Abstract
Main conclusion
Transgenic mustard plants ( Brassica juncea ) expressing non-allergenic and biologically safe RiD peptide show higher tolerance against Lipaphis erysimi.
Rorippa indica defensin (RiD) has previously been reported as a novel insecticidal protein derived from a wild crucifer Rorippa indica. RiD was found to have an effective insecticidal property against mustard aphid, Lipaphis erysimi. In the present study, RiD was highly upregulated in R. indica during aphid infestation initiating a defense system mediated by jasmonic acid (JA), but not by salicylic acid (SA)/abscisic acid (ABA). RiD has also been assessed for biosafety according to the FAO/WHO guideline (allergenicity of genetically modified foods; Food And Agriculture Organisation of the United Nations, Rome, Italy, 2001) and Codex Alimentarius Guideline (Guidelines for the design and implementation of national regulatory food safety assurance programme associated with the use of veterinary drugs in food producing animals. Codex Alimentarius Commission. GL, pp 71–2009, 2009). The purified protein was used to sensitize BALB/c mice and they showed normal histopathology of lung and no elevated IgE level in their sera. As the protein was found to be biologically safe and non-allergenic, it was used to develop transgenic Brassica juncea plants with enhanced aphid tolerance, which is one of the most important oilseed crops and is mostly affected by the devastating pest—L. erysimi. The transgene integration was monitored by Southern hybridization, and the positive B. juncea lines were further analyzed by Western blot, ELISA, immunohistolocalization assays and in planta insect bioassay. Transgenic plants expressing RiD conferred a higher level of tolerance against L. erysimi. All these results demonstrated that RiD is a novel, biologically safe, effective insecticidal agent and B. juncea plants expressing RiD are important components of integrated pest management.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The yield and production of Brassica juncea are severely affected by the devastating pest Lipaphis erysimi (Dutta et al. 2005a; Bandopadhyay et al. 2013). The aphids suck phloem sap from the plants leading to nutrition deficiency and subsequent wilting. They also help in transmitting viruses during sap sucking, which hence causes various diseases such as mosaics (Moran and Thompson 2001; Moran et al. 2002). In spite of the conventional agricultural practices, this particular pest reduces the yield of the crops leading to huge financial loss to the farmers (Dutta et al. 2005a; Sarkar et al. 2016). Therefore, several attempts have been undertaken to identify aphid-tolerant varieties from the gene pool, but without any success. No resistant genotype or R-genes have been found within the Brassica germplasm against L. erysimi (Yadava and Singh 1999; Dutta et al. 2005a; Bandopadhyay et al. 2013). This led to the attempts in developing transgenic Brassica-carrying genes for aphid tolerance from distant plant relatives. A number of insecticidal agents like proteases (Rahbe et al. 2003) and lectins (Kanrar et al. 2002; Dutta et al. 2005a; Mondal et al. 2006) have been used to develop transgenic B. juncea lines which help in reducing aphid colonization. But there is a need for new insecticidal agents to be discovered due to behavioral reorientation of the insects and induced resistance in due course of time (Das et al. 2013; Sarkar et al. 2016). Lack of resistant genotypes against L. erysimi led to the identification of a wild crucifer, Rorippa indica, which was reported to be tolerant toward this pest (Mandal and Sikdar 2003; Bandopadhyay et al. 2013). It survives the pest attack even when there is heavy colonization. A novel defensin peptide, Rorippa indica defensin (RiD), derived from this plant was noted to be effective against the mustard aphid, L. erysimi (Bandopadhyay et al. 2013; Sarkar et al. 2016).
Plant defensins are generally pathogenesis-related proteins (PR-12) which are part of the innate immune response against pests and pathogens (Sels et al. 2008). They are small cysteine-rich peptides responsible for host defense. Some plant defensins show potent insecticidal activities apart from antifungal and bactericidal properties (Chen et al. 2002; Melo et al. 2002), one of which is BrD1, a defensin gene from Brassica rapa that confers resistance toward brown planthopper (Nilaparvata lugens) in transgenic rice (Choi et al. 2009). R. indica defensin (RiD), a peptide from the wild crucifer, has been previously reported to have high insecticidal activity against L. erysimi with an LC50 value of 9.099 ± 0.621 µg/mL. RiD is secreted into the apoplastic region indicating its role in inhibiting nutrient uptake by aphids, which occurs mainly via the extracellular route to pierce through the cells (Sarkar et al. 2016).
Sap sucking by aphids also induces various signaling mechanisms in plants mediated by hormonal cross talk between different phytohormones such as jasmonic acid (JA), salicylic acid (SA), abscisic acid (ABA) or ethylene (ET) (Morkunas et al. 2011). Feeding and piercing damage of plants by aphids or any phloem feeders triggers the synthesis of jasmonates which in turn regulates the expression of PR proteins and defense-related proteins along with the biosynthesis of secondary metabolites (Cooper and Goggin 2005). As a result, constitutive as well as transient expression of jasmonate-regulated genes has been seen to regulate plant defense (Manners et al. 1998; Penninckx et al. 1998; Moran et al. 2002; Kusnierczyk et al. 2011). Expression of plant defensins is also induced by jasmonates as part of defense (Kiba et al. 2003; Tesfaye et al. 2013). Thus, induction and overexpression of jasmonate-related genes such as defensins which have insecticidal properties might defend aphid colonization through the development of transgenics.
The suitability of RiD as an effective insecticidal agent against L. erysimi indicates that it can be a potent candidate for integrated pest management. But there has always been a myriad of concerns about the impact of transgenes for food derived from genetically modified crops. It is therefore necessary to assess the transgene for allergenicity before introducing it into a crop plant, especially when it is derived from wild plants (Islam 2008; Ghosh et al. 2013). According to the FAO/WHO guidelines (2001) and Codex Alimentarius guidelines (2009), prediction of an allergen requires extensive sequence homology search across known databases of allergens and the identification of motifs responsible for a specific allergen activity. The next important strategy is to characterize the stability of the protein, which includes thermal stability assays, pepsin digestibility and allergenecity assessment of the recombinant proteins using animal models (Nieuwenhuizen et al. 2006; Plundrich et al. 2015; Toomer et al. 2015).
In the aforesaid background, we assessed the risk factors and allergenecity of RiD for transgenic application. As RiD was found to be effective against L. erysimi and biologically safe, we tried to develop transgenic B. juncea plants expressing RiD to control aphid population on the said plant.
Materials and methods
Plant material, and insect and bacterial strains used
Brassica juncea cv. B-85 seeds were obtained from Berhampur Pulse Research Station, West Bengal, India, and used for plant transformation. Seeds of both Rorippa indica (Hiern) and B. juncea were surface sterilized with 2% sodium hypochlorite (NaOCl) solution. Plants were grown in Soilrite (KEL, Bangalore, India) and kept in growth chambers with a temperature of 25 ± 2 °C under a 16 h photoperiodic white light of 3000 lx for the bioassays. The mustard aphid, Lipaphis erysimi (Kalt.), was reared from Madhyamgram experimental farm, Bose Institute, Kolkata, according to Gavkare and Gupta (2013). DH5α strain of Escherichia coli and Agrobacterium AGL-1 cell line were used for cloning and plant transformation purposes, respectively.
Aphid bioassay
For the choice assay, an infested leaf with L. erysimi was placed at the center of 60-day-old B. juncea and R. indica plants kept in the same chamber and the plants were observed for 7 days. The number of aphids on each plant was calculated each day. Each experiment was carried out in triplicate. The significance in aphid survivability in this choice assay was analyzed by Student’s t test.
