Abstract
The arthropod head is a complex metameric structure. In insects, orthodenticle (otd) functions as a ‘head gap gene’ and plays a significant role in patterning and development of the anterior head ectoderm, the protocerebrum, and the ventral midline. In this study, we characterize the structure and developmental deployment of two otd paralogs in the amphipod crustacean, Parhyale hawaiensis. Photd1 is initially expressed at gastrulation through germband stages in a bilaterally symmetric, restricted region of the anterior head ectoderm and also in a single column of cells along the ventral midline. Late in embryogenesis, Photd1 is expressed within the developing anterior brain and the expression along the embryonic midline has become restricted to a stereotypic group of segmentally reiterated cells. The second ortholog Photd2, however, has a unique temporal–spatial expression pattern and is not detected until after the head lobes have been organized in the developing ectoderm of the germband during late germband stages. Anteriorly, Photd2 is coincident with the Photd1 head expression domain; however, Photd2 is not detected along the ventral midline during formation of the germband and only appears in the ventral midline late in embryonic development in a restricted group of cells distinct from those expressing Photd1. The early expression of Photd1 in the anterior head ectoderm is consistent with a role as a head gap gene. The more posterior expression of Photd1 is suggestive of a role in patterning the embryonic ventral midline. Photd2 expression appears too late to play a role in early head patterning but may contribute to latter patterning in restricted regions of both the head and the ventral midline. The comparative analysis of otd reveals the divergence of gene expression and gene function associated with duplication of this important developmental gene.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genes that control the anterior head and nervous system of the fly, Drosophila melanogaster, have guided ideas regarding the development and evolution of the arthropod head. In Drosophila, a small number of transcription factor genes play a major role in the specification of the anterior head ectoderm and the anterior-most neuromeres that comprise the supraesophageal ganglion (Younossi-Hartenstein et al. 1997). These ‘head gap genes’ are deployed very early in fly development in overlapping functional domains (Cohen and Jürgens 1990; Schmidt-Ott et al. 1994) and their initially broad anterior domains of expression are progressively restricted and sharpened during the processes of both cellularization and gastrulation (Dalton et al. 1989; Finkelstein and Perrimon, 1990; Walldorf and Gehring 1992; Wimmer et al. 1993; Grossniklaus et al. 1994; Wimmer et al. 1997). The resulting anterior expression domains demarcate specific regions of the developing Drosophila brain (Hirth et al. 1995). Among these genes, orthologs of orthodenticle (otd) have been demonstrated to play a central role in anterior head and brain development in disparate bilaterians (e.g., Acampora et al. 1998; reviewed in Arendt and Nubler-Jung 1996; Sharman and Brand 1998; Lichtneckert and Reichert 2005).
In Drosophila and other insects, the supraesophageal ganglia can be subdivided along the neuroaxis, from anterior to posterior, into three neuromere subunits the protocerebrum (PC), deutocerebrum (DC), and tritocerebrum (TC) (Reichert and Boyan 1997). In Drosophila, otd is initially expressed in a wide anterior circumferential stripe in the blastoderm embryo (Finkelstein and Perrimon 1990; Cohen and Jürgens 1991). As development progresses, the otd expression boundary becomes restricted to the embryonic brain and is detected throughout the developing PC neuromere and in the anterior DC neuromere. Mutant otd alleles delete or severely reduce the size of the PC neuromere and the PC-associated preoral medial commissure (Hirth et al. 1995). otd appears to, in part, functionally act anteriorly in the specification of cell identity via regulation of proneural gene expression (Younossi-Hartenstein et al. 1997), the intrasegmental regulation of the segment polarity genes engrailed (en) and wingless (wg) in embryos (Gallitano-Mendel and Finkelstein 1998), and later in larval and pupal development of medial structures including the ocelli of the adult fly (Wieschaus et al. 1992; Royet and Finkelstein 1995; Hirth et al. 1995). In addition, reciprocal negative regulation between the gene unplugged (unpg) and otd suggests that otd plays an important role in specification of the anterior boundary between the DC and TC neuromeres in Drosophila (Hirth et al. 2003; Lichtneckert and Reichert 2005). Expression of otd is also prominent in the embryonic ventral midline of Drosophila and plays an important role in the patterning of the segmentally reiterated ventral nerve cord (VNC) commissures, particularly the posterior commissure (Finkelstein et al. 1990; Klämbt et al. 1991).
The embryonic development of both the head and ventral midline in Drosophila is distinctive from most arthropods. Unique to the dipteran flies, the anterior head segments involute in late stages of development and the ventral midline is generated from a distinct group of mesectodermal cells. Given the highly divergent mode of embryonic head development in flies, the expression patterns of orthodenticle orthologs have been examined in an insect with less-derived embryonic development, the beetle Tribolium castaneum. In contrast to Drosophila, Tribolium possesses two orthologs of otd, Tcotd-1, and Tcotd-2 (Li et al. 1996). The expression of the two Tribolium otd paralogs are divergent with respect to each other as well as to their putative ortholog in Drosophila (Li et al. 1996). Knockdown of Tcotd-1 function via parental RNAi produces a headless embryonic phenotype that is more severe than the Drosophila gap gene phenotype, which indicates an important anterior determinant role of Tcotd-1 in Tribolium (Schröder 2003). It is notable that the effects of knockdown restricted to zygotic expression of Tcotd-1 were generally more moderate and did not eliminate the entire head but resemble the Drosophila gap gene phenotype (Schröder 2003). Thus, while Tcotd-1 retains an early anterior gap gene patterning function analogous to its ortholog otd in Drosophila, Tcotd-1 has an additional functional role in specifying the entire head region in Tribolium. This observation in Tribolium suggests the possibility of otd serving as an anterior determinant in other arthropods.
In the absence of equivalent information from additional arthropod taxa, these data can only apply to the most recent common ancestor of holometabolous beetles and flies. Thus, it is problematic to generalize details regarding the mode of ancestral arthropod head development exclusively from data produced in flies and other insects. Among arthropod taxa more distantly related to insects, only limited information is currently available. In the chelicerate Archegozetes longisetosus, expression of a single orthodenticle ortholog has been reported and cursory examination of expression during late embryogenesis reveals Alotd in an anterior head domain as well as along the ventral midline similar to Tcotd-1 in Tribolium (Telford and Thomas 1998). Recent work suggests that the crustaceans are sister taxa to the insects (Friedrich and Tautz 1995; Dohle 2001; Giribet et al. 2001, 2005; Hwang et al. 2001; Richter 2002; Cook et al. 2005; Regier et al. 2005). Comparative neuroanatomy of crustaceans (e.g., Hanstrom 1928; Sandeman et al. 1992; Gerberding 1997; Strausfeld 1998; Harzsch 2003) suggests that a similar and presumably homologous supraesophageal neuromere ground plan is shared with the insects (Sandeman et al. 1992). Additional comparative studies between crustaceans and insects have suggested both strong similarities and notable differences in neuronal morphology (Whitington et al. 1993; Whitington 1996). These observations have been extended to suggest homologies among neuronal identities by correlating neuronal morphology with the expression of molecular markers (Duman-Scheel and Patel 1999; Browne et al. 2005). Using these studies, our understanding of insect neurogenesis, in combination with the embryonic staging and cell lineage data available for Parhyale hawaiensis (Gerberding et al. 2002; Browne et al. 2005), provides a framework from which to make a detailed analysis of the expression of head patterning orthologs in a model crustacean. At the level of segment morphology and segment specification, the heads of crustaceans, such as Parhyale, are quite different from the heads of insects, such as Drosophila. For example, insects do not possess paired appendages on the intercalary segment whereas crustaceans possess antennae on the homologous segment (An2). In addition, many aspects of the olfactory systems of insects and crustaceans appear to be considerably divergent (Strausfeld 1998; Strausfeld and Hildebrand 1999). Thus, the a priori expectation would be for gene orthologs controlling the morphology of insects to have divergent temporal and spatial expression in crustaceans.
In this paper, we report detailed expression patterns associated with orthologs of otd in the model amphipod crustacean, P. hawaiensis. To further characterize anterior head and ventral midline development in Parhyale, we cloned otd orthologs and carried out a detailed study of their expression during embryogenesis via in situ mRNA transcript analysis. This analysis, when paired with available sequence and expression data in other taxa, allowed us to assess the relationship between otd orthologs and paralogs among arthropods. Our analysis suggests an evolutionary scenario accounting for the gene duplication and expression characteristics associated with otd and is informative with regard to reconstructing the ancestral state of the most recent common ancestor between crustaceans and insects, a marine arthropod stem species existing during the Cambrian ∼550 mya.
