Abstract
Main conclusion
Cotton cytosolic pyruvate kinase GhPK6 is preferentially expressed in the late stage of fiber elongation process, transgenic experiments indicated that its expression level was negatively correlated to cell expansion rate.
Abstract
Pyruvate kinase (PK) plays vital regulatory roles in rapid cell growth in mammals. However, the function of PK in plant cell growth remains unclear. In allotetraploid upland cotton (Gossypium hirsutum L.), a total of 33 PK genes are encoded by the genome. Analysis of the transcriptome data indicated that only two cytosolic PK genes, GhPK6 and its duplicated gene GhPK26, are preferentially expressed in elongating cotton fiber cells. RT-qPCR and western blot analyses revealed that the expression of GhPK6 was negatively correlated with fiber elongation rate, which well explains the observed sharp increase of cytosolic PK activity at the end of fast fiber elongation process. Furthermore, virus-induced gene silencing of GhPK6 in cotton plants resulted in increased fiber cell elongation and reduced reactive oxygen species (ROS) accumulation. On the contrary, Arabidopsis plants ectopically expressing GhPK6 exhibited ROS-mediated growth inhibition, whereas the addition of ROS scavenging reagents could partly rescue this inhibition. These data collectively suggested that GhPK6 might play an important role in regulating cotton fiber elongation in a ROS-dependent inhibition manner.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The economically important fibers of allotetraploid upland cotton (Gossypium hirsutum L.) are single-celled trichomes that differentiate from the ovule epidermis. These trichomes undergo dramatic and polarized elongation after initiation, followed by secondary wall deposition and maturation, and finally form the mature fibers that are harvested from the seed and used in the textile industry (Lee et al. 2007). In general, >20,000 fibers may form in near synchrony on each seed and typically elongate up to >3.0 cm in length in most commercially important species. During the elongation period [0–20 days post-anthesis (dpa)], extreme cell expansion with peak elongation rates of >2 mm/day is observed in upland cotton (Yang et al. 2008). Furthermore, the cellulose in a single cell accounts for >95 % of the dry weight of mature fiber cells (Kim and Triplett 2001). Cotton fiber cells, therefore, are not only among the fastest-growing and longest cells in the plant kingdom but are also a cellulose synthesis factory, making these cells an excellent model system for studying cell elongation and cellulose biosynthesis.
Fiber cell elongation rate and duration contribute significantly to the final length and morphogenesis of fiber cells and are governed by developmental programs that coordinately regulate the cell turgor, cell wall extensibility and biosynthesis of new cell components (Smart et al. 1998). Significant progress has been made recently in the global identification of genes, proteins and microRNAs that are involved in fiber cell elongation (Arpat et al. 2004; Shi et al. 2006; Kumar et al. 2013; Xue et al. 2013; Zhang et al. 2013). In these studies, a considerable number of identities exhibited stage-specific expression patterns, indicating their probable roles in the cell elongation process. Furthermore, various influencing factors, including structural proteins (Li et al. 2005; Han et al. 2013), carbohydrate-active enzymes (Thaker et al. 1999; Pang et al. 2010), osmoregulation proteins (Ruan et al. 2003; Wang et al. 2010), and phytohormone biosynthetic proteins (Zhang et al. 2011; Han et al. 2014), have been explored for their individual roles played in the process of fiber cell elongation. However, little is known about the effects of metabolic regulation on fiber cell elongation.
As an important metabolic enzyme, pyruvate kinase (PK) catalyzes the final step of glycolysis, transfers the high-energy phosphate group from phosphoenolpyruvate (PEP) to ADP to produce ATP and pyruvate (Fernie et al. 2004). Pyruvate then enters the mitochondria to generate ATP through the tricarboxylic acid (TCA) cycle and oxidative phosphorylation with by-product reactive oxygen species (ROS) (Steffens et al. 2013). There is a growing evidence that PK might play important roles in cell growth (Israelsen et al. 2013). In mammals, rapidly proliferating cells, including cancer cells and stem cells, express the less active M2 isoform of PK (PKM2), which promotes the metabolism of glucose by aerobic glycolysis, and thereby the efficient conversion of glucose into macromolecules that are needed to construct a new cell (Christofk et al. 2008; Vander Heiden et al. 2010).
In plants, PK also plays a critical role in cellular metabolic flux. Plant PKs exist as cytosolic (PKc) and plastidic (PKp) isozymes that show substantial differences in their respective molecular, kinetic and regulatory properties (Ambasht and Kayastha 2002). The function of PKp is related to fatty acid biosynthesis in Arabidopsis seeds (Andre et al. 2007; Baud et al. 2007). Meanwhile, the impacts of down-regulated PKc levels on plant metabolism have also been investigated. Transgenic studies have demonstrated altered morphology with indentations in the fully expanded leaves of leaf PKc-lacking tobacco plants that were grown under low light (Knowles et al. 1998). Such bumpy surfaces in fully expanded leaves may be due to the excessive expansion of certain cells. In another report, the inhibition of PKc expression in potato tubers resulted in respiration inhibition not only by down-regulating the contents of pyruvate and other organic acids that are involved in the TCA cycle but also through the activity repression of alternative oxidases (AOXs) (Oliver et al. 2008). Down-regulated PKc in T-DNA insertion mutant rice implied that PKc might affect the glycolytic pathway, causing dwarfism (Zhang et al. 2012). These studies collectively indicate that PKc may function as a species- and tissue-specific regulator of plant growth and development. However, similar information concerning the role of PKc in plant cell expansion is still deficient.