Treatment with phytohormones and qRT-PCR of marker regulatory genes
Five groups consisting of R. indica and B. juncea plants (60 days old each) were used for this experiment with three replicates. Three groups were sprayed with methyl jasmonate (MeJA) (100 µM), SA (100 µM), and ABA (100 µM), respectively. The fourth group was infested with mustard aphids (30 each) and the fifth group—plants with no treatment—were taken as control. About 1 g of the young infested/treated leaves of each plant was harvested after 1 day for RNA isolation by TRIzol (Invitrogen, Carlsbad, CA, USA) and subsequent cDNA preparation using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). The experiment was done in triplicate. A quantitative real-time PCR (qRT-PCR) analysis was carried out for the expression of defensin upon exogenous hormone application as well as for the MeJA, SA and ABA modulating genes upon aphid infestation. The primers used are given in Supplementary Table S1. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal reference gene. The PCR efficiencies were evaluated by the standard curve method (Pfaffl 2001) using five serial dilutions of cDNA samples for each primer pair. The correlation coefficients (R 2) and slopes were obtained from the standard curves and the PCR efficiency (E) for each primer pair was calculated according to the equation, E = (10−1/slope − 1). Primer pairs with amplification efficiency 0.96–1.00 were used. Differential gene expression data of qRT-PCR were analyzed using 2−ΔΔCT quantitation methods (Livak and Schmittgen 2001) and the significant difference was analyzed using Student’s t test.
Sequence homology and bioinformatics study of RiD
The full length coding sequence of Rorippa indica defensin (RiD), 243 bp (GenBank accession—KP893333) has been previously isolated and cloned from R. indica (Sarkar et al. 2016). The allergenicity of the RiD protein was analyzed by comparing the deduced amino acid sequence with all available protein sequences in the allergenic protein databases. The sequence of RiD was searched against the following databases—Structural Database of Allergenic Proteins (SDAP) (Ivanciuc et al. 2003), AllerTOP (Dimitrov et al. 2013) and Allergen Online database (Goodman et al. 2016) with full FASTA alignment on an 80 amino acid frame. A match of contiguous 8 amino acid with a threshold sequence identity of >35% was referred to be an allergen. Moreover, as allergenicity also depends on specific epitopes (Lorenz et al. 2015), the presence of IgE specific epitopes of RiD was searched in the AlgPred database (Saha and Raghava 2006). Potential pepsin cleavage sites of RiD amino acid were also analyzed in ExPASy PeptideCutter (Gasteiger et al. 2005).
In vitro pepsin digestibility of RiD
Simulated gastric fluid (SGF) reaction buffer was prepared (122.8 mg of NaCl to 59.2 mL of distilled water, pH to 1.2) with pepsin (Sigma-Aldrich, St. Louis, MO, USA) at a concentration of 3.2 mg/mL (approximately, 3460 U activity/mg). Purified RiD (Sarkar et al. 2016) was allowed to incubate in SGF at a concentration of 0.50 mg/mL to each reaction vial and the digestion was carried out at 37 °C for time periods of 0, 2, 5, 15, 30, 60 and 120 min. Only RiD without pepsin was used in the control setup. The reaction was stopped by adding 5 N NaOH. Each sample was then analyzed in 12% SDS-PAGE followed by Western blotting using anti-RiD antibody.
Thermal stability assay
CD analysis for monitoring the protein stability at different temperatures
Far UV-CD (circular dichroism) was used to measure the extent of secondary structural changes during different temperature treatments using Jasco Corp. J-815 Circular dichroism spectroscopy (CD) spectropolarimeter with a temperature controller within a wavelength range of 190–260 nm and a scan rate of 100 nm/min. RiD with concentration of approximately 0.2 mg/mL in PBS (phosphate-buffered saline), pH 7.0, was taken in a quartz cuvette with a 0.1-cm path length and was measured at different temperatures in the range of 30–90 °C at intervals of 10 °C. Spectra were obtained after cooling at an average of ten scans on a degree ellipticity scale.
Functional stability of RiD at different temperatures
Insecticidal activity of RiD against L. erysimi was studied using artificial diet incubated at different temperatures (30–90 °C). Nine µg/mL of RiD (LC50 value) was used in each set and 25 °C was considered to be the control temperature. The total mortality (P′) at each temperature-treated diet was calculated according to the formula, P′ = C + P (1 − C), considering a population (P) which would have survived in the absence of the insecticidal agent and where C is the natural mortality without the agent (Abbott 1925).
Targeted sera screening
Sera of ten allergic patients suffering from bronchial asthma, allergic rhinitis and dermatitis (age 21–50 years) were used for IgE-specific ELISA (enzyme-linked immunosorbent assay) following the method described by Ghosh et al. (2013). Sera were collected from an allergy clinic (Sinha Patho Lab, Dhanbad, India). Smokers and patients with chronic illness were excluded from this study. This experiment was approved by the ethics committee of the institute. Sera of healthy individuals without any history of allergenicity were used as the negative control. U-bottom microtiter plates (Tarsons, Kolkata, India) were used for this study. Each sample was measured thrice. The wells were coated with 50 μg of pure RiD protein in PBS and incubated overnight at 4 °C coated with a coating buffer (15 mM sodium carbonate, 35 mM sodium bicarbonate, 3 mM sodium azide; pH 9.6). The wells were next washed thrice (15 min each) with PBST (PBS–Tween 20), pH 7.0 and blocked with 50 μL of 1% BSA (Sigma-Aldrich) in PBST for 3 h at 4 °C. The wells were again washed with PBST three times and incubated with 50 μL of individual patient’s sera diluted (1:5) with blocking solution (PBST–BSA) at 25 °C for 16 h. The wells were then incubated with 50 μL of monoclonal mouse anti-human IgE–alkaline phosphatase conjugate (Sigma-Aldrich), diluted 1: 1000 times in blocking solution at 25 °C for 3 h following washing with PBST. Finally, 50 μL of liquid substrate, p-nitrophenyl phosphate (pNPP) system for ELISA (Sigma-Aldrich) was added and incubated in the dark at room temperature for 10–15 min for color development. The reaction was stopped by adding 3 N NaOH. The absorbances of each well were measured at 405 nm with an ELISA reader (POLARstar OPTIMA multidetection microplate reader, BMG Labtech, Ortenberg, Germany). The P/N value (ratio of average OD of individual patient sera with respect to the control group) greater than 3.5 for a particular serum was considered to be potentially IgE reactive (Chakraborty et al. 2005; Ghosh et al. 2013).
Histological studies using BALB/c mice
Histological studies were carried out taking healthy 8- to 10-week-old female BALB/c mice (22 ± 2 g) following the methods described by Ghosh et al. (2013). Standard conditions of controlled temperature (22 ± 2 °C), humidity (55 ± 16%) and a 12:12 h light:dark cycle were maintained and the mice were fed with a commercially available mannose-free pellet diet and water for 1 week for acclimatization to laboratory conditions. The mice were sensitized by the intraperitoneal (ip) route. Three groups were made of five mice each. Group-I mice were sensitized with 100 μL of PBS, group-II were sensitized with RiD and group-III with ovalbumin (OVA, HiMedia, Mumbai, India) (100 μg of protein in 100 μL PBS), by ip (intraperitoneal) route, once a day for 2 weeks. OVA is considered to be a potent allergen and is thus used as a positive control. On day 15, mice were killed for the collection of lung and gut tissue. Blood from each mice was collected in EDTA vials for further IgE estimation. The Animal Ethics Committee of Bose Institute approved the study protocol. The challenged mice were killed by cervical dislocation. The lung tissues were removed from the killed mice of each group, fixed immediately in 10% neutral buffered formaldehyde (v/v) (0.1 M phosphate buffer, pH 7.4) and embedded in paraffin. The tissues were cut into 3–5 μm sections, deparaffinized with xylene and graded ethanol, and stained with hematoxylin and eosin (H&E). To determine any increased mucus production and high concentration of carbohydrates or glycoproteins (Hauber and Zabel 2009), the lungs sections were also stained with periodic acid–Schiff (PAS) stain. Histopathological assessment was done under a light microscope and photographed using an in-line camera (Leica Microsystem DN1000; Camera DFC450C).
