Abstract
Main conclusion
This is an original study focus on ERF gene response to alkaline stress. GsERF6 functions as transcription factor and significantly enhanced plant tolerance to bicarbonate (HCO 3 − ) in transgenic Arabidopsis .
Alkaline stress is one of the most harmful, but little studied environmental factors, which negatively affects plant growth, development and yield. The cause of alkaline stress is mainly due to the damaging consequence of high concentration of the bicarbonate ion, high-pH, and osmotic shock to plants. The AP2/ERF family genes encode plant-specific transcription factors involved in diverse environmental stresses. However, little is known about their physiological functions, especially in alkaline stress responses. In this study, we functionally characterized a novel ERF subfamily gene, GsERF6 from alkaline-tolerant wild soybean (Glycine soja). In wild soybean, GsERF6 was rapidly induced by NaHCO3 treatment, and its overexpression in Arabidopsis enhanced transgenic plant tolerance to NaHCO3 challenge. Interestingly, GsERF6 transgenic lines also displayed increased tolerance to KHCO3 treatment, but not to high pH stress, implicating that GsERF6 may participate specifically in bicarbonate stress responses. We also found that GsERF6 overexpression up-regulated the transcription levels of bicarbonate-stress-inducible genes such as NADP-ME, H +-Ppase and H +-ATPase, as well as downstream stress-tolerant genes such as RD29A, COR47 and KINI. GsERF6 overexpression and NaHCO3 stress also altered the expression patterns of plant hormone synthesis and hormone-responsive genes. Conjointly, our results suggested that GsERF6 is a positive regulator of plant alkaline stress by increasing bicarbonate ionic resistance specifically, providing a new insight into the regulation of gene expression under alkaline conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adverse environmental conditions, such as soil saline-alkalinity, drought, low temperature, and mineral deficiency severely affect crop yield and quality worldwide, resulting in great economic losses. Plants have developed complex mechanisms to deal with salt stress, such as the well-known SOS signaling pathway (Mahajan et al. 2008) and MAP kinase signal transduction (Nakagami et al. 2005). Comparatively little attention was paid to alkaline stress, and not much information was reported about how plants respond to alkaline stress, although it imposes much more severe damage than other stresses on plant growth and development (Yang et al. 2008).
Alkaline soils are usually classified by low availability of nutrients, high concentration of HCO3 − and CO3 2−, and high pH (Misra and Tyler 1999). Bicarbonate (HCO3 −) and carbonate (CO3 2−) are the principal contributors to alkalinity, whereas hydroxide, borate, ammonia, organic bases, phosphates, and silicates are considered as minor contributors (Petersen 1996). Many common commercial crops are reported to be severely affected by HCO3 − (Hajiboland et al. 2005; Valdez-Aguilar and Reed 2010; Rouphael et al. 2010). The bicarbonate also reduces the uptake and utilization of iron (Fe2+), leading to Fe deficiency and lime-induced leaf chlorosis (Chaney et al. 1992). Some early studies compared the effects of neutral salts (NaCl or Na2SO4) and alkaline salts (NaHCO3 or Na2CO3) on growth, photosynthesis, ion balance and osmotic adjustment in different species of plants such as barley (Yang et al. 2009), cotton (Chen et al. 2011), Chenopodium glaucum (Shasha 2012) and wheat (Guo et al. 2015), aiming to elucidate the mechanisms of alkali stress damage and the physiological adaptive mechanisms of plants to alkali stress. High concentration of NaCl (>300 mM) seriously delayed seed germination of C. glaucum, however, the inhibitory effect of NaHCO3 stress on radicle and hypocotyl elongation was greater than NaCl stress even at lower concentration (100 mM) (Shasha 2012). NaHCO3 stress also inhibits the absorption of some inorganic anions such as H2PO4 −, Cl− and NO3 −, which greatly decreases the selective absorption ratio of K+/Na+, and breaks the ionic balance (Yang et al. 2007). Organic acids and SO4 2− might play important roles to maintain intracellular ion balance and pH stabilization under alkali stress, while, inorganic ions are major contributors to ion balance under salt stress (Yang et al. 2009). Moreover, the seedlings exposed to NaHCO3 generated higher levels of reactive oxygen species (ROS, O2 −, H2O2), and showed increased activities of antioxidant enzymes (SOD, POX) even at lower concentration of NaHCO3 (Shasha 2012). All of these studies suggest the alkaline salts could generate more severe effects on plants than the neutral salts, and the researchers speculate it might be due to the additive influence of high pH. Until now, most studies about alkaline stress have focused on the physiological impact on plants, while its molecular mechanism is rarely reported.