Materials and methods
Amphipod culture
P. hawaiensis is maintained as reported in Browne et al. (2005) with the following modifications. The breeding colony is kept in interconnected shallow plastic trays at ∼30°C at the Kewalo Marine Lab. Recirculating seawater is provided via magnetic drive pump (Iwaki). Fresh, filtered seawater is added at regular intervals. Animals are fed a liquefied mixture of algae, plankton, fatty acids (Selco), and vitamins (Kent Marine).
Cloning and sequence analysis
Total RNA was isolated with TRIzol Reagent (Gibco BRL) from a pool of mixed-stage P. hawaiensis embryos. First strand cDNA was generated with the SuperScript Preamplification System (Gibco BRL). Initial degenerate PCR was completed using conserved nested primers to aligned otd proteins (forward primer 5′-GAR MGN CAN CAN TTY AC-3′, reverse outer primer 5′-NCK NCK RTT YTT RAA CCA-3′, and reverse inner primer 5′-CAR YTN GAY GTN YTN GA-3′).
For isolation of additional 5′ sequences, polyA RNA was isolated using the Micro Poly(A)Pure Small Scale mRNA Purification Kit (Ambion) from a pool of P. hawaiensis embryos. First strand cDNAs were generated with the SMART PCR cDNA Synthesis Kit (BD Biosciences). Initial degenerate PCR was completed using the forward primer 5′-CAG MGG MGG GAR MGI ACI ACI TTY AC -3′ and reverse primer 5′-GC CCK CCK RTT YTT RAA CCA IAC YTG-3′. Sequences 5′ of the homeodomain were obtained via 5′RACE (SMART RACE cDNA Amplification Kit, BD Biosciences) with nested sequence-specific primers obtained from the initial degenerate PCR. Sequences 3′ of the homeodomain were obtained by performing 3′RACE (Gibco BRL) with sequence-specific nested primers obtained from the initial degenerate PCR (primers available upon request).
Phylogenetic analysis
Maximum parsimony (MP) and maximum likelihood (ML) analyses were executed with PAUP* 4.0Beta (Swofford 2003). Bayesian phylogenetic inference analysis was executed with MrBayes 3.1.1 (Ronquist and Huelsenbeck 2003). Relevant orthodenticle sequences were selected from those available via genbank; all available arthropod sequences were used. Searches were rooted with the D. melanogaster paired gene and resulting trees were visualized with TreeView X (Page 1996).
A mutational saturation plot (Philippe et al. 1994; Philippe and Forterre 1999) was generated using a MP analysis with ten random sequence addition MP heuristic searches. The best tree was used to generate a patristic distance matrix. In additional parsimony analyses, MP heuristic searches were subjected to 2,000 bootstrap replicates, each with a random addition MP heuristic search.
For ML analyses, 1,000 random sequence addition ML heuristic searches were run. Searches were then subjected to 2,000 bootstrap replicates, each with a random addition ML heuristic search. The molecular evolution model, HKY + G, was selected using Model-Test 3.7 (Posada and Crandall 1998) for ML analysis.
For Bayesian analyses, all parameters were unlinked except topology. Two different codon partition schemes were generated: (1) codon position partition 1, 2, 3 or (2) codon position partition 1+2, 3. Multiple searches were run from 2 million generations to 5 million generations and trees were sampled every 100 generations. Posterior estimates from runs were analyzed by eye with Tracer v1.3 (Rambaut and Drummond 2003, Tracer v1.3, http://www.evolve.zoo.ox.ac.uk) to determine the number of generations to burn in and to assess convergence of data sets. Consensus trees from independent runs within each partition scheme were compared to assess convergence and topology congruence of data sets.
In situ hybridization
P. hawaiensis embryos were removed from the ventral brood pouch and allowed to develop to the desired stage at 26°C (Browne et al. 2005), then dissected with tungsten wire needles and fixed for 60 min in filtered seawater mixed 9:1 with 37% formaldehyde at room temperature. Fixed embryos were then washed with PT (1× PBS, pH 7.4, and 0.1% Triton X-100) and stored in 100% MeOH at −20°C. Whole-mount in situ hybridization using digoxigenin-labeled RNA was performed as reported in Finnerty et al. (2003), with the following changes. Before hybridization, fixed embryos were incubated for 60 min in a sodium dodecyl sulfate (SDS)-based detergent solution [1% SDS, 0.5% Tween-20, 50 mM Tris–HCl (pH 7.5), 1.0 mM EDTA (pH 8.0), and 150 mM NaCl] at 65°C. After hybridization, embryos were intensively washed with 2× SSC solution at 65°C to remove hybridization buffer and free probe. After the 65°C washes, embryos were then washed at room temperature in PBT (1× PBS containing 0.1% bovine serum albumin and 0.1% Triton X-100). Embryos were then incubated in sheep anti-digoxigenin-AP (1:3,000) overnight at 4°C and subsequently washed with PBT before performing BCIP/NBT color reaction. After color reaction, embryos were washed with PT and incubated overnight at 4°C in 50% glycerol + 1 mg/ml Hoechst dye. Embryos were stored in 70% glycerol at −20°C. Images were generated from dissected and mounted embryos using an Axioskop2 (Zeiss) and Axio Imager Z1 (Zeiss) and photographed with either an Axiocam (Zeiss) or an ORCA ER (Hamamatsu) using MAC-compatible Openlab and Velocity (Improvision) imaging software.
Antibody staining
Staged P. hawaiensis embryos (Browne et al. 2005) were fixed in the same way as for in situ but for a shorter period of time (15 min). Antibody detection was performed as reported in Patel (1994). All antibody incubations were done overnight at 4°C at the following concentrations: mouse anti-acetylated tubulin primary antibody (Hybridoma Bank, clone E7) 1:1 with PBT, alexa-594 conjugated goat anti-mouse secondary antibody (Molecular Probes) 1:250 with PBT, and HRP conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch). Embryos were then washed and incubated overnight at 4°C in 50% glycerol + 1 mg/ml Hoechst solution and stored in 70% glycerol at −20°C. Images were generated from dissected and mounted embryos as described above.
Results
Isolation and characterization of P. hawaiensis otd orthologs
Two P. hawaiensis otd sequences, Photd1 and Photd2, were recovered from repeated degenerate PCR screens on independent pools of Parhyale cDNA. The isolated fragments (103 bp) were sufficient for unambiguous orthology identification. Nested non-overlapping primers were designed to the two initial degenerate PCR fragments and used to amplify the remaining mRNA transcript sequences both 5′ and 3′ of the initial homeodomain fragments. All of the Photd1 sequences are identical with minor variations in the length of the recovered transcripts. The longest recovered complete Photd1 cDNA was 2 kb. In contrast, the organization of the Photd2 paralog is significantly more complex. The Photd2 gene has multiple splice variants both 5′ and 3′ of an invariant homeodomain core sequence (supplemental data). RACE yielded five unique open reading frame sequences upstream of the homeodomain and three unique open reading frame sequences downstream of the homeodomain. Therefore, the Photd2 gene appears to be capable of generating potentially 15 unique transcripts; however, the true number of Photd2 transcripts produced from these recovered alternative splice variants is unknown.
Phylogenetic analysis
The orthodenticle homeodomains show extreme amino acid conservation whereas regions both upstream and downstream of the homeodomain are too variable among orthologs to reliably align in distantly related taxa. In addition, due to Photd2 possessing multiple splice variants both upstream and downstream of the homeodomain, we restricted our phylogenetic analyses of orthodenticle genes in disparate taxa to the homeodomain region. Our survey of metazoan taxa revealed different complements of otd paralogs in different groups (Fig. 1). In cases in which two paralogs exist, the paralogs appear more similar to each other than to orthologs in other species. We propose two readily distinguishable modes of gene expansion are likely to have occurred to generate the range of otd paralogs across metazoans. In the lineage leading to the vertebrates, whole-genome duplication has generated additional paralogous genes (Germot et al. 2001) which, in most cases, are found on different chromosomes. In contrast, in the cnidarian (Finnerty et al., submitted for publication) and arthropod lineages for which genomic information is available, tandem gene duplication appears to have generated paralogous genes in close proximity to one another. However, it is unclear whether duplications occurred independently in each taxon or at particular nodes in the metazoan tree. Specifically in the arthropod lineage leading to the Tetraconata (crustaceans + insects) (Dohle 2001), it is likely that a single duplication of an ancestral otd gene occurred to give rise to two paralogs in descendant taxa (Fig. 1). Within any given lineage arising after this duplication, otd paralogs are likely to have been subjected to the effects of two homogenizing phenomena: (1) increased probability of gene conversion (mechanism by which mismatched heteroduplexed DNA basepairs between gene copies are repaired) due to the proximity of the paralogs and (2) the effects of mutational saturation. Both of these phenomena would contribute to otd paralogs becoming more similar to one another than to otd orthologs in related taxa. As a test for mutation saturation among otd paralogs, we generated a mutational saturation plot (supplemental data). The presence of a mutational plateau indicates that saturation for mutational changes has occurred, therefore indicating a loss of phylogenetic signal necessary to accurately resolve branch lengths (Philippe et al. 1994; Philippe and Forterre 1999). Thus, node relationships could not be recovered to generate a comprehensive, reliable, gene tree indicating orthology among otd paralogs using traditional phylogenetic tools (data not shown). In summary, the orthology of the two Parhyale genes to orthodenticle is clear; however, whether multiple independent gene duplications have occurred in different lineages or whether an early metazoan ancestor already had separate otd paralogs cannot be ruled out based on available evidence (Fig. 1).