IgE estimation in mice
Sera were collected from the blood samples of all the four sets of mice and IgE levels were estimated according to the method described by Proust et al. (2009) with some modifications. Wells of U-bottom microtiter plates (Tarsons) were coated with the sera along with the coating buffer (15 mM sodium carbonate, 35 mM sodium bicarbonate, 3 mM sodium azide; pH 9.6) overnight at 4 °C. Next day, the wells were washed thrice (15 min each) with PBST, pH 7.0, and blocked with 50 μL of 1% bovine serum albumin (BSA) (Sigma-Aldrich) in PBST for 3 h at 4 °C. The wells were then washed again with PBST three times and incubated with 50 μL of anti-mouse IgE secondary antibody, HRP conjugate (Thermo Fisher Scientific, Waltham, MA, USA) diluted 1:1000 times in blocking solution at 25 °C for 3 h following washing with PBST. For color development, 100 μL of liquid substrate, OPD (o-phenylenediamine dihydrochloride, Sigma-Aldrich) was used. The absorbances of each well were measured at 450 nm with an ELISA reader (POLARstar OPTIMA multidetection microplate reader, BMG Labtech). IgE from each sera sample was measured in triplicate. The ratio of average OD of individual sera with respect to the control group was calculated where control was the average OD values of the sera of all healthy mice.
Construction of plant transformation vector and B. juncea transformation
The full length coding sequence of RiD (243 bp) amplified using gene-specific primers containing BamHI and SacI sites (underlined) (F-5′ TTTTGGATCCATGGCTAAGTTTGCT 3′ and R-5′AAAAGAGCTCTTAACATGGGAAGTAAC 3′) was introduced into the multiple cloning site (MCS) of modified pBI221 vector digested with the said restriction enzymes. The CaMV35S–RiD–NOS terminator cassette was excised out from modified pBI221 vector and was finally inserted into HindIII and EcoRI restriction sites of pCAMBIA1301 plant transformation vector. The final vector was named as pCAMBIA1301–CaMV35s–RiD. Finally, the recombinant vector plasmid containing RiD coding sequence was introduced into competent Agrobacterium tumefaciens AGL-1 cell line for plant transformation. pCAMBIA1301 vector not containing the coding sequence of RiD was used to regenerate vector control plants.
Agrobacterium-mediated B. juncea transformation was carried out according to Dutta et al. (2005b) with few modifications. The shootlets were regenerated in the presence of selection media containing 30 mg/L hygromycin and subsequently transferred to root inducing media, RIM (2 mg/L IBA) with 30 mg/L hygromycin. After well development of roots, the plantlets were transferred to Soilrite (KEL) for hardening. All cultures were maintained in culture room at a temperature of 25 ± 2 °C under a white light of 3000 lx for 16 h photoperiod. Another set of plants was used without Agrobacterium transformation, regenerated under similar condition in the absence of selection media (hygromycin) and used as untransformed control plants.
PCR analyses of the transgenics
Genomic DNA of all the putative transformants and untransformed plants were isolated using the CTAB method (Doyle and Doyle 1987). PCR analysis was carried out using β-glucuronidase (GUS) and hygromycin-specific primers (Supplementary Table S2).
β-Glucuronidase (GUS) analysis
The histochemical GUS assay of the leaves of the transgenic plants was carried out using 5-bromo-4-chloro-3-indolyl-beta-d-glucuronide (X-gluc) (Duchefa Biochemie, Haarlem, Netherlands) as a substrate, according to Jefferson (1987).
Southern hybridization
Stable integration of RiD-containing gene cassette was confirmed using Southern hybridization protocols of Sambrook and Russell (2001). Genomic DNA (20 µg) of each transformant as well as untransformed wild-type plants was digested using HindIII and separated on 0.8% agarose gel by electrophoresis. The gels were subsequently blotted into positively charged nylon membrane (GE Healthcare, Little Chalfont, UK). A 400 bp amplicon of hptII gene radiolabeled with [α32P] dCTP using DecaLabel DNA labeling kit (Fermentas, Thermo Fisher Scientific) was used as a probe for hybridization. Following hybridization, the membranes were washed and exposed to a phosphor imager screen and finally developed using GE Typhoon Phosphorimager.
Immunohistolocalization of RiD in transgenic B. juncea plants
Transverse sections of leaves, stems and roots from both transgenic and vector control plants were primarily incubated in 10% (v/v) TCA (Sigma-Aldrich) at 4 °C for 1 h followed by three to four times wash with ethanol:acetic acid (3:1, v/v) for complete removal of chlorophyll. The tissue sections were subsequently washed through a series of graded ethanol to water (90, 70, 50, 30% (v/v), respectively, each of 10–15 min duration) and then blocked with 3% (w/v) BSA (Merck, Gernsheim, Germany) in phosphate-buffered saline (PBS) at room temperature for 2 h. The tissue samples were then incubated with an anti-RiD antibody (1:8000 dilution) dissolved in BSA overnight at room temperature. After washing with 1× PBS, anti-rabbit IgG–fluorescein isothiocyanate (FITC)-conjugated secondary antibody (1:20,000 dilution) (Sigma-Aldrich) was added to the sections and incubated for 1 h at room temperature. Finally, the slides were examined with an Axioscope Carl Zeiss inverted fluorescent microscope using an excitation filter of 450–490 nm for FITC.
Segregation analysis of the transgene
T1 seeds from the self-pollinated T0 plants were collected and allowed to germinate on 30 µg/mL hygromycin (Duchefa Biochemie). Depending on the number of viable seeds, the segregation pattern of the RiD gene in progeny plants was generated and validated by χ 2 test.
Western-blot analysis
Total soluble protein was isolated from each transgenic line and vector control plants using 20 mM Tris–HCl (pH 7.5) and 0.2 mM PMSF (phenylmethane sulfonyl fluoride) (Sigma-Aldrich). The total soluble protein from R. indica was considered as positive control. The isolated protein from each sample was quantified by Bradford assay (Bradford 1976). Forty micrograms of total soluble protein from each plant sample were loaded onto 12% SDS-PAGE gel and then transferred onto a positively charged Hybond C membrane (GE Healthcare). Subsequently, the membrane was probed with anti-RiD polyclonal primary antibody at 1:8000 dilution followed by anti-rabbit IgG–horse radish peroxidase (HRP) conjugate (Sigma-Aldrich) at 1:20,000 dilutions. The presence or absence of bands was detected using chemiluminescence ECL reagent (Pierce, Thermo Fisher Scientific).
ELISA of total soluble protein
The quantification of the expression of RiD in the transgenic plants was done by ELISA. the wells of microtiter plates (Tarsons) were coated with 50 µg total soluble protein along with coating buffer (15 mM sodium carbonate, 35 mM sodium bicarbonate, 3 mM sodium azide; pH 9.6). The wells were then blocked with BSA and incubated with anti-RiD primary antibody at 1:8000 dilution followed by anti-rabbit IgG–horse radish peroxidase (HRP) conjugate (Sigma-Aldrich) at 1:20,000 dilutions. OPD (o-phenylenediamine dihydrochloride, Sigma-Aldrich) was used as a substrate for color development and the OD was recorded at 450 nm in a microtiter plate reader (POLARstar OPTIMA multidetection microplate reader, BMG Labtech).
