Abstract
Key message
Transgenic Brassica juncea plants expressing Colocasia esculenta tuber agglutinin (CEA) shows the non-allergenic nature of the expressed protein leading to enhanced mortality and reduced fecundity of mustard aphid—Lipaphis erysimi.
Abstract
Lipaphis erysimi (common name: mustard aphid) is the most devastating sucking insect pest of Indian mustard (Brassica juncea L.). Colocasia esculenta tuber agglutinin (CEA), a GNA (Galanthus nivalis agglutinin)-related lectin has previously been reported by the present group to be effective against a wide array of hemipteran insects in artificial diet-based bioassays. In the present study, efficacy of CEA in controlling L. erysimi has been established through the development of transgenic B. juncea expressing this novel lectin. Southern hybridization of the transgenic plants confirmed stable integration of cea gene. Expression of CEA in T0, T1 and T2 transgenic plants was confirmed through western blot analysis. Level of expression of CEA in the T2 transgenic B. juncea ranged from 0.2 to 0.47% of the total soluble protein. In the in planta insect bioassays, the CEA expressing B. juncea lines exhibited enhanced insect mortality of 70–81.67%, whereas fecundity of L. erysimi was reduced by 49.35–62.11% compared to the control plants. Biosafety assessment of the transgenic B. juncea protein containing CEA was carried out by weight of evidence approach following the recommendations by FAO/WHO (Evaluation of the allergenicity of genetically modified foods: report of a joint FAO/WHO expert consultation, 22–25 Jan, Rome, http://www.fao.org/docrep/007/y0820e/y0820e00.HTM, 2001), Codex (Codex principles and guidelines on foods derived from biotechnology, Food and Agriculture Organization of the United Nations, Rome; Codex, Codex principles and guidelines on foods derived from biotechnology, Food and Agriculture Organization of the United Nations, Rome, 2003) and ICMR (Indian Council of Medical Research, guidelines for safety assessment of food derived from genetically engineered plants, http://www.icmr.nic.in/guide/Guidelines%20for%20Genetically%20Engineered%20Plants.pdf, 2008). Bioinformatics analysis, pepsin digestibility, thermal stability assay, immuno-screening and allergenicity assessment in BALB/c mice model demonstrated that the expressed CEA protein from transgenic B. juncea does not incite any allergenic response. The present study establishes CEA as an efficient insecticidal and non-allergenic protein to be utilized for controlling mustard aphid and similar hemipteran insects through the development of genetically modified plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rapeseed-mustards are the third most important oil crops after soybean and oil palm (Zhang and Zhou 2006; Shekhawat et al. 2012). India is the fifth largest vegetable oil economy in the world (Jha et al. 2012) possessing 5.53 million hectares of land under mustard cultivation (Shekhawat et al. 2012). A major constraint on the productivity of this economically important crop is the attack of Lipaphis erysimi (commonly known as mustard aphid), the second most important biotic factor after Alternaria blight (Grover and Pental 2003). L. erysimi is a hemipteran insect and is the most important pest of B. juncea causing yield loss of 10–90% depending upon the severity of infection and crop stage (Rana 2005). Nymphs and adults damage the crop by sucking the phloem sap (Verma and Singh 1987). Additionally, this aphid causes further damage to the crop by acting as vector for several different plant viruses (Wang et al. 1998; Dombrovsky et al. 2005; Rana 2005).
Conventional practice to control this insect pest is the application of chemical insecticides which is detrimental to the environment, additionally, usage of insecticides provoke the emergence of resistant insect biotypes. Conventional breeding approach was found to be inapplicable to develop insect resistance for this crop due to the lack of resistance gene in the wild germplasm (Yadava and Singh 1999). The most popular bio-control agent reported till date, namely Bt-toxin from Bacillus thuringiensis remained ineffective against sap-sucking pests (Rao et al. 1998; Porcar et al. 2009; Li et al. 2011). However, insecticidal efficacy of mannose-binding lectins in controlling hemipteran insects have been demonstrated by many groups in both artificial diet-based bioassays (Sauvion et al. 1996; Bandyopadhyay et al. 2001; Powell 2001; Banerjee et al. 2004) and in planta insect bioassays on transgenic plants expressing those lectins (Ramesh et al. 2004; Dutta et al. 2005a, b; Hossain et al. 2006; Saha et al. 2007; Sadeghi et al. 2007, 2008; Chakraborti et al. 2009).
Identification and exploitation of a novel insecticidal agent is necessary in crop improvement as insects might develop resistance against a particular insecticidal agent due to their behavioral reorientation (Chen 2008). This may help defend building up of such resistance. Novel insecticidal agents are also important regarding gene stacking approach where different insecticidal genes, effective against different insects, can be stacked together for better pest management. Colocasia esculenta tuber agglutinin (CEA) is a GNA-related lectin [previously known as monocot mannose-binding lectin (Van Damme et al. 2007)], isolated by the present group from the tubers of C. esculenta (Roy et al. 2002) and its insecticidal potential has also been demonstrated by the present group against a wide array of hemipteran insects such as—red cotton bug, cowpea aphid, cotton aphid, and mustard aphid (Roy et al. 2002, 2014; Majumder et al. 2004; Das et al. 2013). Binding of CEA to insect midgut was confirmed by confocal microscopic analyses (Roy et al. 2014). For elucidation of mode of action of CEA, Roy et al. (2014) further identified the cognate receptors of CEA in mustard aphid (L. erysimi) and whiteflies. Ligand blot followed by LC MS/MS led to the identification of vacuolar ATP synthase and sarcoplasmic reticulum Ca2+ ATPase as the receptors of CEA from Bemisia tabaci, whereas ATP synthase, clathrin heavy chain and HSP70 were identified as receptors of CEA from L. erysimi. Deglycosylation assay indicated probable glycan-mediated interaction of CEA with the said receptors. Pathway prediction study indicated towards disruption of cellular processes upon such interaction, leading to growth retardation and loss of fecundity of target insects (Roy et al. 2014). Although reports are not available for CEA against Lepidopteran insects, several alike GNA-related lectins such as Pinellia ternata agglutinin (PTA), Allium sativum leaf agglutinin (ASAL) have been found to be strongly antagonistic against Helicoverpa armigera, H. zea, Heliothis virescens and many more (Upadhyay et al. 2010; Jin et al. 2012). CEA has been reported to affect normal growth and development resulting in decreased pupation and immergence of Dipteran pest Bactrocera cucurbitae (Thakur et al. 2013). Hence, for further biotechnological application, the cloned CEA gene has been completely sequenced (GenBank accession no. JX435122, Das et al. 2013) and was utilized to develop L. erysimi-resistant transgenic B. juncea plants in the present study.
On the other hand, food allergy is considered as one of the major biosafety concern for any food derived from genetically modified crops (Fermín et al. 2011). As no single experiment is available yet for the safety assessment of a candidate protein, recommendations by Codex Alimentarius Commission (2003), guidelines by Indian Council of Medical Research (ICMR) (2008) and “Decision Tree Approach” by FAO/WHO (2001) were followed. Considering all those recommendations, in the present work, safety assessment of transgenic B. juncea protein containing CEA was carried out by—bioinformatics analysis, in vitro pepsin digestibility assay, thermal stability assay, immuno-screening and in vivo allergenicity assessment in animal model.
Taking together, the present study aims to utilize the insecticidal potential of CEA in developing transgenic mustard plants resistant to aphid attack and safety assessment of the transgenic protein further for better consumer acceptance.