In this study, we found that two of the 19 PKc genes were preferentially expressed in cotton fibers and the activity of cotton PKc exhibited a negative correlation with the fiber cell elongation rates. Results from virus-induced gene silencing (VIGS) and transgenic experiments further indicated that the differentially expressed PKc gene, GhPK6, negatively regulates cotton fiber and Arabidopsis hypocotyl elongation in a ROS-dependent inhibition manner. These findings demonstrate an important molecular correlation between PKc-regulated cellular metabolism and cell expansion in plants.
Materials and methods
Plant materials
Upland cotton (Gossypium hirsutum) cultivar CRI 35 was grown in a normal agronomic field from April to September in Beijing. Cotton flowers on the day of anthesis (0 dpa) were tagged, and the developing cotton bolls were harvested during selected stages. The fibers and seeds were carefully dissected from each boll, immediately frozen in liquid nitrogen along with other tissues and stored at −80 °C until used to extract protein, RNA and other organic materials. Fresh cotton bolls were used for fiber length and cell wall thickness measurements and enzyme activity assays. Arabidopsis thaliana ecotype Columbia 0 (Col-0) were grown at 22 °C under 16/8-h light/dark conditions.
Database search and sequence retrieval
To identify all the PK genes in cotton (G. hirsutum), multiple database searches were performed. First, the Arabidopsis PK gene sequences were retrieved from the Arabidopsis information resource (http://www.arabidopsis.org) using ontologies/keywords search interface with pyruvate kinase as keyword. Next, coding sequence of these Arabidopsis PK genes were used as queries to perform repetitive local blast searches against the downloaded G. hirsutum genome data (http://www.cgp.genomics.org.cn) (Li et al. 2015). All output genes with an E value <1.0 were collected and translated to proteins, which were further blast searched against protein sequence data repositories at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) to confirm the identity as PK enzymes.
Subcellular localization, phylogenetic and dataset-based gene expression analysis
The subcellular localization of each GhPK protein was predicted using TargetP (http://www.cbs.dtu.dk/services/TargetP) (Emanuelsson et al. 2000). The amino acid sequences were aligned using ClustalX (version 2.1). Phylogenetic trees were then constructed using MEGA software (version 5.05) with the neighbor-joining (NJ) method and 1000 bootstrap replicates. To determine the relative expression levels of the PK genes in cotton tissues, all gene sequences were BLAST searched against the G. hirsutum ovule (0 and 10 dpa), fiber (10 and 20 dpa), root, stem and leaf transcriptome dataset (NCBI Sequence Read Archive SRP059947, which was transformed to FASTA files by NCBI SRA Toolkit) with E value <E −10 and 100 % identity (Ma et al. 2016). The expression level of each GhPK gene was then calculated based on the matched read numbers and gene lengths using the RPKM algorithm (Mortazavi et al. 2008). A complete linkage hierarchical cluster analysis of the GhPK genes was performed through comparison of the RPKM value of each genes using MeV 4.7 software package.
Enzyme activity assay
PKc activity was determined as described by Turner et al. (2005). Briefly, the cotton fibers were homogenized using a mortar and pestle with 50 mM potassium phosphate buffer (pH 7.0) containing 1 mM EDTA and 0.1 % (w/v) phenylmethanesulfonyl fluoride. The homogenate was squeezed through three layers of cheesecloth and then centrifuged at 12,000g for 15 min. The obtained soluble fraction was used for the PKc activity assay. The reaction was coupled to the lactate dehydrogenase reaction and assayed at 25 °C by monitoring the oxidation of NADH at 340 nm using an Ultrospec 3300 Pro spectrophotometer (Amersham Biosciences, Uppsala, Sweden) with a continuous recording function. The assay conditions were 50 mM HEPES–KOH (pH 6.4), 25 mM KCl, 12 mM MgCl2, 2 mM PEP, 1 mM ADP, 1 mM DTT, 5 % (w/v) PEG8000, 0.15 mM NADH and 2 units/ml lactate dehydrogenase (Sigma, Shanghai, China) in a final volume of 500 μL. Protein concentrations were determined using Bradford method, whereas V max and K m values were automatically calculated by the spectrophotometer.
To assay the enzyme activity of AOX and succinate dehydrogenase (SDH), the intact mitochondria was first isolated from fresh cotton fibers using the differential centrifugation method (Laties 1974). The sonicated mitochondria supernatant was used for SDH and AOX activity assays following the methods described by Singer et al. (1973) and Affourtit and Moore (2004), respectively.
ATP and ROS determination
For the quantitative analysis of the intracellular ATP concentration, an ATP Assay System Bioluminescence Detection Kit (FF2000) was used as described by the manufacturer (Promega, Wisconsin, USA). To detect superoxide radicals, histochemical staining with nitro blue tetrazolium (NBT) was performed using the methods of Dong et al. (2009). For the quantitative analysis of the intracellular H2O2 content, the titanium oxidation measurement method was used following the protocol of Li et al. (2007).
Molecular cloning and real-time RT-PCR analyses
The total RNA was extracted from the cotton roots, leaves, stems, flowers, seed and fibers using the RNAprep pure Plant kit (TIANGEN, Beijing, China). Each RNA sample was treated with DNase I after extraction to remove residual DNA. The cDNA was reverse-transcribed from 1 µg of total RNA using oligo-dT primers. The full-length ORF of GhPK6 was PCR-amplified using the primer pairs PK6-F/PK6-R (Table S1). The qRT-PCR assays were performed using the SYBR Premix Ex Taq (TaKaRa, Dalian, China) and a MiniOpticon real-time PCR system (Bio-Rad, California, USA). A cotton ubiquitin gene (UBQ7, DQ116441) was used as a standard control.
