplialen Keimstreifs von Diastylis rathkei (Crustacea, Cumacea) I. Die Bildung der Teloblasten und ihrer Derivate. Z Morphol Tiere 67:307–392
Dohle W (1972) Über die Bildung und Differenzierung des postnauplialen Keimstreifs von Leptochelia spec. (Crustacea, Tanaidacea). Zool Jb Anat 89:503–566
Dohle W (1976) Die Bildung und Differenzierung des postnauplialen Keimstreifs von Diastylis rathkei (Crustacea, Cumacea). II. Die Differenzierung und Musterbildung des Ektoderms. Zoomorphologie 84:235–277
Dohle W (1989) Zur Frage der Homologie ontogenetischer Muster. Zool Beitr N F 32:355–389
Dohle W, Scholtz G (1988) Clonal analysis of the crustacean segment: the discordance between genealogical and segmental borders. Development 104 (Suppl):147–160
Dohle W, Gerberding M, Hejnol A, Scholtz G (2004) Cell lineage, segment differentiation and gene expression in crustaceans. In: G. Scholtz. (eds) Evolutionary developmental biology of Crustacea A. A. Balkema Lisse pp 95–133
Gerberding M, Browne WE, Patel NH (2002) Cell lineage analysis of the amphipod crustacean Parhyale hawaiensis reveals an early restriction of cell fates. Development 129:5789–5801
Hahnenkamp L (1974) Die Bildung und Differenzierung des Keimstreifens der Asseln (Isopoda) und anderer höherer Krebse. Eine vergleichend-embryologische Studie. Zulassungs-arbeit für die erste (wissenschaftliche) Staatsprüfung Berlin
Hejnol A, Scholtz G (2004) Clonal analysis of Distal-less and engrailed expression patterns during early morphogenesis of uniramous and biramous crustacean limbs. Dev Genes Evol 214:473–485
Hejnol A, Schnabel R (2005) The eutardigrade Thulinia stephaniae has an indeterminate development and the potential to regulate early blastomere ablations. Development 132:1349–1361
Hejnol A, Schnabel R (2006) What a couple of dimensions can do for you: comparative developmental studies using 4D-microscopy-examples from tardigrade development. Integ Comp Biol 46:151–161
Lawrence PA (1992) The making of a fly. Blackwell Scientific Publications, Oxford
Maderspacher F, Bucher G, Klingler M (1998) Pair-rule and gap gene mutants in the flour beetle Tribolium castaneum. Dev Genes Evol 208:558–568
Nielsen C, Martinez P (2003) Patterns of gene expression: homology or homocracy? Dev Genes Evol 213:149–154
Patel NH (1994) The evolution of arthropod segmentation: insights from comparisons of gene expression patterns. Development (Suppl):201–207
Patel NH, Kornberg TB, Goodman CS (1989a) Expression of engrailed during segmentation in grasshopper and crayfish. Development 107:201–212
Patel NH, Martin-Blanco E, Coleman KG, Poole SJ, Ellis MC, Kornberg TB, Goodman CS (1989b) Expression of engrailed proteins in arthropods, annelids, and chordates. Cell 58:955–968
Pavlopoulos A, Averof M (2005) Establishing genetic transformation for comparative developmental studies in the crustacean Parhyale hawaiensis. Proc Natl Acad Sci USA
Richter S, Scholtz G (2001) Phylogenetic analysis of the Malacostraca (Crustacea). J Zool Syst Evol Res 39:113–136
Sanetra M, Begemann G, Becker MB, Meyer A (2005) Conservation and co-option in developmental programmes: the importance of homology relationships. Front Zool 2:15
Schnabel R, Hutter H, Moerman D, Schnabel H (1997) Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: variability of development and regional specification. Dev Biol 184:234–265
Scholtz G (1984) Untersuchungen zur Bildung und Differenzierung des postnauplialen Keimstreifs von Neomysis integer LEACH (Crustacea, Malacostraca, Peracarida). Zool Jb Anat 112:295–349
Scholtz G (1990) The formation, differentiation and segmentation of the post-naupliar germ band of the amphipod Gammarus pulex L. (Crustacea, Malacostraca, Peracarida). Proc R Soc Lond B 239:163–211