In planta insect bioassay
The insecticidal activity of RiD in the transgenic plants was analyzed by using in planta insect bioassay cages. Thirty nymphs of L. erysimi of third instar stage were placed on the leaf surface of the plants and the whole leaf was placed inside a cage. Three cages were used for each plant. The bioassay was performed in triplicate for each transgenic line as well as vector control. The aphid survivability within the cages was monitored at every interval of 24 h for 7 days. The fecundity of aphids on the transgenic plants in response to the expressed RiD was monitored by the total number of nymphs produced on each plant at the end of the bioassay experiment. One-way ANOVA followed by Duncan’s multiple range tests (DMRT) were carried out to compare the differences between the vector control and the transgenic plants and P < 0.05 was considered to be statistically significant.
Results
Aphid bioassay
A choice assay was conducted between B. juncea and R. indica to understand the aphid behavior in the presence of both. When given a choice, it was seen that aphids at first were attracted to both the plants, but eventually the population on B. juncea increased with time (Supplementary Fig. S1).
JA and not SA/ABA contributes to RiD-dependent defense mechanism in R. indica
To determine the signaling hormone behind the expression of RiD in R. indica, the plants were sprayed with MeJA, SA and ABA individually. Another plant was infested with aphids and a qRT-PCR was performed before and after each treatment. RiD was seen to be highly expressed when the plants were infested with aphids (33.5-fold) and even more after MeJA treatment (82.12-fold), but not much after the treatment with SA or ABA (Fig. 1a). We also quantified the induction of marker regulatory genes for each signaling pathway (JA, SA and ABA) after aphid infestation using qRT-PCR. As shown in Fig. 1b, the levels of defensin and JA-responsive genes—LOX3, LOX4, OPR1 and OPR2—were seen to be highly upregulated in response to aphid infestation with 33.5-, 52.34-, 63.14-, 42.56- and 62.2-fold increase, respectively. ABA-responsive genes (NCED1 and NCED2) were also seen to be upregulated upon aphid infestation with 19.23- and 21.05-fold, respectively. However, no SA-responsive genes were seen to be induced upon aphid infestation (Fig. 1b).
RiD is a jasmonate-regulated gene. a Quantitative real-time PCR analysis of R. indica defensin (RiD) under the aphid infestation and application of exogenous MeJA, SA and ABA. b Real-time expression analysis of JA marker genes (defensin, LOX3, LOX4, OPR1, OPR2), ABA marker genes (NCED1, NCED2) and SA marker genes (NPR1, PR1) in response to aphid infestation. Bars represent standard error (SE) of three biological replicates (n = 3). The significant changes, P ≤ 0.01 (analyzed by Student’s t test), are marked by asterisk
Bioinformatic analyses
In silico analysis of transgenes is a preliminary step before development of transgenics. According to FAO/WHO (2001) and Codex Alimentarius Guideline (2009), a match of minimum eight identical and contiguous amino acids with a threshold value of 35% between the query protein and the database of allergens is considered to be a possible allergen. For RiD, no significant match was found in the search for short contiguous sequences in the AllergenOnline database (Goodman et al. 2016), SDAP allergenic database (Ivanciuc et al. 2003) and AllerTOP (Dimitrov et al. 2013). Search for IgE cross-reactive epitopes in RiD using AlgPred database (Saha and Raghava 2006) also showed that the amino acid sequence of RiD does not contain any probable IgE binding epitope.
Pepsin digestion
After incubation of RiD in simulated gastric fluid (SGF), the digestion profile of RiD was analyzed in 12% SDS-PAGE stained with Coomassie Brilliant Blue, which determined that RiD was completely digested within 2 min of treatment (Fig. 2a). RiD was also not detectable in the corresponding immunoblot after 2 min of digestion (Fig. 2b). In silico pepsin digestion using ExPASy PeptideCutter also showed 19 cleavage sites in RiD (Supplementary Table S3) corroborating the digestion data.
Pepsin digestion and heat stability assay of RiD. a 12% SDS-PAGE profile of SGF-treated RiD. Lane M molecular weight marker; lane C RiD; lanes 0–120 RiD treated with SGF for 0, 2, 5, 15, 30, 60 and 120 min, respectively, and stained with Coomassie Brilliant Blue. b Corresponding Western blot of the 12% SDS-PAGE using anti-RiD antibody showing degradation of RiD in SGF. c CD spectra of RiD at different temperatures recorded over wavelength 190–260 nm. d Effect of RiD as an insecticidal agent against L. erysimi at different temperatures of 30, 50, 60, 70 and 90 °C for 30 min
Thermal stability assay
CD analysis
In the CD analysis, gradual change in the ellipticity was observed with the increase in temperature. Temperature-induced loss of structural integrity was evident at 50 °C. After heating at 70 °C and onward, the negative ellipticity (λ max) of RiD was completely lost, indicating complete loss of secondary structural folds of RiD due to heat treatment (Fig. 2c). Loss of structure suggests that the protein is not stable at higher temperatures.
Functional activity
The insecticidal efficacy of RiD was also lost at 50 °C and onward, indicating the possibility of disruption of the insecticidal motif in the secondary structure (Fig. 2d).
IgE-specific ELISA
In targeted sera screening, significantly low IgE cross-reactivity (P/N value = 0.56 ± 0.09–1.89 ± 0.21) was detected when RiD was used as probe. In contrast, in response to OVA, a high IgE cross-reactivity was detected (P/N value ≥3.5) using the sera from the allergic patients (Table 1).
Histopathological analysis in animal model
H&E staining
Hematoxylin and eosin (H&E) staining of lung section of OVA-sensitized mice showed severe inflammation characterized by hyperplasia of the inner lining of the bronchial membranes. Extensive infiltration of eosinophils was seen in the peribronchial tissues, whereas RiD-sensitized mice showed normal bronchioles with no inflammation similar to that of PBS-sensitized control mice (Fig. 3a, b). Degranulation of mast cells was well observed in the mice sensitized with OVA, but not in RiD/PBS-sensitized mice lungs.
a, b Hematoxylin and eosin (H&E) staining of all the treated lung sections showing inflammation in the bronchioles of OVA-sensitized mice. Arrows indicate degranulation of mast cells. c PAS staining of lung sections of mice sensitized with PBS, OVA and RiD, showing hyperplasia of goblet cells and mucus accumulation in OVA-sensitized lung section. a–c Bars 100 µm
PAS staining
Visible hyperplasia and high mucus accumulation was observed in OVA-sensitized lung sections, but no such morphology was observed in RiD/PBS-sensitized ones (Fig. 3c).
IgE estimation in mice
Sera from each set of mice were collected for IgE level estimation. Sera from mice sensitized with OVA were seen to have elevated levels of IgE (4.15 ± 0.92) with respect to control, whereas in mice sensitized with RiD no significant level of IgE was detected as it was found in the case of PBS-sensitized mice sera (Supplementary Table S4).
Development of Brassica juncea transgenic plants expressing RiD
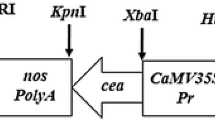
The chimeric plant transformation vector pCAMBIA1301–CaMV35s–RiD (Fig. 4a) in Agrobacterium tumefaciens AGL-1 strain was used to transform B. juncea B-85. A total of 834 explants were transformed out of which only 62 putative Brassica transformants were successfully generated with hygromycin selection (30 mg/L). A representative image is shown in Supplementary Fig. S2a.