Antibody preparation and western blotting
The specific antibodies were prepared by the immunization of rabbits with synthesized protein-specific peptides (Table S2) mixed with Freund adjuvant by Beijing Protein Innovation (Beijing, China). Two-step antigen affinity purifications were further performed to enrich the antibodies to ensure the titers against BSA-coated synthesized peptides larger than 1:12,800 by ELISA. Goat anti-rabbit antibodies that were conjugated to horseradish peroxidase were used as secondary antibodies. For western blotting, 20 µg of proteins for each sample were denatured in 6 × SDS-PAGE sampling buffer by boiling in a water bath for 5 min, then separated by 12 % SDS-PAGE and transferred to a PVDF membrane. The signals were revealed with Lumi-Light western blotting substrate (Roche, Mannheim, Germany).
Protein expression and purification
The full-length ORF of GhPK6 was PCR-amplified from full-length cDNA, digested with BamHI and SacI and cloned into the prokaryotic expression vector pET-28a (Novagen, Wisconsin, USA). The recombinant His-tagged GhPK6 proteins were expressed in Escherichia coli strain BL21 (DE3) and purified using a nickel sulphate resin following the manufacturer’s instructions (Qiagen, Hilden, Germany).
Virus-induced gene silencing in cotton plants
The TRV vectors and Agrobacterium tumefaciens for virus-induced gene silencing (VIGS) were prepared accordingly (Qu et al. 2012). The N-terminal 500-bp fragments of the GhPK6 genes were PCR-amplified and digested with EcoRI and BamHI and then ligated into the pTRV2 plasmid. The constructs were transformed into Agrobacterium tumefaciens GV3101 as described. The PCR-confirmed GV3101 strains incorporating the pTRV1 and pTRV2 vectors or their derivatives were infiltrated into the cotyledons of 10-day-old seedlings of G. hirsutum CRI 35 using a needleless syringe. The seedlings were then grown at 22 °C with a 16-h/8-h light/dark photoperiod in a greenhouse supplemented with 50 ml of 1/2 MS medium once a week. 42 plants with fourth true leaf detected as gene-silenced were further grown to flowering, among which five plants with 15 dpa fiber further detected as gene-silenced (#4, #7, #16, #27, #37) were finally used for phenotype and biochemical analyses. Specifically, cotton bolls grown at the same fruiting position of the seventh sympodial branch of each plants were harvested for fiber length measurement, pyruvate kinase activity analysis and H2O2 content assay.
Arabidopsis transformation and phenotypic analyses
The ORF of GhPK6 was PCR-amplified, digested with BamHI and SacI and cloned into the pBE2113 vector to replace the GUS gene. The construct was then transferred into Arabidopsis ecotype Col-0 by the floral dip method. The positive transformants were selected on MS medium with 50 mg/L kanamycin and were grown until maturation. Totally 23 homozygous lines were obtained and three homozygous lines were used for the phenotypic analyses. The seeds of wild-type (Col-0) and three homozygous lines (#12, #13, #17) of transgenic Arabidopsis were sterilized, vernalized and grown in 1/2 MS medium for analyses of seedling root growth. The seedlings were transferred into soil and grown to flowering for analyses of leaf and stem growth. To analysis hypocotyl growth, the vernalized seeds were vertically grown under dark in 1/2 MS medium containing different concentrations of ascorbate and GSH for 1 week.
Statistical analyses
Unless otherwise specified, all the experiments were at least repeated for three biological and technical replicates. Tukey’s multiple comparison test was used to test the variation of elongation rate, enzyme activity, ATP and ROS level during fiber elongation process. Student’s t test was used to test the effect of GhPK6 expression on cotton fiber and Arabidopsis growth, ATP and ROS level. All the statistical analyses were performed with SPSS 16.0 statistical software package (IBM, 2007). Significance was declared at the 0.05 (*) and 0.01 (**) level of probability.
Accession numbers
The GenBank accession numbers for GhPK6, GhPK26 and Cotton_01025 are KX369413, KX369414 and KX369415, respectively.
Results
PKc activity is negatively correlated with fast elongation of cotton fiber cell
In accordance with previous report (Basra and Malik 1983), fiber length measurements demonstrated that cotton fiber cell continuously elongate during 5–25 dpa, however, the elongation rate gradually decreased due to the deposition of secondary cell wall (Figure S1). Specifically, the elongation rate was higher than 1 mm/day between 5 and 20 dpa, however, it sharply decreased to less than 0.2 mm/day after 20 dpa (Fig. 1a). On the contrary, PKc enzyme activity was invariant from 5 to 15 dpa but significantly increased at 20 dpa (Fig. 1b). Interestingly, the levels of ATP, and the level of oxidative phosphorylation by-product H2O2, the major stable form of ROS, were also both significantly elevated at 20 dpa (Fig. 1c, d), along with the increased activity of PKc. These results indicated that PKc enzyme activity is negatively correlated with fast fiber elongation in cotton.
Cytosolic pyruvate kinase activity in elongating cotton fibers and its relevance to the H2O2 content and fiber cell elongation rate. a Fiber cell elongation rate in the elongation process. b Cytosolic pyruvate kinase activity, c ATP content and d H2O2 contents of cotton fibers at 4 time points during the elongation stage. For each experiment, the means of data from three replicates are presented with SE. Statistical analyses were performed using Tukey’s multiple comparison test. dpa days post-anthesis
GhPK6 and its duplicated gene GhPK26 are preferentially expressed in cotton fibers
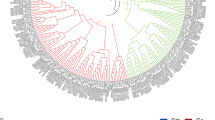
Through Blast search using known PK gene sequences from Arabidopsis as queries, 33 PK genes were identified in the G. hirsutum genome (Fig. 2a; Table S3). Due to the lack of standard annotation designated to the 33 PK genes in G. hirsutum, we named them GhPK1 to GhPK33 according to their chromosomal order. Notably, the GhPK1 to GhPK13 genes are located at the At chromosomes, the GhPK14 to GhPK25 genes are located at the Dt chromosomes, and the remaining 8 PK genes are located at the sequencing scaffolds (Table S3). TargetP prediction indicated that 19 PK genes were PKc isoforms, whereas the remaining 14 genes were PKp isoforms (Table S3). Phylogenetic analysis indicated that the 19 PKc and 14 PKp genes were clustered in two separate groups (Fig. 2a). Interestingly, most of the PK genes were further clustered in different gene pairs. Considering that G. hirsutum is an allotetraploid plant, the homogenous genes clustered in pairs should be the duplicated genes originated from different diploid ancestors (Li et al. 2015).