Methods
Plant materials and bacterial strains used
Brassica juncea cv. B-85 seeds obtained from Berhampur Pulse Research Station, West Bengal, India were used for plant transformation. The DH5α strain of Escherichia coli and the AGL-1 strain of Agrobacterium tumefaciens were used for cloning and plant transformation, respectively.
Construction of chimeric cea gene cassette for plant transformation
Initially the CaMV 35S promoter-multiple cloning site (MCS)—nos terminator cassette from the vector pPZPY (Yamamoto et al. 1998) was cloned within the HindIII and EcoRI sites of the MCS of binary plant transformation vector pCAMBIA 1301 and the resulting vector was named pCAMBIA1301-35S-nos. The 795 bp complete coding sequence of cea (GenBank Accession no. JX435122), earlier reported and cloned by the present group (Das et al. 2013), was PCR amplified using the primers CEAF and CEAR containing XbaI and KpnI restriction sites (underlined), respectively (Table 1), and cloned into the corresponding restriction sites of the pCAMBIA1301-35S-nos plasmid. The resulting pCAMBIA1301-35S-cea gene construct (Fig. 1) was finally mobilized into Agrobacterium strain AGL1 for further plant transformation experiment.
Diagrammatic representation of the T-DNA region of the chimeric pCAMBIA1301-35S-cea gene construct. cea, coding sequence of Colocasia esculenta tuber agglutinin (CEA); hptII, hygromycin phosphotransferase II gene; gus, β-Glucuronidase gene; CaMV35SPr, cauliflower mosaic virus 35S promoter; nos polyA, nopaline synthase terminator; CaMV35S polyA, cauliflower mosaic virus 35S terminator; LB, left border of T-DNA; RB, right border of T-DNA
Mustard transformation
Agrobacterium-mediated transformation and regeneration of mustard were performed according to the method described by Dutta et al. (2005a). Callusing and shoot regeneration were performed in the presence of selection media containing 30 mg/l of hygromycin. Shootlets were rooted in the root induction medium as described by Rajagopal et al. (2007). After root development, plants were hardened and were transferred to soil in glasshouse maintained at 25 °C and 16:8 h light: dark photoperiod. A set of explants were transformed using Agrobacterium strain AGL 1 containing the empty pCAMBIA1301-35S-nos vector (without the cea coding sequence) and allowed to regenerate similarly in the selection media for the regeneration of vector control plants. Another set of explants were allowed to regenerate via tissue culture in the absence of antibiotics to yield untransformed control plants. Each transgenic event was self-pollinated to obtain the T1 and T2 generations.
Screening of putative transformants by PCR
For the preliminary screening of the putatively transformed plants, PCR analysis was performed. Genomic DNA was isolated from the leaves of 1-month-old plants putatively transformed with chimeric cea gene construct as well as the untransformed and vector control plants (Dellaporta et al. 1983). PCR analysis was carried out using the cea gene-specific primers (CEAF and CEAR, Table 1) and hygromycin-resistance gene (hpt)-specific internal primers (HPTF and HPTR, Table 1). PCR cycles were: initial incubation at 94 °C for 5 min followed by 30 cycles of denaturation (94 °C) for 1 min, annealing step of 57.5 °C (for cea) and 58 °C (for hpt) for 45 s and extension (72 °C) for 90 s with a final extension of 10 min at 72 °C in Veriti Thermal Cycler (Applied Biosystems, CA, USA). Hundred to two hundred nanogram-genomic DNA were used as template. The binary vector plasmid used for plant transformation (pCAMBIA1301-35S-cea) was used as positive control and genomic DNA from untransformed and vector control plants were used as negative controls. The PCR amplification products were checked in 1% agarose gel.
Southern blot analysis
Southern blot analysis was carried out following the protocol of Sambrook et al. (1989) with some modifications (Dutta et al. 2005a). Twenty microgram-genomic DNA was digested with EcoRI (New England Biolabs Inc., Ipswich, Massachusetts) from each transformant as well as vector control plants and separated in 0.8% (w/v) agarose gel. Gels were blotted onto positively charged nylon membranes (Hybond-N+; Amersham™ Biosciences, Buckinghamshire, UK). The cea gene coding sequence was amplified by using two internal primers: CEASF and CEASR (Table 1) and the resulting 352 bp amplicon was used as probe. The probe was radio labeled with [α-32P] dCTP using Rediprime II™ Random Prime Labeling System (Amersham™ Biosciences, Buckinghamshire, UK). Membranes were hybridized overnight at 68 °C using the radiolabelled probe and washed thoroughly using 2XSSC, 0.1% SDS at room temperature for 45 min and at 68 °C for another 45 min using 0.1% SSC, 0.1% SDS. Finally, membranes were exposed to Kodak X-ray films for 7 days at − 80 °C and the exposed films were developed thereafter.
Segregation analysis of the transgene
Genomic DNA was isolated from 1-month-old T1 plants (Dellaporta et al. 1983) and PCR analysis was carried out using cea gene-specific primers (CEAF and CEAR, Table 1). The PCR-amplified products were run in 0.8% agarose gel. After separation of the amplified product, segregation pattern of cea gene in progeny plants were calculated and validated by χ2 analysis.
Western blotting
Total soluble protein was extracted from the fresh leaves of 1-month-old transgenic- (T0, T1 and T2 generation) and vector control plants as described by Dutta et al. (2005a) in extraction buffer containing 20 mM of Tris–HCl (pH 7.5) and 0.2 mM of PMSF (phenylmethylsulfonyl fluoride) (Sigma-Aldrich, MO, USA). Protein concentration in each sample was quantified by Bradford assay (1976). Western blotting was performed with 40 µg of total soluble protein using the method as reported earlier (Dutta et al. 2005a). Extracted proteins from the individual lines were separated on 15% SDS–PAGE and electroblotted to positively charged Hybond C membrane (Amersham Biosciences Buckinghamshire, UK). Membrane blocking was carried out with 5% (w/v) skim milk (Merck Millipore, MA, USA) in 20 mM of phosphate buffered saline (PBS, pH 7.5). After blocking, the membranes were probed with anti-CEA polyclonal primary antibody [raised in rabbit following the protocol of Harlow and Lane (1988)] at 1:10,000 dilution followed by incubation in anti-rabbit IgG-horseradish peroxidase (HRP) conjugate (Sigma-Aldrich, MO, USA) (secondary antibody) at 1:20,000 dilutions. Western blot was developed in Kodak film using Immobilon™ Western Chemiluminescent HRP Substrate (Merck Millipore, MA, USA).
ELISA of soluble protein extracts
To determine the level of expression of CEA in the transgenic lines, ELISA was carried out following the method described by Dutta et al. (2005a). Microtiter (Immunomaxi, Switzerland) wells were coated with 50 µg of total soluble protein extracted from the leaves of T2-transgenic- and vector-control plants or purified native CEA serially diluted in the range between 2.5 µg and 25 ng overnight at 4 °C in coating buffer (15 mM of sodium carbonate, 35 mM of sodium bicarbonate, 3 mM of sodium azide, and pH 9.6). After blocking, wells were incubated with anti-CEA primary antibody at 1:10,000 dilutions followed by incubation with HRP-conjugated anti-rabbit secondary antibody (Sigma-Aldrich, MO, USA) at 1:20,000 dilutions. After the addition of OPD (o-phenylenediamine dihydrochloride) peroxidase substrate [SIGMAFAST™ OPD (Sigma-Aldrich, MO, USA)], development of color reaction was recorded at 450 nm in Thermo Scientific Varioskan Flash Spectral Scanning Multimode Reader (Thermo Scientific, MA, USA).