a Diagrammatic representation of T-DNA cassette of plant expression vector pCAMBIA1301–CaMV35s-RiD. CaMV35S cauliflower mosaic virus 35S promoter, CaMV35S poly-A tail cauliflower mosaic virus 35S terminator, RiD Rorippa indica defensin, hptII hygromycin phosphotransferase II, GUS β-glucuronidase, NOS terminator nopaline synthase terminator; LB left border of T-DNA, RB right border of T-DNA. b Southern-blot analysis of the HindIII digested genomic DNA of ten T0 transgenic B. juncea plants. c Southern-blot analysis of HindIII digested genomic DNA from leaves of five T1 progenies of P5 Southern-positive plant showing single-copy events. Amplified 1000 bp hpt gene from pCAMBIA1301 vector was used as positive control (lane 1) and HindIII digested genomic DNA from untransformed control plants as negative control (lane 2). Approximate DNA molecular weight markers are indicated on the left of each blot
Screening for the stable integration and inheritance of RiD
Putative T0 transformants were preliminary screened by GUS and hptII-specific primers using PCR. An amplification of 1009 bp of GUS (Supplementary Fig. S2b) and 1024 bp of hptII (Supplementary Fig. S2c) was identified in RiD transformants, but not in the untransformed control plants. Ten PCR-positive plants were screened by Southern hybridization to check the presence of the transgene. All the plants showed different banding patterns. P22 was a double-copy insertion event and there was absence of any band in plant P21 (Fig. 4b). All independently transformed Southern-positive plants were allowed to self-pollinate. The seeds were subsequently grown into T1 generation plants. Arbitrarily, five independent T1 transgenic lines were chosen for downstream functional and expression analysis. The integration of the transgene in plants of T1 transgenic lines was further confirmed using Southern-blot hybridization. Figure 4c shows a representative blot of five plants from the P5T1 line. The inheritance pattern of the transgene was observed by growing the seeds from the self-pollinated T0 plants as well as untransformed control plants on a hygromycin (30 µg/mL) plate for 1 week (Supplementary Fig. S2d). Segregation of the RiD transgene followed the 3:1 Mendelian segregation pattern and the observed ratio was validated using χ 2 test (Table 2). Further analysis of the expression of the RiD transgene were carried out using five lines of single-copy insertion events (P5T1, P8T1, P12T1, P23T1, P45T1) corresponding to their respective T0 events.
Expression of RiD in the transgenic lines
Preliminary expression of the transgene in the transgenic lines was analyzed using histochemical GUS assay. Leaves from the five T1 transgenic lines (P5T1-1, P8T1-10, P12T1-6, P23T1-12, P45T1-4) as well as vector control plant showed constitutive expression, whereas there was no expression seen in the untransformed leaves (Fig. 5a). Expression of RiD in the T1 transgenics was further validated by Western-blot analysis and ELISA using anti-RiD antibody. Five T0 lines, namely P5, P8, P12, P23 and P45, and their corresponding T1 transgenic plants (P5T1-1, P8T1-10, P12T1-6, P23T1-12, P45T1-4) were subjected to immunoblot assay. A band at ~6 kDa was clearly observed in Western-blot analyses which indicated the presence of expressed RiD in the transgenics and not in the vector control B. juncea (Fig. 5b, c). The quantitative expression of RiD was analyzed by indirect ELISA. The expression levels ranged from 0.27 to 0.86% of total soluble protein with the highest in P23T1-12 and lowest in P5T1-1 and P45T1-4 (Fig. 5d). Inmmunohistofluoresence localization of the expressed RiD was studied by treating the transverse section of leaves, stem and root of the transgenic plants as well as vector control plants with anti-RiD primary antibody, followed by FITC-conjugated anti-rabbit IgG. Constitutive expression of RiD was seen in the transformed plant sections under fluorescence microscopy, whereas no fluorescence, and hence no expression, was observed in the vector control plant sections (Fig. 5e).
Detection of RiD expression in transgenic plants. a Histochemical GUS staining of mature leaves from five T1 transgenic B. juncea plants (P5T1-1, P8T1-10, P12T1-6, P23T1-12, P45T1-4) and one vector control plant showing constitutive GUS expression in all, but no expression in the un-transformed control leaf. b, c Western blot using total soluble protein extract from the five T0 and their corresponding T1 transgenic B. juncea lines expressing RiD. Total soluble protein (40 µg) extracts from leaves of each of the five independent T1 transgenic lines, aphid-infested R. indica plants (+ve control) and vector control plant (negative control) were probed with anti-RiD antibody showing a clear band at 6 kDa. d ELISA analysis for quantitative estimation of RiD in total soluble protein in five T1 transgenic lines. Bars represent SE of three replicates and significant expression was marked by asterisk (P ≤ 0.05). e Immunohistofluorescence assay of transgenic B. juncea plants. Representative image of transverse sections of leaf, stem and root from both vector control and transgenic line P5T1-1 incubated with anti-RiD primary antibody and anti-rabbit IgG–FITC-conjugated secondary antibody. The accumulation of RiD is clearly visible by the green fluorescence in the transgenic plant sections, whereas the transverse sections from control plant showed no green fluorescence. Bar 10 µm
In planta insect bioassay of the transgenic lines
The insecticidal activity of T1 transgenic B. juncea lines expressing RiD was assayed using bioassay cages infested with 30 L. erysimi in each cage (Supplementary Fig. S2e). Survival of the aphid nymphs reduced to 19 ± 0.33 (mean ± SE) (36.67%) from 30 ± 0 (100%) per control plant over the total bioassay period, whereas decline of aphid population on the RiD expressing five T1 transgenic lines varied from 10 ± 0.57 (66.67%) to 16 ± 0.57 (46.67%) at the end of the assay. The fecundity of new nymphs was calculated for each transgenic line. Transgenic lines P23T1 and P12T1 showed maximum reduction in insect survivability and aphid fecundity after 7 days (Fig. 6a, b). The mean number of surviving mustard aphids on the transgenic and control plants were found to be statistically significant (P < 0.05) after 3 days.
In planta insect bioassay. a Percentage survivability of aphids on five T1 transgenic lines and on vector control plant. P23T1 and P12T1 show the highest mortality of aphids of about 66.67% reduction in aphid survivability. b Bar diagram showing fecundity pattern of L. erysimi (mean number of nymphs produced per plant) on vector control and the five T1 transgenic lines. Bars represent SE with n = 3
Discussion
Combating L. erysimi for better production and yield of mustard (Brassica juncea) has always been a concern. Various insecticidal agents have been exploited through the development of transgenic B. juncea plants to effectively reduce aphid colonization (Kanrar et al. 2002; Rahbe et al. 2003; Dutta et al. 2005a; Mondal et al. 2006). As there is no report available for a resistant genotype in the Brassica germplasm, a wild crucifer, R. indica, was explored to identify aphid-tolerant genes using cDNA AFLP analysis (Bandopadhyay et al. 2013). A novel defensin gene (R. indica defensin, RiD) was isolated from the aphid-tolerant R. indica and the said protein was seen to affect the population growth of aphids significantly (Bandopadhyay et al. 2013; Sarkar et al. 2016). Plant defensins have a negative effect on phloem-feeding aphids (Kusnierczyk et al. 2011) and are also widely reported to be a JA-responsive marker gene (Moran and Thompson 2001). Arabidopsis PDF 1 is one of the known defensins which was reported to be upregulated in response to Brevicoryne brassicae (Kusnierczyk et al. 2011). Application of MeJA has also been seen to induce defensins in tomato which helps in the reduction of aphid population growth (Cooper and Goggin 2005). On the contrary, in the Arabidopsis thaliana–Myzus persicae model system, aphid feeding was reported to induce the expression of SA-responsive marker genes (Moran and Thompson 2001; De Vos et al. 2005). To study the possible mode of regulation of RiD in the present study, expression of JA/SA/ABA modulating genes along with RiD was observed using qRT-PCR during aphid infestation. Lipoxygenase (LOX3, LOX4) and 12-oxo-phytodienoic acid reductase (OPR1 and OPR2) are the key regulatory enzymes for the JA biosynthesis pathway (He et al. 2002) which tend to increase in response to herbivory (Arimura et al. 2000). In this study, significant upregulation of RiD (by 33.5-fold) was seen upon aphid infestation and even more (by 82.12-fold) upon exogenous application of MeJA on R. indica (Fig. 1a). Upon aphid infestation, JA-responsive genes were highly upregulated in R. indica. LOX3 and LOX4 were induced by 52.34- and 63.14-fold, respectively. OPR1 and OPR2 were upregulated by 42.56- and 62.21-fold, respectively (Fig. 1b). The ABA-regulatory genes, NCED1 and NCED2 were also seen to be upregulated during aphid infestation with 19.23- and 21.05-fold, respectively (Fig. 1b). This may be a possibility because ABA does not directly play any role, but helps to maintain the physiological conditions of aphid-infested plants (Zhu-Salzman et al. 2004). On the other hand, expression of RiD was not significant in response to exogenous application of ABA/SA (Fig. 1a). Furthermore, the SA-responsive marker genes (PR1 and NPR1) were not significantly expressed during or after aphid infestation (Fig. 1b). All these results indicated that L. erysimi induces the JA-regulatory pathway which in turn induces the expression of RiD and hence mediates the defense response in R. indica.