Phylogenetic and expression profile analysis of cotton pyruvate kinase genes. a Phylogenetic analysis of the 33 cotton pyruvate kinase (GhPK) genes. The phylogenetic tree was constructed using the neighbor-joining method with pairwise deletion and bootstrap analysis with 1000 replicates. b Heat maps of GhPK family members were constructed based on the RPKM values of RNAseq of different tissues of G. hirsutum. dpa days post-anthesis
The expression levels of the 33 PK genes varied greatly. Five genes, including GhPK11, GhPK12, GhPK25, GhPK29 and GhPK31, were weakly expressed in all tissues with a maximum RPKM smaller than 4, whereas 11 PK genes were highly expressed in all tissues with a minimum RPKM larger than 28 (Fig. 2b; Table S4). Among the 11 highly expressed genes, GhPK6, GhPK7, GhPK17, GhPK21, GhPK22, GhPK26, GhPK32 and GhPK33 are PKc isoforms, whereas the remaining three genes, including GhPK19, GhPK27 and GhPK28, are PKp isoforms (Fig. 2a). Some of these highly expressed PK genes, such as GhPK21, GhPK22, GhPK32 and GhPK33, were housekeeping genes because their expression levels were similar in all tissues. However, some of the genes were preferentially expressed in specific tissues (RPKM ratio >2). For example, GhPK6 and its duplicated gene GhPK26 were preferentially expressed in fibers (especially the 20 dpa fiber), GhPK27 and GhPK28 genes were preferentially expressed in ovules, whereas GhPK7 and GhPK17 genes were preferentially expressed in ovules and stems (Fig. 2b; Table S4).
Interestingly, sequence alignment further indicated that GhPK6 and GhPK26 were highly homogenous to the encoding gene of the previously identified PKc protein differentially expressed in fiber elongation process (Figure S2) (Zhang et al. 2013). Consistent with the result, RT-qPCR revealed that GhPK6 transcription was more prominent in cotton fibers than other tissues except flowers (Fig. 3a). Furthermore, both RT-qPCR and western blot analyses demonstrated that GhPK6 mRNA and protein were significantly increased in abundance during fiber cell elongation from 5 to 20 dpa (Fig. 3a, b), synchronizing with the decreasing elongation rates throughout the elongation process. These results demonstrated that GhPK6 were preferentially expressed in cotton fibers, and its expression level was negatively correlated with fiber elongation rate.
Expression analysis of GhPK6. a RT-qPCR analysis of the gene expression of GhPK6 in fibers, seeds, roots, stems, leaves and flowers. The cotton UBQ7 gene was served as a reference. b GhPK6 protein abundance in elongating fibers. Values were obtained from densitometric analyses of the western blot results. The cotton GhHSP70 protein served as a reference. For each experiment, the means of data from three replicates are presented with SE
Inhibition of GhPK6 expression promotes cotton fiber elongation
The biochemical function of GhPK6 protein was firstly analyzed in vitro. In the presence of saturated concentrations of substrates and cofactors, purified His-tag recombinant GhPK6 protein (Fig. 4a) exhibited a broad pH optimum that is centered at approximately pH 7.0 (Fig. 4b), and the Km of GhPK6 for PEP and ADP at pH 7.0 were 106 and 46 μM, respectively (Fig. 4c, d). Similar to known PKc proteins, enzyme activity of GhPK6 was also strongly inhibited by glutamate and citrate (Fig. 4e). These data collectively indicated that GhPK6 is a conserved PKc enzyme with active catalytic activity.
In vitro enzyme activity analysis of recombinant GhPK6 protein. a SDS-PAGE showing the purified recombinant GhPK6 protein. The protein bands were visualized by CBB-staining and the recombinant GhPK6 band was marked using asterisk. Effect of b pH value, c PEP concentration, d ADP concentration and e glutamine and citrate concentration on reaction velocity of recombinant GhPK6 was determined using spectrophotometer. For each experiment, the means of data from three replicates are presented
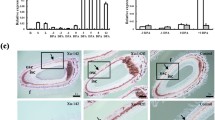
The physiological function of GhPK6 in cotton fiber elongation was analyzed using VIGS. RT-qPCR analysis indicated that the expression of GhPK6 was significantly inhibited in 42 cotton plants agroinfiltrated with TRV vectors; whereas RT-qPCR and western blot analysis collectively indicated that the expression of GhPK6 was further inhibited in the 15 dpa cotton fibers. Specifically, the expression of GhPK6 was almost completely blocked in two plants (Figure S3). Although the plantlet phenotypes were unchanged in GhPK6-VIGS plants, the inhibition of GhPK6 expression in fiber cells significantly decreased the PKc enzyme activity (Fig. 5c) and lengthened the fibers (Fig. 5a, b). Specifically, the mature fibers from GhPK6-VIGS plants were almost 11 % longer than those of the control and mock-infected plants (Fig. 5b). Furthermore, GhPK6-VIGS cotton plants exhibited a significantly decreased ROS level as demonstrated by the H2O2 content assay of the 20 dpa fibers (Fig. 5d) compared to that of the control and mock-infected cotton plants. These data provide solid evidence that the inhibition of GhPK6 expression through VIGS not only decreased the PKc activity and intracellular ROS levels but also promoted fiber cell elongation.