A model proposing evolutionary duplication and loss of orthodenticle paralogs in metazoans. Character legend at the left bottom of figure. Green associated with deuterostome lineages, blue associated with lophotrochozoan lineages, orange associated with ecdysozoan lineages. Cladogram (modified from Adoutte et al. 2000) on the left-hand side indicates evolutionary relationship between species grouped in box on the right-hand side. No implication of polarity is associated with the order of multiple characters occurring on the same branch. For each taxon, orthologs of OTD are included with a schematic of the C-terminal end of each protein that includes the sequence of the short WSP amino acid motif; absence of this motif is indicated with a grey line. Beneath the WSP sequences is the amino acid consensus for the motif. For all taxa, retention of consensus amino acids is indicated with red letters. The WSP motif is detected in at least one orthodenticle paralog of all examined taxa with the notable exception of D. melanogaster. In this model, an ancestral orthodenticle gene was present in the lineage leading to cnidarians + bilaterians and has experienced at least three independent duplication events via two dissimilar duplication processes (black bars). In the lineage leading to the vertebrates, orthodenticle gene duplication occurred via entire genome duplication. In the cnidarian lineage leading to Nematostella vectensis, orthodenticle expansion occurred via local gene duplication generating three closely linked paralogs (NvOtx sequences, K. Pang, personal communication). Available data in the arthropod lineage indicates an orthodenticle expansion also occurred via local gene duplication in the lineage leading to the crustaceans + insects (Tetraconata). In taxa for which data is available, at least one otd paralog retains an identifiable WSP motif. In the insects, T. castaneum and A. mellifera, the duplicate paralogs are known to be in close proximity to one another. The dipteran D. melanogaster has lost the otd paralog containing the WSP motif (crossed black bar). The expression of orthodenticle orthologs in the head occurred before the lineage including the Eubilateria (black oval). This expression character is currently placed after divergence of the Aceolmorpha lineage (ChOtx sequences, A. Hejnol, personal communication). Expression of orthodenticle in the foregut also appears to be an ancient feature, also occurring before the divergence of the deuterostome and protostome lineages (blue oval). Expression of orthodenticle appears in segmentally reiterated neurons in the lineage leading to Lophotrochozoa + Ecdysozoa (red oval). Available data in the arthropod lineage indicates expression of orthodenticle in the ventral midline arose before the divergence of the chelicerate + Tetraconata lineage (grey oval). Foregut expression is lost in the lineage leading to the Tetraconata (crossed blue oval). Additional data indicate that orthodenticle expression in median neuroblast progeny (MNB) predates the divergence Tetraconata lineage (purple oval). Full species names are Apis mellifera, Branchiostoma floridae, Ciona intestinalis, Convolutriloba hastifera, Drosophila melanogaster, Euscorpius flavicaudis, Holopneustes purpurescens, Mus musculus, Nematostella vectensis, Parhyale hawaiensis, Patella vulgata, Platyneris dumerilii, Saccoglossus kowalevskii, and Tribolium castaneum
Expression of the P. hawaiensis Photd1 paralog
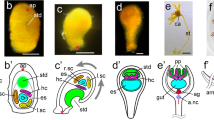
The Photd1 paralog is first detectable in descendants from each of the three ectoderm clones El, Er, and Ep (Gerberding et al. 2002; Browne et al. 2005) at gastrulation in the early stage 8 embryo (Fig. 2a). There is no maternal Photd1 transcript contribution to oocytes nor is there detectable Photd1 zygotic transcription before gastrulation. As the cells that form the germ disc aggregate in the late stage 8 embryo, Photd1 expression spreads, increases in intensity, and is detected anteriorly in an arc as the head lobes begin to condense laterally. The head lobes are composed of cells from the El (ectoderm left) and Er (ectoderm right) ectoderm clones (Gerberding et al. 2002). Photd1 expression also remains present in the future ventral midline cells of the Ep (ectoderm posterior) clone as they converge into a single, tightly associated cluster, at the posterior end of the germ disc (Fig. 2b). In the following stage, stage 9, the anterior lateral-most Photd1-expressing cells are beginning to organize into the characteristic bilaterally symmetric head lobe arcs. A persistent arc of less intense staining connects the head lobes across the midline. This staining is associated with cells at the anterior-most region of the developing germ disc. Posteriorly, the medial cluster of Photd1-expressing cells have organized as a half circle with a sharply demarcated anterior boundary (Fig. 2c). The formation of this boundary occurs as the posterior ectoderm of the germband begins to organize into a characteristic gridded array (Browne et al. 2005). As parasegment precursor rows (PPRs) begin to organize in the developing germband in the stage 11 embryo, anterior lateral Photd1-expressing cells have condensed into two bilaterally symmetric groups coincident with the developing head lobes; the weaker anterior medial arc of expression has begun to diminish (Fig. 2d). Posteriorly, Photd1-expressing cells begin to form the ventral midline by converging medially and extending along the developing ventral midline as a single column of cells (Fig. 2d). The anterior-most Photd1-expressing midline cell is positioned at PPR1, this corresponds to the anterior limit of the Ep clone midline and is coincident with the position of the future first maxillary segment, Mx1. The first ectodermal row to form in the germband is PPR0 which corresponds to the future Mn segment (yellow arrowhead, Fig. 2d). As germband extension continues in the stage 12 embryo, the anterior medial arc of Photd1 expression is no longer detectable. Posteriorly, Photd1 expression within the extending single cell ventral midline column is retained via both maintenance of expression in PPR progeny and by convergence and intercalation of cells from more posterior PPRs as they organize at the posterior-most extreme of the ventral midline column (black arrowhead, Fig. 2e,f). Throughout germband development, Photd1 expression in the ventral midline is confined to cells derived from the Ep blastomere. The cells of the Ep midline lineage are distinct from all other ectodermal cells and are observed to both bisect and maintain a precise physical boundary between the posterior El and Er clones in the developing germband from PPR1-posterior (Fig. 2f; Gerberding et al. 2002; Browne et al. 2005). El and Er blastomere progeny intermix considerably across the midline in regions anterior of the Ep midline lineage (Gerberding et al. 2002).