Moreover, as food allergenicity has become one of the major concerns for food/food products derived through genetic engineering, the safety evaluation of RiD was necessary before transgenic application. From the present study, it is evident that RiD has no sequence similarity with any known allergen. In silico analysis also showed the absence of significant IgE epitopes in RiD. As many protein structures which are stable and resistant toward digestion act as a potent allergen (Astwood et al. 1996; Apostolovic et al. 2016), another important assessment of allergenic proteins is its stability in pepsin digestion (Plundrich et al. 2015; Toomer et al. 2015; Hirano et al. 2016). RiD was seen to be digested within 2 min of treatment with SGF (Fig. 2a, b), supporting the in silico results (Supplementary Table S3). CD analysis showed that the secondary structure of RiD starts to denature at 50 °C and gets distorted at higher temperatures, rendering it heat labile (Fig. 2c). The functional activity of RiD was also disrupted at 50 °C and above, which was evident from the loss of its insecticidal property as shown in Fig. 2d. As allergens are important IgE-sensitizing agents, sera IgE estimation is also considered to be the most advanced approach to detect food allergens (Erwin et al. 2010; Ogunlade et al. 2012). Allergenic proteins show high IgE cross-reactivity with the sera specimens from the allergic patients (Hauber and Zabel 2009; Hancock et al. 2012; Goodman et al. 2013). In the present study, targeted sera-specific ELISA from allergenic patients was conducted to observe the IgE cross-reactivity with RiD protein. A significantly low IgE cross-reactivity was seen using RiD as probe compared to a very high cross-reactivity while using OVA (Table 1). This confirms that RiD does not have IgE-specific epitope as suggested by the in silico analysis and does not sensitize IgE in human sera.
Allergenicity assessment is incomplete without animal model studies. BALB/c mice are the most widely used animal models for allergenicity assessment for their high similarity with humans with respect to many important immunological mechanisms (Nieuwenhuizen et al. 2006; Ghosh et al. 2013). Many allergens such as Anisakis (Nieuwenhuizen et al. 2006), OVA (Hutson et al. 1988), Ara h (Bernard et al. 2014; Apostolovic et al. 2016; Chen et al. 2016) induce lung inflammation in mice models, whereas ω -HXTX-Hv1a (Hvt) from the Australian funnel web spider Hadronyche versuta (Naeem et al. 2015), mutated form of ASAL from Allium sativum (Ghosh et al. 2013), bacterial codA (Singh et al. 2006) and many more have been studied for risk assessment and consequently used as a transgene (Singh et al. 2006; Ghosh et al. 2016). In the present study, three sets of five mice each were sensitized with RiD, PBS (as negative control) and OVA (as positive control), respectively. Lung histopathology was studied in all the sets as it is considered to be one of the most relevant approaches for studying allergenic response in vivo (Koth et al. 2004; Brandenberger et al. 2014). The difference in the allergenic reaction in response to OVA and RiD/PBS was well observed in the H&E and PAS-stained lung sections (Fig. 3). An evident hyperplasia of alveoli and mucus deposition was observed in the OVA-treated lung sections, whereas no such observation was found in the RiD/PBS-treated sections (Fig. 3). The sera of each group of mice treated with OVA, RiD and PBS were also analyzed for IgE levels. Elevated levels of IgE were detected in OVA-treated sera (4.15 ± 0.92), but not in RiD/PBS-treated sera (Supplementary Table S4). The results from targeted sera screening as well as IgE detection in mice suggest that RiD is IgE-nonreactive and biologically safe. Hence, RiD was finally utilized for the development of transgenic B. juncea.
Previous studies report that transgenic plants expressing various defensins showing insecticidal and protease activities have been effective against different insects (Bloch and Richardson 1991; Liu et al. 2006; Santos et al. 2010). In this study, the full length coding sequence of RiD was successfully integrated into susceptible B. juncea cultivar B-85. Initially, the presence/absence of the transgene was detected via PCR analysis. Next, the PCR-positive plants were analyzed for single-copy transgene insertion through Southern-blot analysis. Detailed molecular analyses in the T1 plants established stable RiD integration in the transgenic plants. Eight out of ten plants (P1, P5, P8, P12, P18, P23, P32 and P45) were seen to have single-copy insertion in the T0 generation, whereas plant P22 was seen to have double-copy insertion (Fig. 4b). As multiple-copy gene insertion leads to gene silencing (Tang et al. 2007), transgenic plants with single copy of transgene were preferred to those with multiple copies and were used in further analyses. Furthermore, the segregation of the transgene in T1 generation was analyzed using χ2 test and there was no significant difference between the observed and expected Mendelian ratio (3:1) (Table 2).
The expression of RiD in the single-copy transgenic plants was analyzed by GUS, Western-blot analysis and ELISA. GUS expression analysis of the leaves from five T1 transgenic plants (P5T1-1, P8T1-10, P12T1-6, P23T1-12 and P45T1-4) revealed the expression of GUS in all but not in the untransformed control plant (Fig. 5a). Western-blot analysis confirmed the expression of RiD in the five T1 transgenic plants corresponding to the T0 lines, but not in the vector control B. juncea plants (Fig. 5b, c). Quantitative expression of RiD in the T1 transgenic lines was monitored using ELISA. The expression level of RiD in different transformants varied from 0.27 to 0.86% of total soluble protein with the highest in P23T1-12, followed by P12T1-6 and P8T1-10 (Fig. 5d). This difference suggests that the transgene might have randomly integrated at different transcriptionally active sites in the plant genome of each independent transgenic line. Immunohistochemical analysis was also conducted to visualize the expression of RiD in the T1 plant tissues. RiD was seen to be strongly expressed in all the tissues indicating its presence in leaves, stems as well as roots (Fig. 5e). Finally, in planta bioassay results clearly indicated that the constitutive expression of RiD in transgenic plants reduces the insect population significantly varying from 46.67 to 66.67% (Fig. 6a). P23T1 and P12T1 both show maximum reduction of aphid population up to 66.67%, followed by P8T1 showing a reduction of 56.67%, which is comparable to the expression level of RiD in Western blots (Fig. 5c) and ELISA (Fig. 5d) in the said lines. This reduction of insect population was much significant compared to other transgenic lines expressing insecticidal agents such as wheat germ agglutinin (WGA) with 55% mortality (Kanrar et al. 2002), ASAL with 89% (Dutta et al. 2005a), (E)-β-farnesene (Eβf) with 75% (Verma et al. 2015), and GNA (snowdrop lectin gene) with 51–54% (Ramesh et al. 2004) reduction in aphid population. The fecundity of aphids in RiD-expressing transgenic plants was seen to vary between 18 and 45% compared to the control plants (Fig. 6b). This was significantly lower compared to 60–64 and 35–39% in case of 35SASAL- and RSsASAL-expressing B. juncea plants, respectively (Dutta et al. 2005a).