Inhibition of GhPK6 expression promotes fiber cell elongation. a Phenotypes of the mature fibers that were attached to the seeds from separate bolls of different CK (uninfected control), Mock (empty vector control), GhPK6-VIGS cotton plants. Bar 10 mm. b Analysis of the fiber length in the different cotton plants. Fifty values were recorded for each seed. The error bars indicate standard deviations. c PKc enzyme activity and d H2O2 content of 20 DPA fibers from different cotton plants were determined. The means of data from three replicates are presented with SE. Statistical analyses were performed using Student’s t test. *p < 0.05
Ectopic expression of GhPK6 inhibits Arabidopsis hypocotyl elongation
GhPK6-VIGS cotton plants exhibited an increased fiber length and decreased PKc enzyme activity and intracellular ROS levels in fiber cells, suggesting a negative correlation between cell elongation and GhPK6-mediated ROS accumulation. The link between cell elongation and ROS accumulation was further analyzed in GhPK6-ectopically expressed Arabidopsis plants. The growth of Arabidopsis plants ectopically expressing GhPK6 significantly decreased in that the length of the mature stems and jejune roots and the fresh weight of the rosette leaves were only 50–70 % of those of the wild-type plants (Figure S4). Furthermore, the ROS levels in the 3-week-old leaves of transgenic Arabidopsis plants were much higher (7–8 fold) than those of the wild-type plants (Figure S4). Meanwhile, the activity of PKc and the ATP content in the transgenic plants were 27–90 % and 9–35 % higher than those of the wild-type plants, respectively (Figure S4). These data indicated that the inhibition of Arabidopsis growth via the overexpression of GhPK6 increased PKc activity, ATP and ROS accumulation.
In agreement with the above results, the hypocotyl lengths of 1-week-old GhPK6-overexpressing Arabidopsis seedlings were 20–24 % shorter that those of the wild-type plants (Fig. 6). However, the hypocotyl elongation deficiency of GhPK6-overexpressing Arabidopsis plants could be partly rescued through the addition of the ROS-scavenger ascorbate and GSH (Fig. 6). The effects of ascorbate and GSH on hypocotyl elongation were concentration-dependent. The hypocotyls of GhPK6-overexpressing Arabidopsis plants that were grown with 10 mM ascorbic acid and GSH for 1 week were 5–10 % longer than those of the plants that were grown in 1/2 MS alone. When the concentrations of ascorbic acid and GSH increased to 25 mM, the hypocotyl lengths of the GhPK6-overexpressing Arabidopsis plants further increased by approximately 10–15 %, almost equivalent to those of the wild-type plants (Fig. 6). These data strongly suggest that GhPK6 inhibits Arabidopsis hypocotyl elongation in a ROS-mediated manner.
Ectopic expression of GhPK6 inhibit Arabidopsis hypocotyl elongation. Left representative photos of hypocotyl phenotypes of the three lines of 35S::GhPK6 transgenic Arabidopsis plants and wild-type Arabidopsis (Col-0). Right measurement and statistical analysis of the hypocotyl lengths of different Arabidopsis plants under different growth conditions. The means of data from three replicates are presented with SE. Statistical analyses were performed using Student’s t test. *p < 0.05; **p < 0.01
Discussion
Glycolysis regulation in plants are much more complex than that of animals not only because of the existence of plastid glycolysis branch pathway and PEPC-mediated alternative pathway, but also derived from the synergistic source–sink relationships between photosynthesis leave tissues and energy-consuming organs such as roots and seeds (Plaxton 1996; Zhang 2015). As a rate-limiting enzyme of glycolysis pathway, PKc was determined to play important roles in regulating carbon flux from source to sink, making PKc an important regulator in plant organ development (Ambasht and Kayastha 2002). However, the function of PKc in plant cell growth remains unclear. In this study, we found that two PKc genes, GhPK6 and its duplicated gene GhPK26, were preferentially expressed in elongating cotton fibers. The expression level of GhPK6 was negatively correlated with fast fiber elongation, whereas inhibition of GhPK6 expression could promote the fiber elongation. Results of our study also suggested that the inhibition of cotton fiber and Arabidopsis hypocotyl elongation by GhPK6 was possibly mediated by its long-range downstream by-product ROS. These results collectively suggested that PKc might play an important regulatory role in plant growth at the cellular level.
The activity of PKc was relatively low at 5–15 dpa when the fiber cells had a fast elongation rate and then sharply increased at 20 dpa when the fast elongation of cotton fiber cells ceased (Fig. 1a, b). This result was in line with previously reported PK activity assay data that a peak activity could be observed at late stage (27 dpa) of cotton fiber elongation process (Thaker et al. 1999). Interestingly, the variation of PKc activity was similar to the previously observed activity change of malic enzyme (ME), which catalyzes the conversion of malate to pyruvate (Basra and Malik 1983). In contrast, the activity of phosphoenolpyruvate carboxylase (PEPC) and malate dehydrogenase (MDH), which catalyze the PEP transforming to oxaloacetate and oxaloacetate to malate, were both maintained at high level during 5–15 dpa and sharply decreased at 20 dpa (Basra and Malik 1983; Li et al. 2010). The synergic change of the activity of PKc, ME, PEPC and MDH at 20 dpa could simultaneously enhance the transformation of PEP/malate to pyruvate and reduce the synthesis of malate from PEP, thereby efficiently reduce the intracellular concentration of malate. Because malate is the major accumulated organic solute to drive cotton fiber elongation (Dhindsa et al. 1975), the reduction of malate could inevitably slow down the elongation of cotton fiber.