Expression of P. hawaiensis otd1, gastrulation through germband formation. All embryos are mounted ventral side up with the anterior end oriented towards the top. For each panel, a brightfield image is on the left (Photd1 label is black), and a Hoechst label of the same embryo is on the right (nuclei are blue, Photd1 is false-colored red). In panels d–f, the yellow arrowhead marks the position of the Mn segment. a Early stage 8 embryo (S8), Photd1, is present in descendants of El, Er, and Ep blastomeres at gastrulation. b Late Stage 8 embryo (S8); anteriorly and medially, the field of El and Er cells expressing Photd1 has expanded, forming an arch (asterisk). Posteriorly, the few Ep cells expressing Photd1 have converged medially into a tightly associated cluster. c Stage 9 embryo (S9); anteriorly, the lateral-most Photd1 expressing cells are beginning to organize into the characteristic bilaterally symmetric head lobe arcs (black arrows). Posteriorly, the medial cluster of Photd1-expressing cells organizes with a sharply demarcated anterior boundary. Formation of this boundary occurs as the posterior ectoderm of the germband begins to organize into a stereotypic gridded array. d Stage 11 embryo (S11); Photd1-expressing cells have condensed anteriorly into two bilaterally symmetric groups largely coincident with the head lobe arcs (black arrows); the more anterior medial expression is fading (asterisk). Posteriorly, Photd1-expressing cells begin to form the ventral midline by converging into a single column of cells medially. The anterior boundary of the Photd1-expressing midline cells is at the level of PPR1 which is immediately posterior of the Mn segment (yellow arrowhead) and corresponds with the first maxillary segment, Mx1. e Stage 12 embryo (S12); anterior medial expression of Photd1 is undetectable. Posteriorly, as the germband extends, expression in the single cell ventral midline column is maintained via both cell division within forming parasegments as well as convergence and intercalation of additional cells at the posterior extreme of the midline as new PPR rows organize in the region of the wedge-shaped cluster of Photd1-expressing cells (black arrowhead). f Stage 13 embryo (S13), view of the posterior germband. The posterior-most wedge of Photd1 cells are indicated with an arrowhead. The ventral midline cells of the single cell column are all derived from the Ep blastomere and express Photd1 and bisect and maintain the boundary between the posterior El and Er clones in the developing germband
By stage 21, segmentation is complete and the stomodeum is visible medially in the anterior region of the An2 segment (Fig. 3f,i). Expression of Photd1 appears strongly in the anterior head ectoderm and developing brain, medially in the labrum, and Photd1 expression in the ventral midline has expanded into ectoderm of the mandibular (Mn) segment anterior of Ep clone descendant cells (Fig. 3a). The region of medial ectoderm between the labrum and the Mn segment has invaginated to form the stomodeum and the lining of the foregut (Fig. 3a; Browne et al. 2005). In the developing anterior head, Photd1 expression is detected across the midline in the region associated with the formation of the preoral protocerebral commisure (PCC) and the future protocerebral bridge (black arrowhead Fig. 3d,e; white arrowhead Fig. 3g,i).
P. hawaiensis otd1 brain expression. All panels (except panel a in which embryo anterior is left) are oriented anterior towards the top. a–i Stage 21 (S21) embryos; j–r stage 23 (S23) embryos. d–f and m–o Nomarski images though progressively more dorsal focal planes of a S21 embryo and a S23 embryo, respectively, with Photd1 label in black. g–i and p–r Matching focal plane overlays for Hoechst DNA counterstain (nuclei are blue) and Photd1 (false-colored red). a Segmentation is complete by S21; anteriorly, Photd1 appears strongly in the anterior head ectoderm (black arrow) and medially in the labrum (Lb). Ventral midline expression has expanded anteriorly to include the mandibular segment (Mn) (expansion into the mandibular segment is first detected at stage 18, data not shown). The region of medial ectoderm between the labrum and the Mn segment invaginates to form the opening of the stomodeum and lumen of the foregut (asterisk). Continuity of ventral midline expression is maintained to the posterior extreme of the embryo where Photd1 is expressed strongly in the ectoderm of the developing proctodeum (Pr) and hindgut. b A schematic of the Parhyale supraesophageal ganglia showing the neuromere position of the major neuropil substructures in a S21 embryo; the midline is indicated by a vertical dashed line and the stomodeum (ST) is indicated in blue and surrounded by a circum-oral neuropil ring of axon tracts (in black). The protocerebral neuromere (PC) is indicated in green and includes the preoral protocerebral commissure (PCC) medially, followed by the hemiellipsoid body (Hb) and lateral lobe (Ll, orange arrow) and by the medulla complex (Md) laterally. The deutocerebral neuromere (DC) is indicated in yellow and includes the antennal lobe (AL) neuropil. The tritocerebral neuromere (TC) is indicated in red and includes the antenna 2 neuropil (AN). c S21 embryo stained for acetylated tubulin to detect axon tract morphology in the developing brain. Medially, the PCC is detected. Lateral of the PCC and projecting anteriorly along the neuroaxis, axon tracts are detected innervating the developing bilateral Hb. Posterior of the Hb are axon tracts innervating the developing lateral lobes of the PC (orange arrowhead, also in b, f, i). Just posterior and lateral of the PCC, the AL of the anterior DC can be seen projecting axons into the circumoesophageal ring. The two major axon tracts associated with the labral stomatogastric nervous system (Lb SNS) are visible as a prominent medial decending tract, the nervus connectivus, from the PCC and a more posterior lateral tract, the labral nerve, innervating the TC. For panels d–i, in the developing anterior head, Photd1 expression (black staining) crosses the midline in the region associated with the formation of the future protocerebral bridge (black arrowhead in d, e and white arrowhead in g, h). Bilaterally near the base of the labrum, at the level of the An2 segment and the tritocerebrum (TC), Photd1 is expressed in a field of four to six cells (orange double-arrow in d, g and e, h) associated with the development of components of the labral stomatogastric nervous system (Lb SNS). Ventral midline expression of Photd1 in the ectoderm is now present in the Mn segment (purple arrowhead in e, h and f, i). Photd1 is also present in the distal medial-most region of the developing Lb (d, g) and distal tips of the developing mandibles (Mn) (purple arrow in d, g). Photd1 expression in the Md of the PC is detected throughout the proximal most region, the medulla terminalis (Mt) (e, h and f, i), and also in the medulla interna (Mi) (e, h and f, i). Photd1 expression is particularly strong in cell bodies of the forming lateral lobes of the PC (orange arrowheads in f, i). Lateral expression of the anterior Photd1 expression domain extends posteriorly to the proximal base of the antennae (An1) that innervate the anterior DC (yellow arrowheads in e, h). j–l Nomarski (j), Hoechst DNA counterstain (k, nuclei are blue), and matching focal planes for Hoechst and acetylated tubulin (l, nuclei in blue, acetylated tubulin in red) to show morphology and position of axon tracts in the developing S23 Parhyale brain. Focal plane bisects the neuroaxis of the supraesophageal ganglia. The anterior-most supraesophageal neuromere, the protocerebrum (PC) is composed of a number of distinct substructures. At the anterior midline is the protocerebral bridge (PB); this is followed posteriorly by a thick bundle of axons forming the protocerebral commissure tract (PCT). Immediately posterior of the PCT is the central body (CB) (l). Proceeding laterally, the first anterior bilateral neuropils are the hemiellipsiod bodies (Hb) (j–l). Posterior of the PCT are the lateral lobe neuropils (orange arrowhead) (k, l). The lateral-most structure of the PC is the medulla complex (Md) (j–l). The Md complex can be further subdivided into the proximal-most medulla terminalis (Mt), lateral medulla interna (Mi), and distal-most medulla externa (Me) neuropils (l). The second supraesophageal neuromere, the deutocerebrum (DC), is positioned anteriorly and laterally of the circumesophageal ring surrounding the stomodeum (ST) and includes a thick bundle of axons forming the deutocerebral commissure tract (DCT) (l). Lateral and posterior of the DCT are the antennal lobes (AL) that receive nerves from the first antennae (An1) (j–l). The third supraesophageal neuromere, the tritocerebrum (TC), is positioned along the lateral posterior portion of the circumesophogeal ring and receives nerves from the second antennae (An2) (l). m–r Photd1 expression in the Md of the PC has retracted to the proximal-most region of the Mt in cells lying adjacent to the lateral border of the Hb and Photd1 expression remains pervasive throughout the developing Mi. Expression also remains pervasive in developing lateral lobes of the PC (orange arrowhead, n, q and o, r). A transverse ectodermal band of Photd1 is also detected at the anterior base of the labrum (black arrow m, white arrow p). Photd1 expression near the base of the labrum, at the level of the TC, associated with the development of the stomatogastric nervous system (SNS) is retained (orange double-arrow; m, p)
The Parhyale supraesophageal ganglia are composed of the protocerebral neuromere (PC), the deutocerebral neuromere (DC), and the tritocerebral neuromere (TC) (Fig. 3b). These three neuromeres together constitute the brain of Parhyale. Expression of Photd1 is clearly regionalized within the developing PC (Fig. 3d–f). In the medulla complex of the PC, Photd1 expression is detected throughout the medulla terminalis (Mt), in the medulla interna (Mi), and strongly in a few large cell bodies forming the anlagen of the lateral lobes at the proximal base of the Medulla complex (Fig. 3e,f,h,i). The anterior domain of Photd1 extends into the proximal base of the An1 antennal appendages that innervate the DC (yellow arrowhead, Fig. 3e,h). Photd1 is detected neither in the developing anterior medial hemiellipsoid bodies (Hb) nor in the medulla externa (Me). More posteriorly, a de novo domain of Photd1 is detected in a small bilateral cluster of four to six cells at the base of the labrum and flanking the stomodeum that appear to be positionally associated with receiving/sending axons to the labral nerve of the stomatogastric nervous system (SNS) via the TC neuromere (Fig. 3d,e,g,h). At the posterior extreme of the ventral midline, Photd1 is also expressed strongly in the developing proctodeum and hindgut ectoderm (Fig. 3a).