Scholtz G (1992) Cell lineage studies in the crayfish Cherax destructor (Crustacea, Decapoda): germ band formation, segmentation, and early neurogenesis. Roux’s Arch Dev Biol 202:36–48
Scholtz G (1997) Cleavage, germ band formation and head segmentation: the ground pattern of the Euarthropoda. In: Fortey RA, Thomas RH (eds) Arthropod relationships. Chapman & Hall, London, pp 317–332
Scholtz G (2005) Homology and ontogeny: pattern and process in comparative developmental biology. Theory Biosci 124:121–143
Scholtz G, Dohle W (1996) Cell lineage and cell fate in crustacean embryos—a comparative approach. Int J Dev Biol 40:211–220
Scholtz G, Wolff C (2002) Cleavage, gastrulation, and germ disc formation of the amphipod Orchestia cavimana (Crustacea, Malacostraca, Peracarida). Contrib Zool 71:9–28
Scholtz G, Patel NH, Dohle W (1994) Serially homologous engrailed stripes are generated via different cell lineages in the germ band of amphipod crustaceans (Malacostraca, Peracarida). Int J Dev Biol 38:471–478
Scholtz G, Dohle W, Sandeman RE, Richter S (1993) Expression of engrailed can be lost and regained in cells of one clone in crustacean embryos. Int J Dev Biol 37:299–304
Schram F (1984) Relationships within eumalacostracan Crustacea. Trans S Diego Soc Nat Hist 20:301–312
Schram F (1986) Crustacea. Oxford University Press, New York, Oxford
Schram FR, Koenemann S (2001) Developmental genetics and arthropod evolution: part 1, on legs. Evol Dev 3:343–354
Siewing R (1951) Besteht eine engere Verwandtschaft zwischen Isopoden und Amphipoden? Zool Anz 147:166–180
Vehling D (1994) Die Entwicklung des postnaulialen Keimstreifs von Porcellio scaber. Eine zellgenealogische Studie. Diplomarbeit, Freie Universität Berlin
Vincent JP, O’Farrell PH (1992) The state of engrailed expression is not clonally transmitted during early Drosophila development. Cell 68:923–931
Wolff C (2004) Zur Beinentwicklung des amphipoden Krebses Orchestia cavimana (Peracarida, Malacostraca)—eine zellgenealogische Studie, Humboldt Universität Berlin
Acknowledgements
We thank Nipam Patel and Harald Saumweber for the monoclonal Antibody 4D9 and Craig Magie for improving the manuscript. This work was supported by the DFG (Scho 442/5-2,3).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Roth
Electronic supplementary material
Below is the link to the electronic supplentary material.
Time lapse of the gastrulation in Porcellio scaber. 30 h in 60 s. View on the cells of the germ disc. The cells divide in two division waves and start to sink into the gastrulation center on the left. The future anterior orientated to the top/left. (MOV 22 MB)
Time lapse of the division of the cells in row A and B in Porcellio scaber. 32 h in 60 s. The SIMI°BioCell reconstruction in the left upper corner. Every sphere represents the cells described as in the publication. Anterior is to the left. (MOV 26 MB)
Time lapse of the division of the cells in the genealogical unit E(2) and E(3) in Porcellio scaber. 20 h in 42 s. The SIMI°BioCell reconstruction in the right corner. The ectoteloblasts give off their derivates to anterior (left). (MOV 11 MB)
Time lapse of the differential divisions in the right half of a thoracal segment in Porcellio scaber. 33 h in 80 s. During the movie, a reconstruction of the differential cleavages with SIMI°BioCell shows up in the lower left corner. The movie ends during limb bud formation. Anterior is to the top. (MOV 22 MB)
Rights and permissions
About this article
Cite this article
Hejnol, A., Schnabel, R. & Scholtz, G. A 4D-microscopic analysis of the germ band in the isopod crustacean Porcellio scaber (Malacostraca, Peracarida)—developmental and phylogenetic implications. Dev Genes Evol 216, 755–767 (2006). https://doi.org/10.1007/s00427-006-0105-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-006-0105-4