Hemagglutination assay using transgenic B. juncea protein containing CEA
Hemagglutination assay was performed with the total soluble protein extracted from the T2-transgenic mustard plants expressing CEA as well as the vector control plants following the method described by Bala et al. (2013). One millilitre rabbit blood was drawn using a hypodermic syringe pre-filled with 1 ml of 0.9% NaCl solution. After thorough mixing and washing, the sedimented erythrocytes were resuspended in 0.9% saline solution and dispensed into the wells of the microtiter U-bottom plate. B. juncea transgenic plant proteins were dispensed to each well at a twofold serial dilution starting from 250 µg to 15.6 µg/well. The U-plate was incubated for 1 h at 37 °C and then agglutination was monitored visually.
In planta insect bioassay
Mustard aphids were maintained on mustard plants in insect houses (maintained at 25 °C, 70% relative humidity and 16:8 h light:dark conditions). In planta insect bioassay was conducted on CEA expressing T2-transgenic lines and empty vector-transformed plants of B. juncea as described by Bala et al. (2013). One-end open cages were used for insect bioassay. Twenty nymphs of L. erysimi of third instar stage were transferred to the leaf surface of the plants which were previously placed in the cages. Subsequently, the open end of the cage was sealed with adhesive tape. Three such cages were used per individual transgenic and control plant and three replications were performed for each individual line. The survival of insects within the cages was monitored at an interval of every 24 h upto 9 days. The mean data per plant was expressed as the percentage of the total aphids surviving on the respective days. Effect of CEA on fecundity of the insect was monitored by counting the total number of nymphs produced per individual transgenic plants at the end of the bioassay period. ANOVA followed by Duncun’s multiple range tests were conducted to calculate the significance of differences observed between the transgenic and control plants in the insect bioassay.
Safety assessment of CEA
Sequence homology search of CEA
The amino acid sequence of CEA was searched in the databases to find out homology with the known allergenic proteins. The following databases were searched: Structural Database of Allergenic Proteins (SDAP; https://fermi.utmb.edu/) (Ivanciuc et al. 2003) and Allergen Online database (http://www.allergenonline.com/databasefasta.shtml) (Goodman et al. 2016). Search was performed using FASTA alignments for an 80 amino acids sliding window with a threshold sequence identity of > 35% to be referred as allergen. Exact match for eight contiguous amino acids with known allergic protein was also studied. Mapping of IgE-specific epitopes for CEA was done using the AlgPred database (AlgPred; http://www.imtech.res.in/raghava/algpred/submission.html) (Saha and Raghava 2006).
Digestibility of CEA in simulated gastric fluid (SGF)
The digestibility of purified CEA isolated from the tubers of C. esculenta was examined in simulated gastric fluid (SGF), as described by Singh et al. (2006) with some modifications as described by Ghosh et al. (2013). Purified CEA (50 µg) was dissolved in 50 µl of simulated gastric fluid (SGF) (0.32% of w/v pepsin, 0.03 M of NaCl, pH 1.2) in each set. Digestion was carried out at 37 °C for 0, 2, 5, 15, 30, 60, and 120 min. Only CEA was used as control. The reaction was stopped immediately by adding 5 N of NaOH. Each sample was loaded into 15% SDS–PAGE gel followed by western blotting using anti-CEA antibody.
Thermal stability assay of CEA
The stability of the transgenically expressed CEA was monitored by incubating the T2-transgenic B. juncea protein containing CEA at different temperatures and assessing its ability to hemagglutinate rabbit erythrocytes as described by Bala et al. (2013). Rabbit erythrocytes were diluted at a final concentration of 5% (v/v) in 0.9% NaCl. The amount of total soluble protein required for complete hemagglutination of rabbit erythrocytes from each of the selected CEA-expressing plant lines was determined before assessing the stability of CEA. Hence, 250 µg of the total soluble protein from all the selected CEA-expressing T2 plants was dissolved in phosphate buffered saline (PBS) and incubated separately at 4, 25, 37, 55, 75 and 95 °C for 30 min. After incubation, each sample was immediately transferred to ice for rapid cooling and agglutination activity was observed as described earlier.
In vitro IgE-specific ELISA
IgE-specific ELISA was performed following the method described by Ghosh et al. (2013) with the sera of ten allergic patients (aged 21–50 years) having a history of food allergy with any one or more than one of the following symptoms: bronchial asthma, rhinitis and dermatitis. Sera were collected from a referral allergy clinic (Sinha Patho Lab, Dhanbad, India). Sera collected from healthy individuals without any history of allergy was used as negative control. Blood samples (sera) were collected from patients with their written consent. The entire study was approved by the Human Ethics Committee of Bose Institute. Microtiter wells were coated with coating buffer (15 mM of sodium carbonate, 35 mM of sodium bicarbonate, 3 mM of sodium azide and, pH 9.6) containing 250 µg of mustard protein (corresponding to 1 µg of expressed CEA) from the highest CEA-expressing T2-transgenic plant or same amount of total soluble protein from B. juncea vector control plant. One microgram of ovalbumin (OVA; Ovalbumin from chicken egg white, Himedia, India) was used for coating the wells in the positive control set up. Microtiter plates were incubated overnight at 4 °C. After blocking, wells were incubated overnight with 50 µl of individual patient’s sera diluted (1:5) with blocking solution [1% bovine serum albumin (Sigma-Aldrich, MO, USA) in PBST {phosphate buffered saline and Tween 20 (0.5% v/v), pH 7.3}] at 37 °C. Subsequently, wells were incubated with mouse monoclonal anti-human IgE—alkaline phosphatase conjugate (Sigma-Aldrich, MO, USA) at 1:1000 dilutions. After final wash, p-nitrophenyl phosphate (pNPP) liquid substrate for ELISA (Sigma-Aldrich, MO, USA) was added to the wells. Development of color reaction was measured at 405 nm in Thermo Scientific Varioskan Flash Spectral Scanning Multimode Reader (Thermo Scientific, MA, USA). The P/N value (ratio of average OD of individual patient sera with respect to that of the control group) of individual patient sera was calculated. The control was the average OD values of sera from all healthy individuals. A P/N value greater than 3.5 for a particular serum was considered to be potentially IgE reactive (Chakraborty et al. 2005; Ghosh et al. 2013).
Evaluation of the allergenic potential of CEA in BALB/c mice
Histopathological analysis of allergenic potential of CEA was conducted in BALB/c mice following the protocol described by Ghosh et al. (2013). The Animal Ethics Committee of Bose Institute approved the study protocol. Healthy 8–10-week-old female BALB/c mice (22 ± 2 g) were randomly segregated into four groups of five mice each (i.e., five replicates/experimental set-up) and sensitized by the intraperitoneal (ip) route. Group 1 mice were sensitized with 100 µl of PBS daily by ip injection. Group 2, 3 and 4 mice were sensitized by ip route, once a week for 7 weeks with control mustard protein, ovalbumin (OVA; Ovalbumin from chicken egg white, Himedia, India) and transgenic mustard protein from T2 B. juncea plants, respectively, (100 µg of protein in 100 µl PBS in each case). On day 50, mice were sacrificed for the collection of lung and gut tissue. The tissues were fixed immediately in 10% neutral-buffered formaldehyde (v/v) (0.1 M phosphate buffer, pH 7.4) and embedded in paraffin. Three to five micrometer tissue sections were prepared, de-paraffinized with xylene and graded ethanol and finally stained with hematoxylin and eosin (H&E). Histopathological observations were recorded by a light microscope connected with an in-line camera (Leica Microsystem DN1000; Camera DFC450C).