Blast search indicated that 33 PK genes were encoded by the Gossypium hirsutum genome (Fig. 2a; Table S3). The number of PK genes in G. hirsutum is much higher than that of Arabidopsis (14), tomato (10), rice (11) and diploid Asian cotton G. arboreum (18) (Andre et al. 2007; Oliver et al. 2008; Zhang et al. 2012; Zhang 2015), suggesting that the PK gene family are expanded by genome duplication and polyploidization events in the evolutionary process of cotton plants. Analysis of the transcriptome data further indicated that 33 PK genes have different expression levels. Notably, some of the genes were constitutively expressed with similar expression level in all tissues, whereas the others were preferentially expressed in specific tissues (Fig. 2b; Table S4). Size-exclusion chromatography revealed that both plant PKc and PKp are tetrameric proteins, either heterotetramer or homotetramer, composed of four subunits with different or similar molecular mass (Ambasht and Kayastha 2002). The differential expression of cotton PK genes in different tissues strongly suggested that different heterotetramer or homotetramer of PKc and PKp could be formed in specific tissues. Specifically, four PKp genes (GhPK1, GhPK5, GhPK27 and GhPK28) were preferentially expressed in cotton ovules (Fig. 2b). The four genes encode three different proteins (GhPK27 and GhPK28 encode the same protein, Table S3), suggesting three subunits of PKp existed in cotton ovules, which is in full agreement with the previous findings that three subunits of PKp are involved in seed oil synthesis in Arabidopsis (Andre et al. 2007; Baud et al. 2007).
Transcriptome analysis and RT-qPCR both indicated that GhPK6 gene was preferentially expressed in cotton fibers (Figs. 2b, 3a). Western blot analysis further demonstrated that protein abundance of the 55 kDa PKc subunit encoded by GhPK6 gradually increased in cotton fiber elongation process (Fig. 3b). It is understandable that the abundance change of GhPK6 is more prominent than the activity variation of PKc because other PKc genes were also constitutively expressed in fibers (Fig. 2b). Interestingly, the change of protein abundance of GhPK6 is also more prominent than that of mRNA (Fig. 3a, b), suggesting specific posttranslational modifications might further regulate its protein abundance. The 55 kDa PKc subunit was determined being regulated by phosphorylation and C-terminal degradation in soybeans (Tang et al. 2003). Similar posttranslational modifications of PKc might also exist in cotton. It is also noteworthy that recombinant His-tag GhPK6 protein exhibited typical catalytic activity and regulation characteristic of plant PKc (Fig. 4b, e), implying that the 55 kDa PKc subunit encoded by GhPK6 could form homotetramer in vitro. However, the situation is more complex in vivo because as many as 19 PKc genes coexist in cotton plants so that multiple heterotetramers containing the GhPK6 subunit could be formed.
As the stable form of ROS, H2O2 plays dual functions in cotton fiber elongation. Low concentration of H2O2 during early fiber development promotes initiation and early fiber cell elongation (Zhang et al. 2010). In contrast, the peak level of H2O2 corresponds to the time of fast fiber elongation termination (Potikha et al. 1999). Coincidently, the exogenous addition of low concentrations of H2O2 (2–20 μM) could promote fiber cell elongation of in vitro cultured cotton ovules, whereas the addition of a high concentration of H2O2 (approximately 50 μM) severely inhibits fiber cell elongation (Tang et al. 2014). The ROS scavenging enzyme ascorbate peroxidase was found participated in the regulation of redox homeostasis of cotton fiber cells (Li et al. 2007; Guo et al. 2016); however, the detailed mechanism of how the dual functions of H2O2 are regulated in cotton fibers is far from well understood. Based on our observations, down-regulating GhPK6 expression resulted in a decreased H2O2 level (approximately 33.6 %) and longer cotton fibers (approximately 11.8 %) (Fig. 5). More importantly, the ectopic expression of the single GhPK6 gene in Arabidopsis plants readily decreased the growth rate and significantly promoted ROS accumulation (Fig. 6; Figure S4), while the addition of ROS scavenging reagents partly rescued this decrease (Fig. 6). In combination with the observation that the H2O2 level was synchronized with the expression level of GhPK6 in elongating cotton fibers (Figs. 1, 3), our results revealed an important molecular link between the PKc activity, H2O2 level and cell expansion rate.
Our results seemed contradictory to the previous finding that pyruvate generated by PKc could promote the AOX activity to eliminate H2O2 (Hoefnagel et al. 1997). However, as one of the multiple ROS scavenging systems, AOX can only partly slow down the ROS accumulation (Maxwell et al. 1999), but cannot impede the ROS burst promoted by either stress conditions or developmental signals (Rasmusson et al. 2009). In elongating cotton fibers, the AOX activity was always less than 1/20 that of SDH (Figure S5), an indicator of oxidative phosphorylation activity (Gleason et al. 2011). Although the activity of AOX gradually increased in accordance with the PKc activity and free pyruvate, the electron generated by TCA cycle was majorly oxidized by oxidative phosphorylation, which was promoted by high activity of PKc (Figure S5).
In summary, our findings painted a vivid picture regarding the PKc-mediated mechanistic links between cellular metabolism and growth control in cotton fiber cells (Figure S6). In elongating fiber cells, many sucrose molecules are continuously transported through the phloem from photosynthetic leaves (source) into non-photosynthetic fiber cells (sink) and are broken down to yield pyruvate through the glycolytic pathway (Chen et al. 2012; Zhang et al. 2013). The fiber-specific PKc, encoded by GhPK6 and its duplicated gene GhPK26, catalyzes the irreversible synthesis of pyruvate and ATP from PEP and ADP, and is a primary control site of fiber glycolytic flux to the TCA cycle and oxidative phosphorylation (Plaxton 1996). As the transcript and protein abundance of GhPK6 increase, the oxidative phosphorylation by-product ROS gradually accumulates in elongating cotton fibers, and finally inhibits the fiber cell elongation (Fig. 1). Therefore, the abundance/activity of GhPK6 in elongating fiber cells were tightly controlled and exhibited a negative correlation with the fiber cell elongation rate (Figs. 1, 5). As described above, the relative low activity of PKc during the rapid fiber elongation phase could further ensure that PEP is efficiently used by PEPC to synthesis malate to drive cell turgor (Dhindsa et al. 1975; Li et al. 2010). Our model, together with the plasmodesmata reversible gating model (Ruan et al. 2001), linear cell growth model (Shi et al. 2006; Qin and Zhu 2011) and classical turgor-driven cell expansion theory (Kirkham et al. 1972; Smart et al. 1998), contribute to a better understanding of the mysterious elongation process of cotton fibers.