Conclusion
Collectively, the present study was an attempt to enhance the aphid tolerance property in B. juncea by introducing a novel insecticidal protein, RiD, through the development of transgenics. RiD, which is also a JA-marker gene is found to be biologically safe and has been seen to effectively reduce the aphid population in transgenic mustard plants up to 66.67% and this can be well used to instigate sustainable agricultural practices.
Author contribution statement
PS and SRS conceived and designed the experiment. KJ and PS performed the animal model studies. Rest of the experiment was done by PS. PS wrote the manuscript. All authors read and approved the final manuscript.
Abbreviations
- JA:
-
Jasmonic acid
- LC50 :
-
Lethal concentration causing 50% mortality of test population
- MeJA:
-
Methyl jasmonate
- OVA:
-
Ovalbumin
- RiD:
-
Rorippa indica defensin
- SA:
-
Salicylic acid
References
Abbott WS (1925) A method of computing the effectiveness of insecticide. J Econ Entomol 18:265–267
Apostolovic D, Vucinic DS, Jongh HH, Govardus A, Jong H, Mihailovic J et al (2016) Conformational stability of digestion-resistant peptides of peanut conglutins reveals the molecular basis of their allergenicity. Sci Rep 6:29249
Arimura GI, Ozawa R, Shimoda T, Nishioka T, Boland W, Takabayashi J (2000) Herbivory-induced volatiles elicit defence genes in lima bean leaves. Nature 406:512–515
Astwood JD, Leach JN, Fuchs RL (1996) Stability of food allergens to digestion in vitro. Nat Biotech 14:1269–1273
Bandopadhyay L, Basu D, Sikdar SR (2013) Identification of genes involved in wild crucifer Rorippa indica resistance response on mustard aphid Lipaphis erysimi challenge. PLoS One 8(9):e73632
Bernard H, Ah-Leung S, Drumare MF, Feraudet-Tarisse C, Verhasselt V, Wal JM, Creminon C, Adel-Patient K (2014) Peanut allergens are rapidly transferred in human breast milk and can prevent sensitization in mice. Allergy 69:888–897
Bloch CJ, Richardson M (1991) A new family of small (5 kDa) protein inhibitors of insect α-amylase from seeds of sorghum (Sorghum bicolour (L.) Moench) have sequence homologies with wheat gamma-thionins. FEBS Lett 279:101–104
Brandenberger C, Li N, Jackson-Humbles DN, Rockwell CE, Wagner JG, Harkema JR (2014) Enhanced allergic airway disease in old mice is associated with a Th17 response. Clin Exp Allergy 44(10):1282–1292
Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chakraborty P, Ghosh D, Chowdhury I, Roy I, Chatterjee S, Chanda S et al (2005) Aerobiological and immunochemical studies on Carica papaya L. pollen: an aeroallergen from India. Allergy 60:920–926
Chen KC, Lin CY, Kuan CC, Sung HY, Chen CS (2002) A novel defensin encoded by a mungbean cDNA exhibits insecticidal activity against bruchid. J Agric Food Chem 50:7258–7263
Chen X, Negi SS, Liao S, Gao V, Braun W, Dreskin SC (2016) Conformational IgE epitopes of peanut allergens Ara h 2 and Ara h 6. Clin Exp Allergy 46:1120–1128
Choi MS, Kim YH, Park HM, Seo BY, Jung JK, Kim ST et al (2009) Expression of Br D1, a plant defensin from Brassica rapa, confers resistance against brown planthopper (Nilaparvata lugens) in transgenic rices. Mol Cells 28:131–137
Codex Alimentarius (2009) Guidelines for the design and implementation of national regulatory food safety assurance programme associated with the use of veterinary drugs in food producing animals. Codex Alimentarius Commission. GL, pp 71–2009
Cooper WR, Goggin FL (2005) Effects of jasmonate-induced defenses in tomato on the potato aphid, Macrosiphum euphorbiae. Entomol Exp Appl 115:107–115
Das A, Roy A, Hess D, Das S (2013) Characterization of a highly potent insecticidal lectin from Colocasia esculenta tuber and cloning of its coding sequence. Am J Plant Sci 4:408–416
De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ et al (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant-Microbe Interact 18:923–937
Dimitrov I, Flower DR, Doytchinova I (2013) AllerTOP—a server for in silico prediction of allergens. BMC Bioinfo 14(Suppl 6):S4
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Dutta I, Majumdar P, Saha P, Ray K, Das S (2005a) Constitutive and phloem specific expression of Allium sativum leaf agglutinin (ASAL) to engineer aphid (Lipaphis erysimi) resistance in transgenic Indian mustard (Brassica juncea). Plant Sci 169:996–1007
Dutta I, Saha P, Majumdar P, Sarkar A, Chakraborti D, Banerjee S, Das S (2005b) The efficacy of a novel insecticidal protein, Allium sativum leaf lectin (ASAL), against homopteran insects monitored in transgenic tobacco. Plant Biotech J 3:601–611
Erwin EA, James HR, Gutekunst HM, Russo JM, Kelleher KJ, Platts-Mills TA (2010) Serum IgE measurement and detection of food allergy in pediatric patients with eosinophilic esophagitis. Ann Allergy Asthma Immunol 104:496–502
FAO/WHO (2001) Allergenicity of genetically modified foods; food and agriculture organisation of the United Nations, Rome, Italy
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook. Humana, Humana Press, New York, USA, pp 571–607
Gavkare O, Gupta S (2013) An improved method for rearing green peach aphid Myzus persicae (Sulzer). Internat J Plant Protec 6(2):492–493
Ghosh P, Roy A, Chakraborty J, Das S (2013) Biological safety assessment of mutant variant of Allium sativum leaf agglutinin (mASAL), a novel antifungal protein for future transgenic application. J Agric Food Chem 61(48):11858–11864
Ghosh P, Sen S, Chakraborty P, Das S (2016) Monitoring the efficacy of mutated Allium sativum leaf lectin in transgenic rice against Rhizoctonia solani. BMC Biotech 16:24
Goodman RE, Panda R, Ariyarathna H (2013) Evaluation of endogenous allergens for the safety evaluation of genetically engineered food crops: review of potential risks, test methods, examples and relevance. J Agric Food Chem 61(35):8317–8332
Goodman RE, Ebisawa M, Ferreira F, Sampson HA, van Ree R, Vieths S et al (2016) AllergenOnline: a peer-reviewed, curated allergen database to assess novel food proteins for potential cross-reactivity. Mol Nutr Food Res 60:1183–1198
Hancock DB, Romieu I, Chiu GY, Monge JS, Li H, Navarro BE, London SJ (2012) STAT6 and LRP1 polymorphisms are associated with food allergen sensitization in Mexican children. J Allergy Clin Immunol 129(6):1673–1676
Hauber HP, Zabel P (2009) PAS staining of bronchoalveolar lavage cells for differential diagnosis of interstitial lung disease. Diagn Pathol 4:13
He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128(3):876–884
Hirano K, Hino S, Oshima K, Nadano D, Urisu A, Takaiwa F, Matsuda T (2016) Evaluation of allergenic potential for rice seed protein components utilizing a rice proteome database and an allergen database in combination with IgE-binding of recombinant proteins. Biosci Biotechnol Biochem 80(3):564–573
Hutson PA, Church MK, Clay TP, Miller P, Holgate ST (1988) Early and late-phase bronchoconstriction after allergen challenge of nonanesthetized guinea pigs: I. The association of disordered airway physiology to leukocyte infiltration. Am Rev Respir Dis 137(3):548–557
Islam A (2008) Preliminary risk assessment of a novel antifungal defensin peptide from chickpea (Cicer arietinum L.). Appl Biosaf 13(4):222–230