By stage 23, the embryonic CNS is well developed, the VNC possesses a full complement of segmentally reiterated anterior and posterior commissures, and many structural landmarks are clearly identifiable in the developing brain and VNC (Figs. 3j–l and 4a–c; Browne et al. 2005). In the developing medulla complex of the PC, Photd1 expression has retracted (compare Fig. 3e,n). Photd1 is diminished in the Mt and only remains prominent in cells along the proximal medial boundary adjacent to the Hb (Fig. 3m–r). In contrast, expression of Photd1 remains strong in the Mi and in the Ll of the PC as these clusters of cells continue to expand (compare Fig. 3f,wk–l,n–r). Photd1 expression is absent in the Hb and anterior DC (Fig. 3m–r). A narrow band of weak ectodermal expression is detected associated with the anterior border of the labrum (Fig. 3m,p). Photd1 expression remains detectable in the small bilateral cluster of Lb SNS cells flanking the stomodeum (Fig. 3m,p). These clusters most likely innervate the TC.
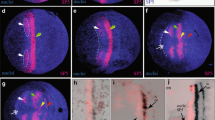
Ventral nerve cord expression of P. hawaiensis otd1. All panels are dorsal views bisecting a stage 23 embryonic VNC and are oriented anterior towards the top. a–c Nomarski (a), matching planes for nomarski and acetylated tubulin (b, acetylated tubulin in red), and a VNC schematic (c). d and e Nomarski with Photd1 in black (d) and matching planes for Hoechst DNA counterstain and Photd1 (e, nuclei are blue; Photd1 is false-colored red). The position of the longitudinal axon tracts is indicated with a green arrow. The position of the anterior commissure (ACom) and of the posterior commissure (PCom) are indicated (a–e). The position of the segmental nerves (SN) is indicated in c and d. The position of the intersegmental nerves (ISN) is indicated in c. A group of cells located immediately anterior of the ACom that express Photd1 can be identified (blue arrows, d). Photd1 is detected in glia ensheathing the ACom (red arrow) and in the cluster of median neuroblast (MNB) progeny between the anterior and posterior commissures (purple arrowhead)
Expression of the P. hawaiensis Photd1 paralog in the VNC
Expression of Photd1 at stage 23 in the VNC includes a defined group of segmentally reiterated cells (Fig. 4d,e). These Photd1-expressing cells are associated with the anterior commissure (ACom) and the region between the ACom and posterior commissure (PCom) (Fig. 4c–e). The presence of several Photd1-expressing cells arrayed along the anterior ACom appears to be a unique attribute not yet reported in any other arthropod (Fig. 4d). The midline glial cells that ensheath the ACom (Gerberding et al. 2001) appear to express Photd1 (Fig. 4c–e). In Drosophila, the positionally analogous glia would be the MGM and MGA glia (Klämbt et al. 1991). The MGM and MGA glia in Drosophila do not express otd. However, the array of Photd1-positive VNC cells shares some features with that of Drosophila otd VNC expression; for example, the central cluster of Photd1 cells between the ACom and the PCom are median neuroblast (MNB) progeny (Fig. 4d,e; Browne et al. 2005). Photd1 expression in both the MNB progeny and the ACom glia are predictable based on PPR midline lineage studies in a related amphipod (Gerberding et al. 1999, 2001). The expression of Photd1 in cells associated with the anterior border of the ACom most likely represents de novo expression flanking the ventral midline lineage. It is also noteworthy that not all ventral midline cell lineages continue to express Photd1; for example, expression is absent in the midline glia that ensheath the PCom (Gerberding et al. 2001). Thus, Photd1 expression is not maintained in all progeny generated by the Parhyale Ep blastomere ventral midline lineage.
In summary, the expression of the Photd1 gene is zygotic, begins during gastrulation, and is associated with the initial organization of the anterior head and ventral midline (Fig. 2). Later in development, Photd1 becomes restricted anteriorly to defined regions of the PC, anterior DC, and a SNS cell cluster that correlates positionally with innervation of the labral nerves via the TC at the lateral junction of the labrum and stomodeum (Fig. 3). In the developing VNC, Photd1 is associated with a subset of ventral midline progeny giving rise to glia ensheathing the ACom, the MNB progeny neurons, and de novo expression in a group of cells arrayed along the anterior border of the ACom (Fig. 4d).
Expression of the P. hawaiensis otd2 paralog
The temporal and spatial expression dynamics of the second otd paralog, Photd2, are very different from that of Photd1, and many aspects have no direct parallel in other arthropod taxa. Three in situ probes corresponding to unique Photd2 splice variants were made, one to a 5′ splice variant and two to unique 3′ splice variants. No variation in expression profile was detected between probes to different splice variants. Expression is shown for the 3′ splice variant possessing the WSP motif (Fig. 5). Photd2 first appears at stage 12 during germband formation in two bilaterally symmetric anterior domains coincident with Photd1 (compare Fig. 2e with Fig. 5a,b). The initiation of Photd2 expression, after the anterior ectoderm has been patterned into distinct head lobes, implies that Photd2, in contrast to Photd1, does not play a major role in patterning the anterior head ectoderm. No Photd2 expression is detected in the formation ventral midline cell lineage of the Ep clone during germband development (Fig. 5a,b). Later in embryonic development (beginning at stage 22), anterior expression of Photd2 is highly restricted in the anterior head and only appears in the developing PC in a small number of cells associated with the base of the Md complex (Fig. 5c,d). Photd2 also begins to appear in a restricted set of ventral midline cells (Fig. 5c–g). At the level of the Mn segment, Photd2 expression is largely coincident with Photd1 midline expression (Fig. 5c,d). However, in the Mx1 and all posterior segments, ventral midline expression of Photd2 is mutually exclusive of Photd1 expression (compare Fig. 4 with Fig. 5e–g). Photd2 cells in the ventral midline appear to have two distinct fates (Fig. 5e,f). At stage 22, the two ventral-most Photd2-expressing cells occur in the middle of developing segments and appear to be ectodermal (Fig. 5e), whereas more dorsally, three Photd2-expressing cells at the midline junctions of each segment may be glia (Fig. 5f; Gerberding et al. 2001).
Expression of P. hawaiensis otd2. All embryos are mounted with the anterior end oriented towards the top. a, b A stage 12 embryo is mounted ventral side up with a nomarski image on the left and Photd2 signal in black. The image on the right is overlay of matching focal planes for Hoechst DNA counterstain with Photd2 (nuclei are blue; Photd2 is false-colored red). Photd2 is first detected at this stage anteriorly in the condensation of cells in the head lobes (black arrows) and is coincident with the anterior Photd1 domain at this stage of development (compare with Fig. 2e). The yellow arrowhead marks the position of the Mn segment. Photd2 is not expressed in the ventral midline lineage until much later in stage 22 embryos. c–g Photd2 expression in a stage 22 embryo. c, d Embryo mounted dorsal side up, with nomarski image on the left and Photd2 signal in black. The image on the right is overlay of matching focal planes for Hoechst DNA counterstain with Photd2 on the right (nuclei are blue; Photd2 is false-colored red). In the developing anterior head, Photd2 expression is restricted to a small number of cells at the base of the Md. Ventral midline expression of Photd2 can now be detected in the midline ectoderm of the Mn segment (purple arrowhead). e–g A stage 22 embryonic thoracic segment mounted ventral side up. e Ventral nomarski focal plane with Photd2 signal in black. f Dorsal nomarski focal plane with Photd2 signal in black. g Collapsed and merged ventral and dorsal focal planes for Hoechst and Photd2 (nuclei are blue; Photd2 is false-colored red). Photd2 is expressed in VNC midline cells exclusive of Photd1 (compare with Fig. 4). The two midline cells in e appear to be ectodermal. The three slightly more dorsal cells in f are at the segmental border and may be glial cells
In summary, Photd2 is expressed zygotically and first appears after the formation of the head lobes in an anterior domain in germband embryos (Fig. 5a,b). Late in embryogenesis, the anterior Photd2 domain has retracted to a few cells at the base of the medulla (Fig. 5c,d). No expression of Photd2 is detected in the ventral midline until late in embryogenesis. Ventral midline expression of Photd2 is mutually exclusive of its paralog Photd1. The fates of these Photd2-expressing midline cells are likely to be both ectodermal in the middle of each segment and glial in more dorsal cells at segment boundaries (Fig. 5e–g).