Thus, understanding the molecular and physiological mechanism of plant responses to alkaline stress and mining key regulatory genes in the stress responses are very important for crop improvement. As a typical salt and alkaline resistant species, wild soybean Glycine soja G07256 was used as a model plant in this study. We have reported a comprehensive and pioneer transcriptome analysis of wild soybean root given by salt and alkaline challenges using Affymetrix® Soybean GeneChip® (Ge et al. 2010), and functionally characterized some key regulatory genes in alkaline stress responses, such as GsCBRLK and GsTIFY10 (Yang et al. 2010; Zhu et al. 2011). Using high throughput RNA sequencing technique, we sequenced the transcriptome of G. soja roots under NaHCO3 stress on the Illumina Genome Analyzer IIx (GAIIx) platform (DuanMu et al. 2015), providing a broad description of gene expression pattern and gene transcriptional dynamics under alkaline stress in G. soja. One of the intriguing candidates is an AP2/ERF transcription factor (TF) which was highly induced by NaHCO3 and may be a putative alkali-responsive gene.
The AP2/ERF family is a large and plant-specific transcription factor family. It can be further classified into four major subfamilies (AP2, DREB, ERF and RAV) based on the similarity of their conserved domains. AP2 subfamily members contain double AP2/ERF domains, and RAV subfamily members contain one AP2/ERF domain and an additional B3 DNA-binding domain, while DREB and ERF subfamily members contain only one single AP2/ERF domain (Sakuma et al. 2002). The DREB and ERF subfamilies can be divided into six subgroups, A-1 to A-6 and B-1 to B-6 (Nakano et al. 2006), respectively. In Arabidopsis, an ERF subfamily gene RAP2.2 can be induced by hypoxia in an ethylene dependent manner, and overexpression or knockout of RAP2.2 resulted in improving or impairing the survival of plants under hypoxic stress, respectively, and RAP2.2 and its downstream genes have a transcriptional cascade and play an important role under hypoxic stress or submergence conditions (Hinz et al. 2010; Bui et al. 2015). A recent study about ERF6 protein in Arabidopsis has shown that ERF6 can modulate oxidative gene expression by interacting with MPK6 (Wang et al. 2013). The expression of ERF6 can be rapidly induced by pathogen, SA, ROS and cold stress but suppressed by water deficit, heat and ABA (Wang et al. 2013). These data indicate that ERF6 has important roles and is required in biotic and abiotic stress signaling during plant growth and development.
In recent years, the identification of some soybean ERF genes and their roles in abiotic stress tolerance has been attempted. A member of ERF family from Glycine max has been reported to function as a transcriptional activator of downstream defense genes in both soybean and Arabidopsis, and mediates the expression of defense-related genes regulated by ethylene (ET), jasmonate (JA) and salicylic acid (SA) (Mazarei et al. 2007). Moreover, GmERF3 also functions as a transcriptional activator activating the expression of reporter genes and is able to bind with both the GCC box and DRE/CRT motifs in yeast cells. Overexpression of GmERF3 can influence plant resistance to both biotic and abiotic stresses such as bacterial, fungal, viral pathogens, salt and drought stresses (Zhang et al. 2009). Similarly, expression of GmERF4 could be induced by both biotic stimuli such as ethylene, JA, SA, soybean mosaic virus and abiotic stresses such as cold, salt and drought, but repressed by ABA. Constitutive expression of GmERF4 in tobacco increased tolerance to salt and drought stresses but did not exhibit detectable tolerance to bacterial infection (Zhang et al. 2010).