Author contribution statement
BZ and JL conceived and designed research. BZ conducted experiments and analyzed data. BZ and JL wrote the manuscript. Both authors read and approved the manuscript.
References
Affourtit C, Moore AL (2004) Purification of the plant alternative oxidase from Arum maculatum: measurement, stability and metal requirement. Biochim Biophys Acta 1608:181–189
Ambasht PK, Kayastha AM (2002) Plant pyruvate kinase. Biol Plant 45:1–10
Andre C, Froehlich JE, Moll MR, Benning C (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19:2006–2022
Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, Main D, Wood T, Leslie A, Wing RA, Wilkins TA (2004) Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol 54:911–929
Basra AS, Malik CP (1983) Dark metabolism of CO2 during fibre elongation of two cottons differing in fiber lengths. J Exp Bot 34:1–9
Baud S, Wuillème S, Dubreucq B, de Almeida A, Vuagnat C, Lepiniec L, Miquel M, Rochat C (2007) Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J 52:405–419
Chen A, He S, Li F, Li Z, Ding M, Liu Q, Rong J (2012) Analyses of the sucrose synthase gene family in cotton: structure, phylogeny and expression patterns. BMC Plant Biol 12:85
Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC (2008) The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumor growth. Nature 452:230–234
Dhindsa RS, Beasley CA, Ting IP (1975) Osmoregulation in cotton fiber: accumulation of potassium and malate during growth. Plant Physiol 56:394–398
Dong CH, Zolman BK, Bartel B, Leea BH, Stevenson B, Agarwal M, Zhu JK (2009) Disruption of Arabidopsis CHY1 reveals an important role of metabolic status in plant cold stress signaling. Mol Plant 2:59–72
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Fernie AR, Carrari F, Sweetlove LJ (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7:254–261
Gleason C, Huang S, Thatcher LF, Foley RC, Anderson CR, Carroll AJ, Millar AH, Singh KB (2011) Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc Natl Acad Sci USA 108:10768–10773
Guo K, Du X, Tu L, Tang W, Wang P, Wang M, Liu Z, Zhang X (2016) Fibre elongation requires normal redox homeostasis modulated by cytosolic ascorbate peroxidase in cotton (Gossypium hirsutum). J Exp Bot 67:3289–3301
Han LB, Li YB, Wang HY et al (2013) The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell 25:4421–4438
Han J, Tan J, Tu L, Zhang X (2014) A peptide hormone gene, GhPSK promotes fibre elongation and contributes to longer and finer cotton fiber. Plant Biotechnol J 12:861–871
Hoefnagel M, Rich PR, Zhang Q, Wiskich JT (1997) Substrate kinetics of the plant mitochondrial alternative oxidase and the effects of pyruvate. Plant Physiol 115:1145–1153
Israelsen WJ, Dayton TL, Davidson SM et al (2013) PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 155:397–409
Kim HJ, Triplett BA (2001) Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol 127:1361–1366
Kirkham MB, Gardner WR, Gerloff GC (1972) Regulation of cell division and cell enlargement by turgor pressure. Plant Physiol 49:961–962
Knowles VL, McHugh SG, Hu Z, Dennis DT, Miki BL, Plaxton WC (1998) Altered growth of transgenic tobacco lacking leaf cytosolic pyruvate kinase. Plant Physiol 116:45–51
Kumar S, Kumar K, Pandey P, Rajamani V, Padmalatha KV, Dhandapani G, Kanakachari M, Leelavathi S, Kumar PA, Reddy VS (2013) Glycoproteome of elongating cotton fiber cells. Mol Cell Proteomics 12:3677–3689
Laties GG (1974) Isolation of mitochondria from plant material. Method Enzymol 31:589–600
Lee JJ, Woodward AW, Chen ZJ (2007) Gene expression changes and early events in cotton fibre development. Ann Bot 100:1391–1401
Li XB, Fan XP, Wang XL, Cai L, Yang WC (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17:859–875
Li HB, Qin YM, Pang Y, Song WQ, Mei WQ, Zhu YX (2007) A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytol 175:462–471
Li XR, Wang L, Ruan YL (2010) Developmental and molecular physiological evidence for the role of phosphoenolpyruvate carboxylase in rapid cotton fibre elongation. J Exp Bot 61:287–295
Li F, Fan G, Lu C et al (2015) Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol 33:524–530
Ma L, Wang Y, Yan G, Wei S, Zhou D, Kuang M, Fang D, Xu S, Yang W (2016) Global analysis of the developmental dynamics of Gossypium hirsutum based on strand-specific transcriptome. Physiol Plantarum. doi:10.1111/ppl.12432
Maxwell DP, Wang Y, McIntosh L (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96:8271–8276
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Oliver SN, Lunn JE, Urbanczyk-Wochniak E, Lytovchenko A, van Dongen JT, Faix B, Schmälzlin E, Fernie AR, Geigenberger P (2008) Decreased expression of cytosolic pyruvate kinase in potato tubers leads to a decline in pyruvate resulting in an in vivo repression of the alternative oxidase. Plant Physiol 148:1640–1654
Pang CY, Wang H, Pang Y, Xu C, Jiao Y, Qin YM, Western TL, Yu SX, Zhu YX (2010) Comparative proteomics indicates that biosynthesis of pectic precursors is important for cotton fiber and Arabidopsis root hair elongation. Mol Cell Proteomics 9:2019–2033
Plaxton WC (1996) The organization and regulation of plant glycolysis. Ann Rev Plant Biol 47:185–214