Ivanciuc O, Mathura V, Midoro-Horiuti T, Braun W, Goldblum RM, Schein CH (2003) Detecting potential IgE-reactive sites on food proteins using a sequence and structure database. SDAP-food. J Agric Food Chem 51(16):4830–4837
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Kanrar S, Venkateswari J, Kirti PB, Chopra VL (2002) Transgenic Indian mustard (Brassica juncea) with resistance to the mustard aphid (Lipaphis erysimi Kalt.). Plant Cell Rep 20:976–981
Kiba A, Tomiyama H, Takahashi H, Hamada H, Ohnishi K, Okuno T et al (2003) Induction of resistance and expression of defense-related genes in tobacco leaves infiltrated with Ralstonia solanacearum. Plant Cell Physiol 44:287–295
Koth LL, Rodriguez MW, Bernstein XL, Chan S, Huang X, Charo IF, Rollins BJ, Erle DJ (2004) Aspergillus antigen induces robust Th2 cytokine production, inflammation, airway hyperreactivity and fibrosis in the absence of MCP-1 or CCR2. Respir Res 5:12
Kusnierczyk A, Tran DH, Winge P, Jorstad TS, Reese JC, Troczyńska J, Bones AM (2011) Testing the importance of jasmonate signalling in induction of plant defences upon cabbage aphid (Brevicoryne brassicae) attack. BMC Genom 12:423
Liu YJ, Cheng CS, Lai SM, Hsu MP, Chen CS, Lyu PC (2006) Solution structure of the plant defensin VrD1 from mung bean and its possible role in insecticidal activity against bruchids. Proteins 63:777–786
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408
Lorenz AR, Scheurer S, Vieths S (2015) Food allergens: molecular and immunological aspects, allergen databases and cross-reactivity. Chem Immunol Allerg 101:18–29
Mandal P, Sikdar SR (2003) Plant regeneration from mesophyll protoplasts of Rorippa indica (L.) Hiern, a wild crucifer. Curr Sci 85:1451–1454
Manners JM, Penninckx IA, Vermaere K et al (1998) The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol Biol 38:1071–1080
Melo FR, Rigden DJ, Franco OL, Mello LV, Ary MB, Grossi de Sa MF, Bloch C Jr (2002) Inhibition of trypsin by cowpea thionin: characterization, molecular modelling, and docking. Proteins 48:311–319
Mondal HA, Maiti MK, Basu A, Sen S, Ghosh AK, Sen SK (2006) Transgenic expression of onion leaf lectin in Indian mustard offers protection against aphid colonization. Crop Sci 46:2022–2032
Moran P, Thompson GA (2001) Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol 125:1074–1085
Moran P, Cheng Y, Cassell JL, Thompson GA (2002) Gene expression profiling of Arabidopsis thaliana in compatible plant–aphid interactions. Arch Insect Biochem Physiol 51:182–203
Morkunas I, Mai VC, Gabrys B (2011) Phytohormonal signaling in plant responses to aphid feeding. Acta Physiol Plant 33:2057–2073
Naeem MA, Mukhtar Z, Khan QM, Khalid ZM, Rasool A, Mansoor S (2015) Biosafety studies of transgenic cotton expressing insecticidal gene from Australian funnel web spider (Hadronyche versuta). Pak J Agri Sci 52(3):685–690
Nieuwenhuizen N, Lopata AL, Jeebhay MF, Herbert DR, Robins TG, Brombacher F (2006) Exposure to the fish parasite Anisakis causes allergic airway hyperreactivity and dermatitis. J Allergy Clin Immunol 117(5):1098–1105
Ogunlade OA, Ige OM, Arinola OG, Onadeko BO (2012) Allergen-specific immunoglobulin E (IgE) antibodies and skin test reactivity in patients with asthma in Nigeria. Clin Immunol Immunopathol Res 4(3):25–28
Penninckx IA, Thomma BP, Buchala A, Metraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10:2103–2113
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Plundrich NJ, White BL, Dean LL, Davis JP, Foegedingc EA, Lila MA (2015) Stability and immunogenicity of hypoallergenic peanut protein–polyphenol complexes during in vitro pepsin digestion. Food Funct 6:2145–2154
Proust B, Astier C, Renaudin JM et al (2009) A murine model of cow milk protein-induced allergic reaction: use for safety assessment of hidden milk allergens. Eur Ann Aller Clin Immunol 41:85–94
Rahbe Y, Deraison C, Bonade-Bottino M, Girard C, Nardon C, Jouanin L (2003) Effects of the cysteine protease inhibitor oryzacystatin (OC-I) on different aphids and reduced performance of Myzus persicae on OC-I expressing transgenic oilseed rape. Plant Sci 164:441–450
Ramesh S, Nagadhara D, Reddy VD, Rao KV (2004) Production of transgenic indica rice resistant to yellow stem borer and sap-sucking insects using super- binary vectors of Agrobacterium tumefaciens. Plant Sci 166(4):1077–1085
Saha S, Raghava GPS (2006) AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res 34:202–209
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, vol 1, issue 6. Cold Spring Harbor Laboratory, Cold Spring Harbor, Protocol 8
Santos IS, Carvalho AO, Filho GA, Nascimento VV, Machado OLT, Gomes VM (2010) Purification of a defensin isolated from Vigna unguiculata seeds, its functional expression in Escherichia coli, and assessment of its insect α-amylase inhibitory activity. Protein Expr Purif 71:8–15
Sarkar P, Jana J, Chatterjee S, Sikdar SR (2016) Functional characterization of Rorippa indica defensin and its efficacy against Lipaphis erysimi. SpringerPlus 5:511
Sels J, Mathys J, De Coninck BM, Cammue BP, De Bolle MF (2008) Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem 46:941–950
Singh AK, Mehta AK, Sridhara S, Gaur SN, Singh BP, Sarma PU, Arora N (2006) Allergenicity assessment of transgenic mustard (Brassica juncea) expressing bacterial codA gene. Allergy 61:491–497
Tang W, Newton RJ, Weidner DA (2007) Genetic transformation and gene silencing mediated by multiple copies of a transgene in eastern white pine. J Exp Bot 58:545–554
Tesfaye M, Silverstein KAT, Nallu S, Wang L, Botanga CJ et al (2013) Spatio-temporal expression patterns of Arabidopsis thaliana and Medicago truncatula defensin-like genes. PLoS One 8(3):e58992
Toomer OT, Do AB, Fu TJ, Williams KM (2015) Digestibility and immunoreactivity of shrimp extracts using an in vitro digestibility model with pepsin and pancreatin. J Food Sci 80(7):1633–1639
Verma SS, Sinha RK, Jajoo A (2015) (E)-β-farnesene gene reduces Lipaphis erysimi colonization in transgenic Brassica juncea lines. Plant Signal Behav 10(7):e1042636
Yadava JS, Singh NB (1999) Proceedings of the 10th international rapeseed congress, Canberra, Australia
Zhu-Salzman K, Salzman RA, Ahn JE, Koiwa H (2004) Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol 134(1):420–431
Acknowledgements
PS acknowledges the Bose Institute for fellowship and infrastructural facilities. The authors acknowledge Prof. Sampa Das for allowing the use of Real-Time PCR Detection System and for providing monoclonal mouse anti-human IgE–alkaline phosphatase conjugate and Dr. Debabrata Basu for providing the vectors, modified pBI221 and pCAMBIA1301. PS is thankful to Mr. Utpal Roy for maintaining the transgenic plants and to Mr. Swarup Biswas for his technical help in performing the CD experiment.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sarkar, P., Jana, K. & Sikdar, S.R. Overexpression of biologically safe Rorippa indica defensin enhances aphid tolerance in Brassica juncea . Planta 246, 1029–1044 (2017). https://doi.org/10.1007/s00425-017-2750-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-017-2750-4