Discussion
The problems imposed on animals by both their environment and respective genetic history have resulted in a diversity of differences in nervous system organization. Considerable variation is observed in the development and overall organization of the brain and nervous systems among insects alone (e.g., Condron and Zinn 1994; Reichert and Boyan 1997; Urbach and Technau 2003b). In insects and other arthropods, understanding the evolution, development, and segmental homologies of the head and brain has posed long-standing problems in the fields of morphology, paleontology, evolution, and molecular genetics, resulting in what has been described as ‘the endless debate’ (e.g., Rempel 1975; Scholtz and Edgecombe 2005; Browne et al. 2005; Maxmen et al. 2005). In this study, we provide new data from a crustacean that helps clarify the organization and development of the arthropod head and brain.
Expression of paralogous Parhyale otd genes in the anterior head
Available evidence suggests that a single orthodenticle gene was likely expressed in the head and foregut of the eubilaterian ancestor (Fig. 1; Bruce and Shankland 1998; Harada et al. 2000; Arendt et al. 2001; Lowe et al. 2003). In the lineage leading to the Lophotrochozoa and Ecdysozoa, orthodenticle expression was acquired in segmentally reiterated neurons along the anterior–posterior axis of the body (Fig. 1; Bruce and Shankland 1998). At some point before the divergence of the Tetraconata, the orthodenticle gene experienced a number of changes. The number of orthodenticle genes expanded via a tandem duplication event (Fig. 1; Li et al. 1996), expression was gained in MNB progeny neurons, and expression was lost in the foregut (Fig. 1). We expect that more comprehensive analysis of orthodenticle ortholog expression in taxa diverging before the Tetraconata, including the lophotrochozoans, may reveal that some of these general expression attributes may have first appeared at deeper evolutionary nodes.
Gene duplication is often accompanied by divergence in both gene expression and function. The expression of the Parhyale otd paralogs are clearly divergent from otd orthologs in both Tribolium and Drosophila. In contrast to the earliest expression in Tribolium, the earliest expression in Parhyale (and Drosophila) is zygotic and has no maternal component (Fig. 2; Finkelstein and Perrimon 1990). Neither paralog in Tribolium is expressed in the DC or TC (Li et al. 1996). The Parhyale Photd1 paralog is clearly expressed in the anterior DC (as is Drosophila otd) as well as in a restricted region of the TC (Fig. 3). This is a feature, thus far, unique to Parhyale. The restriction of the de novo TC domain to cell bodies associated with innervation of the labral SNS, coupled with the presence of a prominent nervus connectivus linking the labral SNS to the PCC (Fig. 3c), makes it tempting to speculate that ancestral labral innervation was restricted to the PC in combination with an anterior stomodeum (Rempel 1975; Eriksson and Budd 2000; Browne et al. 2005; Maxmen et al. 2005). This is important because the positional origin of the labrum, as the most prominent anterior ectodermal structure in most arthropods, has played a central role in attempts to understand underlying segmental patterning of the arthropod head. Expression analysis of otd orthologs in onychophorans would prove particularly useful in helping to resolve questions regarding ancestral features of head patterning and brain development in the Ecdysozoa and may settle the debate regarding the positional origin and innervation patterns associated with the appendicular labrum.
A universal mechanism for patterning both the invertebrate and vertebrate brain has been proposed to account for their tripartite brain organization (Hirth et al. 2003). Based on expression patterns in Drosophila, this model proposes that the interface between the posterior boundary of orthodenticle expression and anterior expression domain of the gene unpg maintains the boundary between the DC and TC via mutual repression. In Tribolium, neither otd paralog is expressed in the DC during brain development. In Parhyale, Photd1 is expressed only transiently in the anterior DC (Fig. 3e,h). In light of this variation in otd expression, it would be informative to validate this mechanism by observing the relationship between Photd1 and Parhyale unpg orthologs in the maintenance of gene expression boundaries correlating with the DC–TC neuromere interface.
The expression of Photd2 in the head is radically divergent both from its paralog Photd1 as well as from otd orthologs in divergent arthropod taxa. The initial anterior domain of Photd2 is coincident with Photd1 but does not appear until after the head lobes are organized (Fig. 5). As development progresses, the anterior Photd2 expression domain decays and, late in development, is only detectable in a few cells of the PC (Fig. 5). Photd2 is consistently expressed at a much lower level than Photd1, a phenomenon also observed in Tribolium.
Expression of paralogous Parhyale otd genes in the ventral midline
Current data suggest orthodenticle expression was acquired in the ventral midline in arthropods, (Figs. 1, 2, 4, and 5; Finkelstein et al. 1990; Li et al. 1996; Telford and Thomas 1998). In Parhyale, as in Drosophila and Tribolium, expression of otd in the midline cells is apparent at an early stage of development (Fig. 2). The ontological origin of Tribolium midline cells are undescribed; however, midline cell lineages are known in Drosophila (Klämbt et al. 1991; Bossing and Technau 1994; Schmid et al. 1999), the grasshopper Schistocerca (Condron and Zinn 1994), as well as in a related amphipod crustacean Orchestia (Gerberding and Scholtz 1999, 2001). Our data suggests some similarities with otd expression in insect midline lineages as well as a number of significant differences.
The ectodermal ventral midline cell lineage in the closely related amphipod, Orchestia, generates a specific set of glial and neuronal progeny. For each segment, two glial cells are produced that ensheath the anterior commissure, two glial cells are produced to ensheath the posterior commissure, two to three glia are produced that are positioned at the midline of the intersegmental border, and ∼10 median neuroblast progeny are produced that cluster between the ACom and PCom (Gerberding and Scholtz 2001). The midline of Parhyale develops in an analogous fashion to that of Orchestia. In Drosophila, the midline develops from a unique arrangement of mesectodermal cells. The midline lineage produces both the MNB progeny neural cluster and the glia that ensheath the anterior commissure (Klämbt et al. 1991). In insects, the PCom is pioneered ahead of the ACom (Klämbt et al. 1991; Myers and Bastiani 1993), whereas in Parhyale the ACom is pioneered ahead of the PCom. Photd1-positive midline cells appear in both the MNB progeny neural cluster and in the glia that ensheath the ACom, expression features shared with both Drosophila and grasshopper (Fig. 4c–e). The position of the Parhyale MNB neural cluster is displaced anteriorly relative to Drosophila and grasshopper, but this is likely a pleiotropic effect due largely to the observed heterochrony in commissure formation (reversed order of commissure formation).
In addition to the conserved features, Photd1 is also uniquely expressed in a group of cells arrayed along the anterior border of the ACom (Fig. 4d). The presence of Photd1 in these cells represents de novo expression flanking the ventral midline lineage, as well as expression in a group of cells not yet observed in other arthropods. In addition, the late embryonic expression of Photd2 in a group of midline cells mutually exclusive of Photd1 has no parallel in other taxa. At least three of these reiterated cells that are found dorsally at the midline junction of segments may represent glial descendants of the ventral midline lineage (Fig. 5e–g; Gerberding and Scholtz 2001).
Implications regarding the evolution and duplication of the orthodenticle gene
Understanding the history of orthodenticle gene duplication during the course of metazoan evolution presents an interesting dilemma. Expansion of otd genes within the vertebrates via genome amplification is straightforward (Germot et al. 2001). However, in all other instances of duplication, otd paralogs are in close proximity, suggesting tandem duplication, and the paralogs appear more similar to one another than to otd orthologs in other species. This suggests two possible scenarios: (1) otd paralogs in each case represent independent duplications or (2) otd duplications are very ancient, occurred at deeper nodes, and paralogs in each lineage appear to be different from their orthologs. The history of otd duplication could be sufficiently complex that we will not be able to determine the relationships between paralogs from different lineages.
We suggest that two well-known genetic mechanisms, gene conversion and mutational saturation, can be used as support for scenario 2 based on currently available genomic information in the taxa analyzed. Gene conversion is a process by which paralogous genes acquire identical nucleotide sequences via mismatch repair of heteroduplexed DNA (Radding 1982). The likelihood of gene conversion increases with the proximity of the paralogs. Gene conversion would become a problem when attempting to reconstruct the orthology relationships between paralogs due to regions between paralogs becoming more similar to each other than to their orthologs in other lineages and, thus, loosing useful phylogenetic signal. The second mechanism, mutational saturation, also results in misleading phylogenetic signal among paralogous sequences (Philippe and Forterre 1999). A manifestion of mutational saturation, for example, is the process of codon usage bias governing the pool of available tRNAs, again acting to make paralogs more similar to one another than to their respective orthologs in other lineages, in this instance via purifying selection (Nei and Kumar 2000).