Results
Development of transgenic Brassica juncea using chimeric cea gene construct
The pCAMBIA1301-35S-cea gene construct (Fig. 1) was used for Agrobacterium-mediated transformation of B. juncea for constitutive expression of CEA. Figure S1 shows representative image of the different stages of regeneration of transgenic B. juncea. The putatively transformed T0 plants were initially examined by cea gene-specific PCR analysis. All the putative-transformed plants showed the presence of both 795 bp amplicon of cea transgene and 1005 bp amplicon of hpt gene, whereas the vector control plants were devoid of cea transgene. Untransformed control plants were devoid of both hpt and cea transgene (Fig. S2a and Fig. S2b).
Stable integration of cea transgene
Genomic DNA was extracted from both T1 and T2 progenies of respective T0 plants, digested with EcoRI and hybridized using radiolabelled cea gene-specific probe. Hybridization of one representative T1 progeny from each corresponding T0 line has been shown in Fig. 2. Seven T1 plants (P1T13, P2T15, P3T14, P4T12, P5T19, P6T16 and P9T18) showed single-copy transgene integration. P7T11 found to be southern-negative and P8T14 found to be a double-copy transgene-integration event. Plants transformed with null vector (C) showed the absence of cea transgene (Fig. 2). Five single-copy, southern positive T1 lines [P2T15, P3T14, P4T12, P6T16 and P9T18] were randomly chosen for further transgene integration and expression studies. T1 progeny plants were allowed to self-pollinate to yield T2 plants which were initially checked for transgene integration by cea gene-specific PCR and then those PCR-positive plants were further confirmed by southern hybridization. Figure S3 shows southern hybridization of five such PCR-positive T2 progeny plants from each of the selected T1 lines, namely—P2(T15)T22, 3, 5, 7, 9; P3 (T14) T23, 4, 5,7, 11; P4(T12) T25, 8, 9, 10, 11; P6(T16) T23, 5, 6, 7, 8 and P9(T1 8) T22, 4, 5, 9, 10. All the PCR-positive T2 plants displayed the presence of the transgene in the southern blot analysis.
Southern blot analysis of T1 transgenic plants of B. juncea. Lanes P1T13, P2T15, P3T14, P4T12, P5T19, P6T16, P7T11, P8T14, P9T18, EcoRI digested genomic DNA from nine T1-transformed plants of B. juncea; lane +ve, 795 bp cea coding sequence used as positive control; lane C, genomic DNA from empty vector-transformed control plant as negative control; 352 bp internal amplicon of the cea gene was used as probe; approximate molecular weight markers are indicated at left
Segregation analysis of the cea transgene
PCR screening for the presence/absence of the cea transgene using genomic DNA isolated from the selected five T1 lines demonstrated that the cea transgene followed 3:1 Mendelian segregation pattern. The difference between the observed and the expected ratio (of cea+ve: cea−ve plants) was verified by χ2 analysis and was found to be not significant at 0.05 level of significance (Table 2).
Expression of CEA in transgenic mustard plants
Western blot analysis was carried out using total soluble protein from the five T0 (P2, P3, P4, P6 and P9) and corresponding T1 (P2T15, P3T14, P4T12, P6T16 and P9T18) and T2 [P2(T15)T22, P3 (T14) T27, P4(T12)T25, P6(T16)T23, P9(T1 8)T24] progeny plants. All of the T0, T1 and T2 plants tested showed identical bands of CEA at ∼ 12.5 kDa region, corresponding to the positive control (Fig. 3a–c), whereas no such band was observed in case of total soluble protein extracted from the leaves of null vector-transformed control mustard plant.
Expression of CEA in transgenic mustard plants. a Western blot analysis of the T0 B. juncea plants. Lanes P2, P3, P4, P6 and P9, Total soluble protein isolated from five T0 plants. b Western blot analysis of the T1 plants. P2T15, P3T14, P4T12, P6T16 and P9T18, Total soluble protein isolated from five T1 plants corresponding to each T0 lines. c Western blot analysis of the T2 plants. P2(T15)T22, P3 (T14) T27, P4(T12)T25, P6(T16)T23, P9(T1 8)T24, Total soluble protein isolated from five T2 B. juncea plants corresponding to the T1 lines. +ve, ~ 1.5 µg purified native CEA used as positive control; C, total soluble protein from vector control plant used as negative control; RbcL: SDS–PAGE profile of Rubisco large subunit as loading control. d ELISA analysis. Graph showing level of CEA expression in T2 B. juncea plants P2(T15)T22, P3 (T14) T27, P4(T12)T25, P6(T16)T23, P9(T1 8)T24 and vector control plant (C) of B. juncea
To quantify the level of expression of CEA in the selected lines, ELISA was performed with the total soluble protein extract from the selected T2-transgenic and null-vector-transformed control B. juncea plants (Fig. 3d). The level of expression of CEA varied from 0.2 to 0.47% of the total soluble protein among the transgenic lines. P6(T16)T23 showed the highest expression of CEA (0.47%), whereas P2(T15)T22 showed the lowest (0.2%).
Rabbit-erythrocyte hemagglutination assay of the total soluble protein from CEA expressing transgenic plants
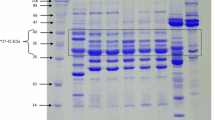
Rabbit-erythrocyte hemagglutination assay was performed using total soluble protein from CEA-expressing T2 plants [P2(T15)T22, P3 (T14) T27, P4(T12)T25, P6(T16)T23, P9(T1 8)T24]. A tight button of rabbit erythrocytes was formed indicating negative reaction in well no. 2—no protein control and well no. 3—containing total soluble protein from empty-vector transformed control plant in panel I–V. Total soluble protein from all the CEA-expressing T2 plants exhibited agglutination of rabbit erythrocytes resulting in the formation of a layer over the wells (well no. 4–8; panel I–V) of the microtiter plate (Fig. 4).
Hemagglutination assay of expressed CEA protein from T2 plants of B. juncea. Rabbit erythrocytes incubated with: panel I–V (wells 4–8), Total soluble protein extracted from CEA-expressing T2 plants P2(T15)T22, P3 (T14) T27, P4(T12)T25, P6(T16)T23 and P9(T1 8)T24, respectively, in a twofold serial dilution started from 250 µg to 15.6 µg/well; well 1 (panel I–V), Positive control (1 µg native purified CEA); well 2 (panel I–V), No protein control; well 3 (panel I–V), total soluble protein from empty vector transformed control B. juncea plant
In planta insect bioassay
To investigate the effect of CEA on survivability of L. erysimi, in planta insect bioassay was carried out in bioassay cages (Fig. 5a) for a total duration of 9 days on five CEA-expressing T2 lines of mustard [P2(T15)T2, P3 (T14) T2, P4(T12)T2, P6(T16)T2, P9(T1 8)T2]. Aphid survivability was recorded with third instar nymphs at an interval of 24 h. Survival of the nymphs declined from 20 ± 0 (mean ± SE) to 13.67 ± 0.33 (68.33%) in case of control plants at the end of the bioassay period, whereas this decline in aphid survival was more significant in the case of CEA-expressing T2 lines of B. juncea varying from 3.67 ± 0.33 (18.33%) to 6 ± 0.58 (30%) (Fig. 5b). The mean number of surviving Lipaphis on control and transgenic plants were found to be statistically significant (P < 0.05) after third day of the bioassay period. After 9 days of the experiment, the CEA-expressing line no. P6(T16)T2 and P4(T12)T2 showed maximum decrease in insect survivability.