Potikha TS, Collins CC, Johnson DI, Delmer DP, Levine A (1999) The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol 119:849–858
Qin YM, Zhu YX (2011) How cotton fibers elongate: a tale of linear cell-growth mode. Curr Opin Plant Biol 14:106–111
Qu J, Ye J, Geng YF, Sun YW, Gao SQ, Zhang BP, Chen W, Chua NH (2012) Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant Physiol 160:738–748
Rasmusson AG, Fernie AR, van Dongen JT (2009) Alternative oxidase: a defence against metabolic fluctuations? Physiol Plantarum 137:371–382
Ruan YL, Llewellyn DJ, Furbank RT (2001) The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansion. Plant Cell 13:47–60
Ruan YL, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15:952–964
Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18:651–664
Singer TP, Oestreicher G, Hogue P (1973) Regulation of succinate dehyrogenase in higher plants: I. some general characteristics of the membrane-bound enzyme. Plant Physiol 52:616–621
Smart LB, Vojdani F, Maeshima M, Wilkins TA (1998) Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated. Plant Physiol 116:1539–1549
Steffens B, Steffen-Heins A, Sauter M (2013) Reactive oxygen species mediate growth and death in submerged plants. Front Plant Sci 4:179
Tang GQ, Hardin SC, Dewey R, Huber SC (2003) A novel C-terminal proteolytic processing of cytosolic pyruvate kinase, its phosphorylation and degradation by the proteasome in developing soybean seeds. Plant J 34:77–93
Tang W, Tu L, Yang X, Tan J, Deng F, Hao J, Guo K, Lindsey K, Zhang X (2014) The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol 202:509–520
Thaker VS, Rabadia VS, Singh YD (1999) Physiological and biochemical changes associated with cotton fiber development. VII. Carbohydrate metabolism. Acta Physiol Plant 21:57–61
Turner WL, Knowles VL, Plaxton WC (2005) Cytosolic pyruvate kinase: subunit composition, activity, and amount in developing castor and soybean seeds, and biochemical characterization of the purified castor seed enzyme. Planta 222:1051–1062
Vander Heiden MG, Locasale JW, Swanson KD et al (2010) Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 329:1492–1499
Wang L, Li XR, Lian H, Ni DA, He YK, Chen XY, Ruan YL (2010) Evidence that high activity of vacuolar invertase is required for cotton fiber and Arabidopsis root elongation through osmotic dependent and independent pathways, respectively. Plant Physiol 154:744–756
Xue W, Wang Z, Du M, Liu Y, Liu JY (2013) Genome-wide analysis of small RNAs reveals eight fiber elongation-related and 257 novel microRNAs in elongating cotton fiber cells. BMC Genom 14:629
Yang YW, Bian SM, Yao Y, Liu JY (2008) Comparative proteomic analysis provides new insights into the fiber elongating process in cotton. J Proteome Res 7:4623–4637
Zhang B (2015) A genome-wide survey of glycolytic genes in diploid Asian cotton (Gossypium arboreum). Plant Gene 4:1–9
Zhang D, Zhang T, Guo W (2010) Effect of H2O2 on fiber initiation using fiber retardation initiation mutants in cotton (Gossypium hirsutum). J Plant Physiol 167:393–399
Zhang M, Zheng X, Song S et al (2011) Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fibre yield and quality. Nat Biotechnol 15:453–458
Zhang Y, Xiao W, Luo L, Pang J, Rong W, He C (2012) Downregulation of OsPK1, a cytosolic pyruvate kinase, by T-DNA insertion causes dwarfism and panicle enclosure in rice. Planta 235:25–38
Zhang B, Yang YW, Zhang Y, Liu JY (2013) A high-confidence reference dataset of differentially expressed proteins in elongating cotton fiber cells. Proteomics 13:1159–1163
Acknowledgments
We are grateful to Professor Yule Liu for the gift of the VIGS vector pTRV1 and pTRV2. We thank members of our laboratory for their helpful discussions. This study was supported by the State Key Basic Research and Development Plan (2010CB126003), the National Natural Science Foundation of China (90608016), the National Transgenic Animals and Plants Research Project (2009ZX08005-026B) and the China Postdoctoral Science Foundation (2014M550074).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2016_2557_MOESM1_ESM.docx
Figure S1. Phenotype of the elongating cotton fibers. Figure S2. Sequence alignment of GhPK6, GhPK26 and the encoding gene of the differentially expressed pyruvate kinase protein identified by 2-DE. Figure S3. Silencing of GhPK6 expression in cotton plants through VIGS. Figure S4. Phenotypes of transgenic Arabidopsis plants ectopically expressing GhPK6. Figure S5. Dynamic of succinate dehydrogenase and alternative oxidase activities in cotton fiber elongation process. Figure S6. A putative model for the GhPK6-mediated mechanistic links between cellular metabolism and growth control in cotton fibers. (DOCX 1812 kb)
425_2016_2557_MOESM5_ESM.xlsx
Table S4. Expression of the pyruvate kinase genes in different cotton tissues based on RNA sequencing data. (XLSX 17 kb)
Rights and permissions
About this article
Cite this article
Zhang, B., Liu, JY. Cotton cytosolic pyruvate kinase GhPK6 participates in fast fiber elongation regulation in a ROS-mediated manner. Planta 244, 915–926 (2016). https://doi.org/10.1007/s00425-016-2557-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2557-8