Among the taxa we sampled, the beetle T. castaneum genome contains two otd paralogs within ∼60 kb of one another (BeetleBase, http://www.bioinformatics.ksu.edu/beetlebase). The hymenopteran Apis mellifera genome contains two otd paralogs within ∼40 kb of one another (BeeBase, http://racerx00.tamu.edu/bee_resources.html). Preliminary information from the crustacean Daphnia pulex genome has revealed the presence of two otd paralogs within ∼24 kb of one another (F. Poulin and N. Patel, personal communication). Sampling among the chelicerates has only revealed a single otd ortholog. Thus, the current evidence suggests a single tandem duplication of an ancestral otd gene in the arthropod lineage leading to the Tetraconata (Fig. 1). Within the Tetraconata, two things appear to have happened to otd paralogs: (1) loss of one otd duplicate in the lineage leading to the dipterans as represented by Drosophila (Fig. 1) and (2) physical proximity of the gene duplicates may have led to sequence homogenization within a given lineage via the two distinct genetic mechanisms outlined above, gene conversion, and/or mutational saturation. Large-scale genomic resources are not yet available for Parhyale; however, we predict that the two P. hawaiensis orthodenticle paralogs are physically linked.
The test for mutational saturation suggests that base pair saturation has played a role in orthodenticle gene divergence in different taxa, making recovery of paralog orthology among the tandem duplicates present in Tetraconata (crustaceans + insects) particularly problematic with currently available phylogenetic tools. Direct evidence of gene conversion could possibly be demonstrated by investigating orthodenticle paralog sequences at the level of populations, revealing instances of allelic variation resulting from heteroduplex DNA mismatch repair. It is tantalizing to hypothesize that the unique retention of the WSP domain in both A. mellifera paralogs may represent evidence for this mechanism downstream of the homeodomain within this lineage (Fig. 1).
In summary, the data presented here represent additional evidence for a single ancestral orthodenticle like gene in the common ancestor of the lineage leading to the cnidarians + bilaterians (Fig. 1; Li et al. 1996). In most cases, we also recover an eight-amino-acid diagnostic sequence, the WSP motif, downstream of the homeodomain in at least one otd paralog (Fig. 1). It is highly probable that this motif existed in the single ancestral orthodenticle gene before the divergence of the cnidarians and the bilaterians. This ancestral orthodenticle gene was independently duplicated in lineages leading to the vertebrates (genome duplication), the cnidarians (three linked copies), and the Tetraconata (tandem duplication). One paralog was subsequently lost in the lineage leading to the dipterans (Fig. 1).
Prospects
Our analysis of otd paralog gene expression in Parhyale has indicated some of the general boundaries/structures associated with the developing supraesophageal neuromeres that comprise the brain. We are hopeful that, when used in combination with other markers, they will contribute to understanding the distribution and number of neural stem cells that initially specify the Parhyale brain (e.g., Urbach and Technau 2003a,b). This type of neuroblast gene expression data will prove to be important in crustaceans as, in sharp contrast to insects, crustacean neuroblasts do not delaminate from the ectoderm and, thus, are often not clearly distinguishable from surrounding non-neural ectodermal cells. In addition, crustacean neuroblasts may switch fates as they divide, generating both neural-fated and non-neural fated ectoderm progeny. Thus, the best means for discriminating between ectodermal cells, neuroblasts, neurons, and glia in crustaceans will be with panels of molecular markers used in concert with neuronal morphology.
Comprehensive characterization of gene expression and, subsequently, genetic function (e.g. Pavlopoulos and Averof 2005) in the Parhyale brain and nervous system will begin to provide us with a framework and new set of tools for exploring diverse aspects related to developmental patterning at three levels: the basic underpinnings of neurogenesis in metazoans, aspects of developmental mechanisms inherent to neurogenesis in arthropods, and specific details related to the unique development of the nervous system of diverse crustaceans.
References
Acampora D, Avantaggiato V, Tuorto F, Barone P, Reichert H, Finkelstein R, Simeone A (1998) Murine Otx1 and Drosophila otd genes share conserved genetic functions required in invertebrate and vertebrate brain development. Development 125:1691–1702
Adoutte A, Balavoine G, Lartillot N, Lespinet O, Prud’homme B, de Rosa R (2000) The new animal phylogeny: reliability and implications. Proc Natl Acad Sci USA 97:4453–4456
Arendt D, Nubler-Jung K (1996) Common ground plans in early brain development in mice and flies. Bioessays 18:255–259
Arendt D, Technau U, Wittbrodt J (2001) Evolution of the bilaterian larval foregut. Nature 409:81–85
Bossing T, Technau GM (1994) The fate of the CNS midline progenitors in Drosophila as revealed by a new method for single cell labelling. Development 120:1895–1906
Browne WE, Price AL, Gerberding M, Patel NH (2005) Stages of embryonic development in the amphipod crustacean, Parhyale hawaiensis. Genesis 42:124–149
Bruce AEE, Shankland M (1998) Expression of the head gene Lox22-Otx in the leech Helobdella and the origin of the bilaterian body plan. Dev Biol 201:101–112
Cohen S, Jürgens G (1990) Mediation of Drosophila head development by gap-like segmentation genes. Nature 346:482–485
Cohen S, Jürgens G (1991) Drosophila headlines. Trends Genet 7:267–272
Condron BG, Zinn K (1994) The Grasshopper median neuroblast is a multipotent progenitor cell that generates glia and neurons in distinct temporal phases. J Neurosci 14:5766–5777
Cook CE, Yue Q, Akam M (2005) Mitochondrial genomes suggest that hexapods and crustaceans are mutually paraphyletic. Proc R Soc Lond B 272:1295–1304
Dalton D, Chadwick R, McGinnis W (1989) Expression and embryonic function of empty spiracles: a Drosophila homeo box gene with two patterning functions on the anterior–posterior axis of the embryo. Genes Dev 3:1940–1956
Dohle W (2001) Are the insects terrestrial crustaceans? A discussion of some new facts and arguments and the proposal of the proper name ‘Tetraconata’ for the monophyletic unit Crustacea plus Hexapoda. Ann Soc Entomol Fr 37:85–103
Duman-Scheel M, Patel NH (1999) Analysis of molecular marker expression reveals neuronal homology in distantly related arthropods. Development 126:2327–2334
Eriksson BJ, Budd GE (2000) Onychophoran cephalic nerves and their bearing on our understanding of head segmentation and stem-group evolution of Arthropoda. Arthropod Struct Dev 29:197–209
Finkelstein R, Perrimon N (1990) The orthodenticle gene is regulated by bicoid and torso and specifies Drosophila head development. Nature 346:485–488
Finkelstein R, Smouse D, Capaci TM, Spradling AC, Perrimon N (1990) The orthodenticle gene encodes a novel homeo domain protein involved in the development of the Drosophila nervous system and ocellar visual structures. Genes Dev 4:1516–1527
Finnerty JR, Paulson D, Burton P, Pang K, Martindale MQ (2003) Early evolution of a homeobox gene: the parahox gene Gsx in the Cnidaria and the Bilateria. Evolut Develop 5:331–345
Friedrich M, Tautz D (1995) Ribosomal DNA phlogeny of the major extant arthropod classes and the evolution of myriapods. Nature 376:165–167
Gallitano-Mendel A, Finkelstein R (1998) Ectopic orthodenticle expression alters segment polarity gene expression but not head segment identity in the Drosophila embryo. Dev Biol 199:125–137
Gerberding M (1997) Germ band formation and early neurogenesis of Leptodora kindti (Cladocera): first evidence for neuroblasts in the entomostracan crustaceans. Invertebr Reprod Dev 32:63–73
Gerberding M, Scholtz G (1999) Cell lineage of the midline cells in the amphipod crustacean Orchestia cavimana (Crustacea, Malacostraca) during formation and separation of the germ band. Dev Genes Evol 209:91–102
Gerberding M, Scholtz G (2001) Neurons and glia in the midline of the higher crustacean Orchestia cavimana are generated via an invariant cell lineage that comprises a median neuroblast and glial progenitors. Dev Biol 235:397–409
Gerberding M, Browne WE, Patel NH (2002) Cell lineage analysis of the amphipod crustacean Parhyale hawaiensis reveals an early restriction of cell fates. Development 129:5789–5801