In planta insect bioassay of transgenic CEA expressing B. juncea on L. erysimi. a Insect bioassay setup on mustard plants. b Graph shows the percentage of survival of mustard aphid on vector control plant and five CEA-expressing T2 lines of mustard [P2(T15)T2, P3 (T14) T2, P4(T12)T2, P6(T16)T2 and P9(T1 8)T2]. c Bar diagram showing fecundity pattern (mean number of nymphs produced per plant) of L. erysimi fed on control and the five T2 lines [P2(T15)T2, P3 (T14) T2, P4(T12)T2, P6(T16)T2 and P9(T1 8)T2]
The effect of CEA on fecundity was determined by counting the total nymphs produced by the adult insects on transgenic and vector control plants at the end of the bioassay period. The number of nymphs produced per plant was reduced by 49.35–62.11% as compared to control plants (Fig. 5c).
Safety assessment of CEA
Sequence homology of CEA
The amino acid sequence of CEA was first analyzed by comparison with allergenic protein databases. In the in silico analysis, no known allergen was found to be similar to CEA by the criteria of more than 35% identity in the amino acid sequence of the query protein, using sliding window of 80 amino acids. The search for exact match for eight contiguous amino acid stretches also yielded no significant match. Mapping of IgE-specific epitopes using Algpred database showed that the CEA protein sequence does not contain any experimentally proven IgE-binding epitope.
Digestibility of CEA in simulated gastric fluid (SGF)
The digestion profile of CEA in SGF was monitored in 15% SDS–PAGE stained with Coomassie Brilliant Blue. The purified CEA was completely digested within 2 min of treatment with SGF (Fig. 6a). CEA was also not detected after 2 min of digestion in SGF in the western blot using anti-CEA antibody (Fig. 6b). In silico pepsin digestion using ExPASy PeptideCutter bioinformatics tool (Supplementary Table S1) also exhibited 66 cleavage sites affirming the SGF digestion data.
Simulated gastric fluid (SGF) digestibility of CEA. a SDS–PAGE profile of SGF-treated CEA. Lane M, protein molecular weight marker; lane 1, untreated CEA; lane 2, pepsin in SGF; lanes 3–9, CEA treated with SGF for 0, 2, 5, 15, 30, 60 and 120 min and resolved in 15% SDS–PAGE and stained with Coomassie Brilliant Blue. b Corresponding western blot of the 15% SDS–PAGE using anti-CEA antibody showing degradation of CEA in SGF. Arrows indicate the positions of pepsin and CEA bands
Thermal stability assay
To check the stability of transgenically expressed CEA, 250 µg of total soluble protein from CEA-expressing T2 plants were initially incubated at different temperatures and then hemagglutination assay was performed. Total soluble protein from all those CEA-expressing plant lines lost their hemagglutination property when they were heated at 37–55 °C (wells 5–6, panel I–V; Fig. 7).
Thermal stability assay of CEA. Panel I–V (wells 3–8), Rabbit erythrocytes incubated with total soluble protein (250 µg) from CEA-expressing T2 plants no. [P2(T15)T22, P3 (T14) T27, P4(T12)T25, P6(T16)T23 and P9(T1 8)T24] pre-incubated at temperatures: 4, 25, 37, 55, 75 and 95 °C, respectively; well 1 (I–V): positive control (1 µg of native purified CEA); well 2 (I–V): no protein control
IgE specific ELISA
In IgE-specific ELISA, significantly low level of IgE was detected when mustard protein from T2-transgenic B. juncea (0.51–1.3) or vector control plant (0.49–1.33) was used as probe. However, higher P/N value (3.59–5.97) was recorded when patients’ sera were probed with the allergen ovalbumin (Table 3).
Histopathological analysis of lungs and gut of sensitized BALB/c mice
Lung histology
Hematoxylin and eosin (H&E)-stained lung sections of ovalbumin (OVA)-sensitized mice showed prominent allergenicity response evident by hyperplasia, destruction as well as exfoliation of bronchiolar lining epithelium. Exfoliated mucosal mass within the bronchiole was also observed (Fig. 8c). Both PBS-treated- and control mustard protein-sensitized mice had normal histological structure of alveoli and bronchioles (Fig. 8a, b). Mice sensitized with mustard protein from T2-transgenic lines also showed normal lung structure with defined alveoli and bronchioles with no evidence of inflammation (Fig. 8d).
Gut histology
Analysis of histo-architecture of gut sections of ovalbumin-treated BALB/c mice revealed destruction of mucosal lining and infiltration of mucosa with inflammatory cells in duodenum, ileum and jejunum villi (Fig. 9c). Whereas histopathological analysis of H&E-stained gut tissues of mice fed with T2-transgenic mustard protein revealed normal structure without any distortion. The gastrointestinal tract segments showed no inflammation and/or infiltration of inflammatory cells into surrounding tissues (Fig. 9d) as in the case of PBS or control B. juncea protein-sensitized mice (Fig. 9a, b, respectively) suggesting normal cellular metabolism.
Discussions
Aphids, the group of plant sap-sucking insects belonging to order hemiptera, account for 13% of the total crop loss worldwide (Chougule and Bonning 2012). Lipaphis erysimi, commonly known as mustard aphid severely affects yield of the economically important crop B. juncea. Since last few decades, mannose-binding snowdrop lectin—Galanthus nivalis agglutinin (GNA) was established as an effective bio-control agent against the sap-sucking hemipteran insects (Gatehouse et al. 1996; Powell et al. 1998). Subsequently, several other GNA-related lectins viz., Pinellia ternata agglutinin (PTA), Allium sativum leaf agglutinin (ASAL), Allium cepa agglutinin (ACA), etc. have been reported from different plant sources and effectiveness of many of them have been demonstrated in transgenic plants (Yao et al. 2003; Zhang et al. 2003; Dutta et al. 2005a, b; Hossain et al. 2006; Saha et al. 2007; Sadeghi et al. 2007, 2008). Colocasia esculenta tuber agglutinin (CEA) was previously isolated by the present group from the tubers of C. esculenta (Roy et al. 2002). Effectiveness of CEA as potent insecticidal agent against a wide array of hemipteran insects including L. erysimi (Roy et al. 2002, 2014; Majumder et al. 2004; Das et al. 2013) encouraged the present group to express the coding sequence of CEA in susceptible cultivars of B. juncea to provide resistance against the pest. The coding sequence of CEA was transformed into B. juncea cultivar cv. B85 by Agrobacterium-mediated transformation. PCR analysis of all the putative transformants with normal phenotype showed the presence of 795 bp cea transgene. As multiple copies of transgene insertion might result in gene silencing (de Carvalho et al. 1992; Matzke and Matzke 1998; Tang et al. 2007), B. juncea lines with single copy transgene integration were selected by southern blot analysis. Seven out of nine randomly selected T1 progeny plants of corresponding T0 lines showed single-copy transgene integration. Stable inheritance of the transgene was also observed in the T2 progenies corresponding to independent southern-positive T1 lines. Segregation analysis confirmed Mendelian 3:1 segregation pattern of the transgene. Western blot analysis of the T0, T1 and T2 transgenic lines confirmed constitutive and stable expression of CEA. Differential quantitative expression (0.2–0.47% of the total soluble protein) of CEA in different T2 plants corresponding to different independent transgenic events suggested random transgene integration at different transcriptionally active sites in the plant genome. The expression level of CEA was found to be comparable with the level of expression of garlic leaf lectin-ASAL, when expressed in transgenic mustard (Dutta et al. 2005a; Bala et al. 2013). Rabbit erythrocyte hemagglutination assay conducted with total soluble protein from T2-transgenic B. juncea plants confirmed retention of hemagglutination property of CEA under transgenic condition. In planta bioassay resolved the mortality of L. erysimi on CEA expressing transgenic plants to be ranging between 70 and 81.67%. The presently observed mortality was comparable/higher than 78–80 and 65–70% documented by Dutta et al. (2005a) and Bala et al. (2013), respectively, when insect-resistant-, transgenic mustard plants were developed expressing ASAL. The observed L. erysimi mortality was comparable with ~ 70% mortality or 80% aphid (L. erysimi) repellency when onion leaf lectin (Allium cepa L. agglutinin, ACA) or (E)-β-farnesene from Mentha arvensis was expressed in B. juncea, respectively, (Hossain et al. 2006; Verma et al. 2015). Reduction of fecundity of L. erysimi in the transgenic B. juncea plants was also comparable as documented earlier by Dutta et al. (2005a), Hossain et al. (2006) and Bala et al. (2013). A very recently identified plant defensin (Rorippa indica defensin) (Sarkar et al. 2016), when expressed in Indian mustard to develop aphid tolerance (Sarkar et al. 2017), could not exhibit such higher mortality or reduction in fecundity as recorded in the current study. Reduction of aphid survivability and fecundity in the transgenic lines, in the present study, were found to be correlated with the level of expression of CEA in the corresponding transgenic lines.
As assessment of allergenicity is a major concern for any food derived from genetically modified plants and as no single experiment is adequate enough for the safety assessment of a protein of interest, a case by case weight of evidence approach was considered for the safety assessment of CEA following FAO/WHO (2001), Codex (2003) and ICMR (2008) recommendations.
Bioinformatics analysis of amino acid sequence of CEA showed no significant match with any known allergen available in the SDAP and AllergenOnline databases. Any IgE-specific epitope also could not be mapped within the amino acid sequence of CEA.
As per joint FAO/WHO expert consultation report (2001) and Codex (2003) guidelines, there is a supposed correlation between the indigestibility of a protein by the alimentary tract enzymes and its potential allergenicity (Fermín et al. 2011). The quick digestion of CEA protein (within 2 min) in simulated gastric fluid, like the other non-allergenic proteins [e.g., spinach ribulose bis-phosphate carboxylase/oxygenase (Astwood et al. 1996), purified choline oxidase from Arthrobacter globiformis (Singh et al. 2006)], exempted it from being allergenic. The presence of 66 pepsin (pH 1.3) cleavage sites according to the ExPASy peptide cutter bioinformatics tool (Gasteiger et al. 2005) further confirmed rapid degradation of CEA in gastrointestinal fluids. As many of the allergenic proteins are resistant to heat (Caballero and Moneo 2004; Scheurer et al. 2004; Suhr et al. 2004; Palacin et al. 2009), the probability of a candidate protein being allergenic may be correlated with its structural stability at high temperature. Hence, in addition to SGF digestion, CEA was also evaluated for thermal stability as a test for persistence during processing. The transgenically expressed CEA was found to be thermo-labile at 37–55 °C—as no agglutination was observed beyond this temperature similar to the case of ASAL expressed in transgenic B. juncea (Bala et al. 2013).
IgE has an important role in type I hypersensitivity (Gould et al. 2003) manifesting various allergic diseases such as allergic asthma, most types of sinusitis, allergic rhinitis, food allergies, specific types of chronic urticaria and atopic dermatitis. In the present experiment, IgE-specific ELISA of transgenic mustard protein showed P/N value well below the threshold level of 3.5 suggesting that CEA is potentially IgE non-reactive (Chakraborty et al. 2005; Ghosh et al. 2013).
For the assessment of the potential allergenicity of novel proteins, the FAO/WHO (2001), Codex (2003) and ICMR (2008) recommendations additionally encourage the use of animal models. As BALB/c strain of mice readily display Th2 responses as compared to other murine strains (Van Gramberg et al. 2013), they are widely used as animal models for such studies (Thang et al. 2011; Bailón et al. 2012). BALB/c mice sensitized with transgenic mustard protein displayed normal appearance of lung and gut tissue, similar to PBS or control mustard protein-treated mice, indicating that CEA has no detrimental effects, whereas ovalbumin sensitized mice showed prominent allergic symptoms and resulted in the loss of normal lung and gut morphology. Similar observations were previously reported by—Singh et al. (2006) in case of transgenic mustard expressing bacterial codA gene and Ghosh et al. (2013) in the biosafety assessment of mutant variant of Allium sativum Leaf Agglutinin (mASAL). Poulsen et al. (2007) and Guo et al. (2015) also, similar to the present experiment, did not observe any adverse effect on the gastrointestinal health of rats in the safety assessment of genetically modified rice expressing snowdrop lectin (GNA) and transgenic maize expressing Cry1Ac, respectively.
The present study establishes the efficacy of a novel lectin, CEA, against L. erysimi by generating transgenic B. juncea plant types. The selected transgenic lines demonstrated significant reduction of survivability and fecundity of the target pest. Moreover, the transgenically expressed CEA protein was found to be non-allergenic as per the tests recommended by FAO/WHO (2001), Codex (2003) and ICMR (2008). Hence, these CEA-expressing transgenic mustard plants might serve as important components for integrated pest management (IPM) programme.
Author contribution statement
SD and AD designed the study. AD and PG performed Mice Histology experiments. All the other experiments were done by AD. AD, PG and SD prepared the manuscript. SD supervised the research. All authors have read and approved the final manuscript.
Abbreviations
- CEA:
-
Colocasia esculenta tuber agglutinin
- MCS:
-
Multiple cloning site
- hpt :
-
Hygromycin phosphotransferase
- ELISA:
-
Enzyme-linked immunosorbent assay
- ANOVA:
-
Analysis of variance
- SDAP:
-
Structural Database of Allergenic Proteins
- SGF:
-
Simulated gastric fluid
- IgE:
-
Immunoglobulin E
- PBS:
-
Phosphate buffered saline
- H&E:
-
Hematoxylin and eosin
References
Astwood JD, Leach JN, Fuchs RL (1996) Stability of food allergens to digestion in vitro. Nat Biotechnol 14(10):1269–1273
Bailón E, Cueto-Sola M, Utrilla P, Rodríguez-Ruiz J, Garrido-Mesa N, Zarzuelo A, Xaus J, Gálvez J, Comalada M (2012) A shorter and more specific oral sensitization-based experimental model of food allergy in mice. J Immunol Methods 381(1–2):41–49
Bala A, Roy A, Das A, Chakraborti D, Das S (2013) Development of selectable marker free, insect resistant, transgenic mustard (Brassica juncea) plants using Cre/lox mediated recombination. BMC Biotechnol 13:88
Bandyopadhyay S, Roy A, Das S (2001) Binding of garlic (Allium sativum) leaf lectin to the gut receptors of homopteran pests is correlated to its insecticidal activity. Plant Sci 161:1025–1033
Banerjee S, Hess D, Majumder P, Roy D, Das S (2004) The interactions of Allium sativum leaf agglutinin with a chaperonin group of unique receptor protein isolated from a bacterial endosymbiont of the mustard aphid. J Biol Chem 279:23782–23789
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Caballero ML, Moneo I (2004) Several allergens from Anisakis simplex are highly resistant to heat and pepsin treatments. Parasitol Res 93(3):248–251
Chakraborti D, Sarkar A, Mondal HA, Das S (2009) Tissue specific expression of potent insecticidal, Allium sativum leaf agglutinin (ASAL) in important pulse crop, chickpea (Cicer arietinum L.) to resist the phloem feeding Aphis craccivora. Transgenic Res 18(4):529–544
Chakraborty P, Ghosh D, Chowdhury I, Roy I, Chatterjee S, Chanda S, Gupta-Bhattacharya S (2005) Aerobiological and immunochemical studies on Carica papaya L. pollen: an aeroallergen from India. Allergy 60(7):920–926
Chen MS (2008) Inducible direct plant defence against insect herbivores: a review. Insect Sci 15(2):101–114