Germot A, Lecointre G, Plouhinec J-L, Le Mentec C, Girardot F, Mazan S (2001) Structural evolution of Otx Genes in Craniates. Mol Biol Evol 18:1668–1678
Giribet G, Edgecombe GD, Wheeler WC (2001) Arthropod phylogeny based on eight molecular loci and morphology. Nature 413:157–161
Giribet G, Richter S, Edgecombe GD, Wheeler WC (2005) The position of crustaceans within Arthropoda—evidence from nine molecular loci and morphology. In: Koenemann S, Jenner RA (eds) Crustacea and arthropod relationships. CRC, Boca Raton, pp 307–352
Grossniklaus U, Cadigan KM, Gehring WJ (1994) Three maternal coordinate systems cooperate in the patterning of the Drosophila head. Development 120:3155–3171
Hanstrom B (1928) Vergleichende Anatomie Des Nervensystems Der Wirbellosen Tiere Unter Berucksichtigung Seiner Funktion. Springer, Berlin Heidelberg New York
Harada Y, Okai N, Taguchi S, Tagawa K, Humphreys T, Satoh N (2000) Developmental expression of the hemichordate otx ortholog. Mech Dev 91:337–339
Harzsch S (2003) Ontogeny of the ventral nerve cord in malacostracan crustaceans: a common plan for neuronal development in Crustacea, Hexapoda and other Arthropoda? Arthropod Struct Dev 32:17–37
Hirth F, Therianos S, Loop T, Gehring WJ, Reichert H, Furukubo-Tokunaga K (1995) Developmental defects in brain segmentation caused by mutations of the homeobox genes orthodenticle and empty spiracles in Drosophila. Neuron 15:769–778
Hirth F, Kammermeier L, Frei E, Walldorf U, Noll M, Reichert H (2003) An urbilaterian origin of the tripartite brain: developmental genetic insights from Drosophila. Development 130:2365–2373
Hwang UW, Friedrich M, Tautz D, Park CJ, Kim W (2001) Mitochondrial protein phylogeny joins myriapods with chelicerates. Nature 413:154–157
Klämbt C, Jacobs JR, Goodman CS (1991) The midline of the Drosophila central nervous system: a model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell 64:801–815
Li Y, Brown SJ, Hausdorf B, Tautz D, Denell RE, Finkelstein R (1996) Two orthodenticle-related genes in the short-germ beetle Tribolium castaneum. Dev Genes Evol 206:35–45
Lichtneckert R, Reichert H (2005) Insights into the urbilaterian brain: conserved genetic patterning mechanisms in insect and vertebrate brain development. Heredity 94:465–477
Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, Gruber CE, Gerhart J, Kirschner M (2003) Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell 113:853–865
Maxmen A, Browne WE, Martindale MQ, Giribet G (2005) Neuroanatomy of sea spiders implies an appendicular origin of the protocerebral segment. Nature 437:1144–1148
Myers PZ, Bastiani MJ (1993) Cell–cell interactions during the migration of an identified commissural growth cone in the embryonic grasshopper. J Neurosci 13:115–126
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford, Oxford
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Patel N (1994) Imaging neuronal subsets and other cell types in whole mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol 44:445–487
Pavlopoulos A, Averof M (2005) Establishing genetic transformation for comparative developmental studies in the crustacean Parhyale hawaiensis. Proc Natl Acad Sci USA 102:7888–7893
Philippe H, Sorhannus U, Baroin A, Perasso R, Gasse F, Adoutte A (1994) Comparison of molecular and paleontological data in diatoms suggest a major gap in the fossil record. J Evol Biol 7:247–265
Philippe H, Forterre P (1999) The rooting of the universal tree of life is not reliable. J Mol Evol 49:509–523
Posada D, Crandall K (1998) ModelTest: testing the model of DNA substitution. Bioinformatics 14:817–818
Radding (1982) Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet 16:405–437
Regier JC, Shultz JW, Kambic RE (2005) Pancrustacean phylogeny: hexapods are terrestrial crustaceans and maxillopods are not monophyletic. Proc R Soc Lond B 272:395–401
Reichert H, Boyan G (1997) Building a brain: developmental insights in insects. Trends Neurosci 20:258–264
Rempel JG (1975) The evolution of the insect head: the endless dispute. Quaest Entomol 11:7–25
Richter S (2002) The Tetraconata concept: hexapod–crustacean relationships and the phylogeny of Crustacea. Org Divers Evol 2:217–237
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Royet J, Finkelstein R (1995) Pattern formation in Drosophila head development: the role of the orthodenticle homeobox gene. Development 121:3561–3572
Sandeman D, Sandeman R, Derby C, Schmidt M (1992) Morphology of the brain of crayfish, crabs, and spiny lobsters: a common nomenclature for homologous structures. Biol Bull 183:304–326
Schmid A, Chiba A, Doe CQ (1999) Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development 126:4653–4689
Schmidt-Ott U, Gonzalez-Gaitan M, Jäckle H, Technau GM (1994) Number, identity, and sequence of the Drosophila head segments as revealed by neural elements and their deletion patterns in mutants. Proc Natl Acad Sci USA 91:8363–8367
Scholtz G, Edgecombe GD (2005) Heads, Hox, and the phylogenetic position of trilobites. In: Koenemann S, Jenner RA (eds) Crustacea and arthropod relationships. CRC, Boca Raton, pp 139–165
Schröder R (2003) The genes orthodenticle and hunchback substitute for bicoid in the beetle Tribolium. Nature 422:621–625
Sharman AC, Brand M (1998) Evolution and homology of the nervous system: cross-phylum rescues of otd/Otx genes. Trends Genet 14:211–214
Strausfeld NJ (1998) Crustacean–insect relationships: the use of brain characters to derive phylogeny amongst segmented invertebrates. Brain Behav Evol 52:186–206
Strausfeld NJ, Hildebrand JG (1999) Olfactory systems: common design, uncommon origins? Curr Opin Neurobiol 9:634–639
Swofford D (2003) PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4: 4.0Beta edn. Sinauer, Sunderland, Massachusetts
Telford MJ, Thomas RH (1998) Expression of homeobox genes shows chelicerate arthorpods retain their deutocerebral segment. Proc Natl Acad Sci USA 95:10671–10675
Urbach R, Technau GM (2003a) Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development 130:3621–3637
Urbach R, Technau GM (2003b) Early steps in building the insect brain: neuroblast formation and segmental patterning in the developing brain of different insect species. Arthropod Struct Dev 32:103–123
Walldorf U, Gehring WJ (1992) Empty spiracles, a gap gene containing a homeobox involved in Drosophila head development. EMBO J 11:2247–2259
Whitington PM (1996) Evolution of neural development in the arthropods. Sem Cell Dev Biol 7:605–614
Whitington PM, Leach D, Sanderman R (1993) Evolutionary change in neural development within the arthropods: axonogenesis in the embryos of two crustaceans. Development 118:449–461
Wieschaus E, Perrimon N, Finkelstein R (1992) Orthodenticle activity is required for the development of medial structures in the larval and adult epidermis of Drosophila. Development 115:801–811
Wimmer EA, Jäckle H, Pfeifle C, Cohen SM (1993) A Drosophila homologue of human Sp1 is a head-specific segmentation gene. Nature 366:690–694
Wimmer EA, Cohen SM, Jäckle H, Desplan C (1997) buttonhead does not contribute to a combinatorial code proposed for Drosophila head development. Development 124:1509–1517
Younossi-Hartenstein A, Green P, Liaw G-J, Rudolph K, Lengyel J, Hartenstein V (1997) Control of early neurogenesis of the Drosophila brain by the head gap genes tll, otd, ems, and btd. Dev Biol 182:270–283
Acknowledgements
This work has been supported by the NSF (WEB, DBI-0310269), the NIH (WEB, NCRR-P20RR16467), the Boehringer Ingelheim Foundation (EAW), the Deutsche Forschungsgemeinschaft (EAW, DFG Wi 1797/2-2), the European Community’s Marie Curie Research Training Network ZOONET under contract MRTN-CT-2004-005624 (EAW). EAW also acknowledges support from the EMBO Young Investigator Programme. We thank Kevin Pang and Andreas Hejnol for Otx sequences, Frank Poulin and Nipam H. Patel for D. pulex otd information, and Casey Dunn for computation advice. We also thank the following for providing critical comments that have significantly improved this communication: Elaine Seaver, Amy Maxmen, Andreas Hejnol, Casey Dunn, Nipam H. Patel, and two anonymous reviewers.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Roth
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Browne, W.E., Schmid, B.G.M., Wimmer, E.A. et al. Expression of otd orthologs in the amphipod crustacean, Parhyale hawaiensis . Dev Genes Evol 216, 581–595 (2006). https://doi.org/10.1007/s00427-006-0074-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-006-0074-7