Chougule NP, Bonning BC (2012) Toxins for transgenic resistance to hemipteran pests. Toxins (Basel) 4(6):405–429
Codex (2003) Codex principles and guidelines on foods derived from biotechnology. Food and Agriculture Organization of the United Nations, Rome
Das A, Roy A, Hess D, Das S (2013) Characterization of a highly potent insecticidal lectin from Colocasia esculenta tuber and cloning of its coding sequence. Am J Plant Sci 4(2A):408–416
de Carvalho F, Gheysen G, Kushnir S, Van Montagu M, Inzé D, Castresana C (1992) Suppression of beta-1,3-glucanase transgene expression in homozygous plants. EMBO J 11(7):2595–2602
Dellaporta SJ, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1(4):19–21
Dombrovsky A, Huet H, Chejanovsky N, Raccah B (2005) Aphid transmission of a potyvirus depends on suitability of the helper component and the N terminus of the coat protein. Arch Virol 150(2):287–298
Dutta I, Majumder P, Saha P, Ray K, Das S (2005a) Constitutive and phloem specific expression of Allium sativum leaf agglutinin (ASAL) to engineer aphid (Lipaphis erysimi) resistance in transgenic Indian mustard (Brassica juncea). Plant Sci 169:996–1007
Dutta I, Saha P, Majumder P, Sarkar A, Chakraborti D, Banerjee S, Das S (2005b) The efficacy of a novel insecticidal protein, Allium sativum leaf lectin (ASAL), against homopteran insects monitored in transgenic tobacco. Plant Biotechnol J 3(6):601–611
FAO/WHO (2001) Evaluation of the allergenicity of genetically modified foods: report of a joint FAO/WHO expert consultation, 22–25 Jan, Rome, Italy. http://www.fao.org/docrep/007/y0820e/y0820e00.HTM
Fermín G, Keith RC, Suzuki JY et al (2011) Allergenicity assessment of the papaya ringspot virus coat protein expressed in transgenic rainbow papaya. J Agric Food Chem 59(18):10006–10012
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) ExPASy Peptide Cutter tool: Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press Inc., Totowa, NJ, pp 571–607
Gatehouse A, Down R, Powell K et al (1996) Transgenic potato plants with enhanced resistance to the peach-potato aphid Myzus persicae. Entomol Exp Appl 79(3):295–307
Ghosh P, Roy A, Chakraborty J, Das S (2013) Biological safety assessment of mutant variant of Allium sativum leaf agglutinin (mASAL), a novel antifungal protein for future transgenic application. J Agric Food Chem 61(48):11858–11864
Goodman RE, Ebisawa M, Ferreira F, Sampson HA, van Ree R, Vieths S, Baumert JL, Bohle B, Lalithambika S, Wise J, Taylor SL (2016) AllergenOnline: a peer-reviewed, curated allergen database to assess novel food proteins for potential cross-reactivity. Mol Nutr Food Res 60(5):1183–1198
Gould HJ, Sutton BJ, Beavil AJ, Beavil RL, McCloskey N, Coker HA, Fear D, Smurthwaite L (2003) The biology of IGE and the basis of allergic disease. Annu Rev Immunol 21:579–628
Grover A, Pental D (2003) Breeding objectives and requirements for producing transgenics for major field crops of India. Curr Sci India 84(3):310–320
Guo QY, He LX, Zhu H, Shang JL, Zhu LY, Wang JB, Li Y (2015) Effects of 90-day feeding of transgenic maize BT799 on the reproductive system in male wistar rats. Int J Environ Res Public Health 12(12):15309–15320
Harlow E, Lane D (1988) Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, pp 60–67
Hossain MA, Maiti MK, Basu A, Sen S, Ghosh AK, Sen SK (2006) Transgenic expression of onion leaf lectin gene in Indian mustard offers protection against aphid colonization. Crop Sci 46:2022–2032
ICMR (2008) Indian Council of Medical Research, guidelines for safety assessment of food derived from genetically engineered plants. http://www.icmr.nic.in/guide/Guidelines%20for%20Genetically%20Engineered%20Plants.pdf. Accessed 2012
Ivanciuc O, Schein CH, Braun W (2003) SDAP: database and computational tools for allergenic proteins. Nucleic Acids Res 31(1):359–362
Jha GK, Pal S, Mathur VC, Bisaria G, Anbukkani P, Burman RR, Dubey SK (2012) Edible oilseeds supply and demand scenario in India: implications for policy. Report Indian Agric Res Inst New Delhi 110:012
Jin S, Zhang X, Daniell H (2012) Pinellia ternata agglutinin expression in chloroplasts confers broad spectrum resistance against aphid, whitefly, Lepidopteran insects, bacterial and viral pathogens. Plant Biotechnol J 10(3):313–327
Li H, Chougule NP, Bonning BC (2011) Interaction of the Bacillus thuringiensis delta endotoxins Cry1Ac and Cry3Aa with the gut of the pea aphid, Acyrthosiphon pisum (Harris). J Invertebr Pathol 107(1):69–78
Majumder P, Banerjee S, Das S (2004) Identification of receptors responsible for binding of the mannose specific lectin to the gut epithelial membrane of the target insects. Glycoconj J 20(9):525–530
Matzke AJ, Matzke MA (1998) Position effects and epigenetic silencing of plant transgenes. Curr Opin Plant Biol 1(2):142–148
Palacin A, Varela J, Quirce S, del Pozo V, Tordesillas L, Barranco P, Fernandez-Nieto M, Sastre J, Diaz-Perales A, Salcedo G (2009) Recombinant lipid transfer protein Tri a 14: a novel heat and proteolytic resistant tool for the diagnosis of baker’s asthma. Clin Exp Allergy 39(8):1267–1276
Porcar M, Grenier AM, Federici B, Rahbé Y (2009) Effects of Bacillus thuringiensis delta-endotoxins on the pea aphid (Acyrthosiphon pisum). Appl Environ Microbiol 75(14):4897–4900
Poulsen M, Kroghsbo S, Schrøder M et al (2007) A 90-day safety study in Wistar rats fed genetically modified rice expressing snowdrop lectin Galanthus nivalis (GNA). Food Chem Toxicol 45(3):350–363
Powell KS (2001) Antifeedant effects of plant lectins towards nymphal stages of the planthoppers Tarophagous proserpina and Nilaparvata lugens. Entomol Exp Appl 99:71–77
Powell KS, Spence J, Bharathi M, Gatehouse JA, Gatehouse AMR (1998) Immunohistochemical and developmental studies to elucidate the mechanism of action of the snowdrop lectin on the rice brown planthopper, Nilaparvata lugens (Stal). J Insect Physiol 44(7–8):529–539
Rajagopal D, Agarwal P, Tyagi W, Singla-Pareek SL, Reddy MK, Sopory SK (2007) Pennisetum glaucum Na+/H+ antiporter confers high level of salinity tolerance in transgenic Brassica juncea. Mol Breeding 19:137–151
Ramesh S, Nagadhara D, Reddy VD, Rao KV (2004) Production of transgenic indica rice resistant to yellow stem borer and sap-sucking insects, using super-binary vectors of Agrobacterium tumefaciens. Plant Sci 166(4):1077–1085
Rana JS (2005) Performance of Lipaphis erysimi (Homoptera: Aphididae) on different Brassica species in a tropical environment. J Pest Sci 78(3):155–160
Rao KV, Rathore KS, Hodges TK et al (1998) Expression of snowdrop lectin (GNA) in transgenic rice plants confers resistance to rice brown planthopper. Plant J 15(4):469–477
Roy A, Banerjee S, Majumder P, Das S (2002) Efficiency of mannose-binding plant lectins in controlling a homopteran insect, the red cotton bug. J Agric Food Chem 50(23):6775–6779
Roy A, Gupta S, Hess D, Das KP, Das S (2014) Binding of insecticidal lectin Colocasia esculenta tuber agglutinin (CEA) to midgut receptors of Bemisia tabaci and Lipaphis erysimi provides clues to its insecticidal potential. Proteomics 14(13–14):1646–1659
Sadeghi A, Broeders S, De Greve H, Hernalsteens JP, Peumans WJ, Van Damme EJ, Smagghe G (2007) Expression of garlic leaf lectin under the control of the phloem-specific promoter Asus1 from Arabidopsis thaliana protects tobacco plants against the tobacco aphid (Myzus nicotianae). Pest Manag Sci 63(12):1215–1223
Sadeghi A, Smagghe G, Broeders S, Hernalsteens JP, De Greve H, Peumans WJ, Van Damme EJ (2008) Ectopically expressed leaf and bulb lectins from garlic (Allium sativum L.) protect transgenic tobacco plants against cotton leafworm (Spodoptera littoralis). Transgenic Res 17(1):9–18
Saha S, Raghava GP (2006) AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res 34(Web Server issue):W202–W209
Saha P, Chakraborti D, Sarkar A, Dutta I, Basu D, Das S (2007) Characterization of vascular-specific RSs1 and rolC promoters for their utilization in engineering plants to develop resistance against hemipteran insect pests. Planta 226(2):429–442
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sarkar P, Jana J, Chatterjee S, Sikdar SR (2016) Functional characterization of Rorippa indica defensin and its efficacy against Lipaphis erysimi. Springerplus 5:511. https://doi.org/10.1186/s40064-016-2144-2
Sarkar P, Jana K, Sikdar SR (2017) Overexpression of biologically safe Rorippa indica defensin enhances aphid tolerance in Brassica juncea. Planta 246(5):1029–1044
Sauvion N, Rahbe´ Y, Peumans WJ, Van Damme EJM, Gatehouse JA, Gatehouse AMR (1996) Effects of GNA and other mannose binding lectins on development and fecundity of the peach-potato aphid Myzus persicae. Entomol Exp Appl 79:285–293
Scheurer S, Lauer I, Foetisch K et al (2004) Strong allergenicity of Pru av 3, the lipid transfer protein from cherry, is related to high stability against thermal processing and digestion. J Allergy Clin Immunol 114(4):900–907
Shekhawat K, Rathore SS, Premi OP, Kandpal BK, Chauhan JS (2012) Advances in agronomic management of Indian mustard (Brassica juncea (L.) Czernj. Cosson): an overview. Int J Agron. https://doi.org/10.1155/2012/408284
Singh AK, Mehta AK, Sridhara S, Gaur SN, Singh BP, Sarma PU, Arora N (2006) Allergenicity assessment of transgenic mustard (Brassica juncea) expressing bacterial codA gene. Allergy 61(4):491–497
Suhr M, Wicklein D, Lepp U, Becker WM (2004) Isolation and characterization of natural Ara h 6: evidence for a further peanut allergen with putative clinical relevance based on resistance to pepsin digestion and heat. Mol Nutr Food Res 48(5):390–399
Tang W, Newton RJ, Weidner DA (2007) Genetic transformation and gene silencing mediated by multiple copies of a transgene in eastern white pine. J Exp Bot 58(3):545–554
Thakur K, Kaur M, Kaur S, Kaur A, Kamboj SS, Singh J (2013) Purification of Colocasia esculenta lectin and determination of its anti-insect potential towards Bactrocera cucurbitae. J Environ Biol 34(1):31–36
Thang CL, Baurhoo B, Boye JI, Simpson BK, Zhao X (2011) Effects of Lactobacillus rhamnosus GG supplementation on cow’s milk allergy in a mouse model. Allergy Asthma Clin Immunol 7:20
Upadhyay SK, Mishra M, Singh H, Ranjan A, Chandrashekar K, Verma PC, Singh PK, Tuli R (2010) Interaction of Allium sativum leaf agglutinin with midgut brush border membrane vesicles proteins and its stability in Helicoverpa armigera. Proteomics 10(24):4431–4440
Van Damme EJ, Nakamura-Tsuruta S, Smith DF et al (2007) Phylogenetic and specificity studies of two-domain GNA-related lectins: generation of multispecificity through domain duplication and divergent evolution. Biochem J 404(1):51–61
Van Gramberg JL, de Veer MJ, O’Hehir RE, Meeusen EN, Bischof RJ (2013) Use of animal models to investigate major allergens associated with food allergy. J Allergy (Cairo) 2013:635695
Verma SN, Singh OP (1987) Estimation of avoidable losses to mustard by the aphid, Lipaphis erysimi (Kalt.) in Madhya Pradesh. Indian J Plant Prot 15(1):87–89
Verma SS, Sinha RK, Jajoo A (2015) (E)-β-farnesene gene reduces Lipaphis erysimi colonization in transgenic Brassica juncea lines. Plant Signal Behav 10(7):e1042636
Wang RY, Powell G, Hardie J, Pirone TP (1998) Role of the helper component in vector-specific transmission of potyviruses. J Gen Virol 79:1519–1524
Yadava JS, Singh NB (1999) In: Proceedings of the 10th international rapeseed congress, Canberra, Australia
Yamamoto YY, Matsui M, Ang LH, Deng XW (1998) Role of a COP1 interactive protein in mediating light-regulated gene expression in arabidopsis. Plant Cell 10(7):1083–1094
Yao J, Pang Y, Qi H, Wan B, Zhao X, Kong W, Sun X, Tang K (2003) Transgenic tobacco expressing Pinellia ternata agglutinin confers enhanced resistance to aphids. Transgenic Res 12(6):715–722
Zhang G, Zhou W (2006) Genetic analyses of agronomic and seed quality traits of synthetic oilseed Brassica napus produced from interspecific hybridization of B. campestris and B. oleracea. J Genet 85(1):45–51
Zhang H, Wu X, Tang K, Wang X, Sun X, Zhou K (2003) A primary study of transferring the Pinellia tenata agglutinin (pta) gene into rice and expression. Acta Genet Sin 30:1013–1019
Acknowledgements
Authors are thankful to Bose Institute for infrastructural facilities. AD is thankful to Council of Scientific and Industrial Research (Grant no. 09/015(0410)/2011-EMR-I) for providing fellowship. Technical assistance of Mr. Swarnava Das, Mr. Sudipta Basu and Mr. Arup Kumar Dey are sincerely acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Informed consent
Blood samples for in vitro IgE-specific ELISA experiment for biosafety assessment of CEA were collected from patients and non-allergic volunteers with informed written consents for participation in the study. The study was approved by the Human Ethics Committee of Bose Institute.
Data availability
All data generated or analyzed during this study are included in this manuscript [and its supplementary information files].
Additional information
Communicated by Tarek Hewezi.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Das, A., Ghosh, P. & Das, S. Expression of Colocasia esculenta tuber agglutinin in Indian mustard provides resistance against Lipaphis erysimi and the expressed protein is non-allergenic. Plant Cell Rep 37, 849–863 (2018). https://doi.org/10.1007/s00299-018-2273-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-018-2273-x