However, little is known about responses of ERF genes to alkaline stress. In the present study, we isolated a putative AP2/ERF gene, GsERF6 from wild soybean. Overexpression of GsERF6 in Arabidopsis conferred deliberately enhanced tolerance to alkali stress, and up-regulated the expression levels of stress-responsive marker genes, and altered the expression levels of biosyntheses and responsive genes of endogenous hormones like ABA, ET and JA. Further analysis showed that GsERF6 overexpression in Arabidopsis displayed particular response to HCO3 − but not high pH stress, suggesting that GsERF6 may play special roles in response to HCO3 − stress. Subcellular localization and transcriptional activation studies showed that GsERF6 is a nuclear protein with transcriptional activation. All the data presented here illustrate the important role of GsERF6 as a regulator of alkali stress tolerance in plants.
Materials and methods
Plant material and stress treatment
The wild soybean line (G07256) was collected from Baicheng in Jilin Province, China as described (Ge et al. 2010). To analyze the expression pattern of GsERF6 under alkaline stress, G. soja G07256 seedlings were grown in 1/4 strength Hoagland’s solution in a culture room. The environmental conditions are as following: 24 °C, 60 % relative humidity and a light regime of 16 h light/8 h dark. The seeds were first shaken in 98 % sulfuric acid for 10–15 min to break dormancy, and then washed with sterile water for at least five times before being placed on wet filter papers in petri dishes in the dark for germination. After 2 days, germinating seeds were then transferred into the growth boxes containing 1/4 strength Hoagland’s solution. The nutrient solution was changed every 3 days. The 21-day seedlings were moved into 1/4 strength Hoagland’s solution with 50 mM NaHCO3 (pH 8.5). Equal amounts of roots were sampled at seven different time points: 0, 1, 3, 6, 9, 12 and 24 h for RNA extraction and qRT-PCR analysis. The root samples were snap frozen in liquid N2 and stored at −80 °C.
Arabidopsis thaliana ecotype Columbia (Col-0) seeds obtained from Nottingham Arabidopsis Stock Centre (NASC) were grown in the chamber at the following conditions: 21–23 °C, 60 % relative humidity, 100 μmol photons m−2 s−1, 16 h light/8 h dark cycles. To analyze the expression of alkali stress-related genes in wild type (WT) and GsERF6 overexpression (OX) lines, the seeds were germinated and grown on 1/2 MS agar plates for 12 days, and then the Arabidopsis plants were moved to the filter paper saturated with normal 1/2 MS solution (control) or 1/2 MS solution with 50 mM NaHCO3. Samples were collected at 0, 6 and 12 h, respectively, after treatment.
RNA isolation, cDNA synthesis and quantitative real-time PCR
Total RNA extractions were done using RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China) and cDNA syntheses were performed by SuperScript™ III Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA) with Oligo(dT)18 reverse primer. qRT-PCR analyses were performed using SYBR Premix ExTaq™ II Mix (TaKaRa, Shiga, Japan). GAPDH (glyceraldehyde-3-phosphate dehydrogenase, accession no. DQ355800) was used as reference in G. soja. Actin2 (ACTIN2) was used as reference in Arabidopsis. The expression levels of all candidate genes were analyzed by the 2−ΔΔCT method. The primers used for qRT PCR were listed in Supplementary Table S1.
GsERF6 gene isolation and sequence analysis
GsERF6 was cloned by homology-based cloning method. GsERF6 was amplified by PCR primer pairs 5′-GATGGCTAACGCTGCTGAAGT-3′ and 5′-TTCGTCAAATGTACAATGTACTCATC-3′. The PCR products were inserted into pEASY-Blunt Simple Cloning vector (Transgen, Beijing, China) and sent for sequencing. Phylogenetic analysis was performed using MEGA 5.0. The NJ tree was constructed by AP2/ERF homologs of G. soja, Selaginella moellendorffii, Physcomitrella patens and Arabidopsis based on the JTT model. Peptide sequence information was obtained from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html). Sequence alignments were performed with ClustalX.
Protein subcellular localization analysis
To analyze subcellular localization of GsERF6, we inserted GsERF6 gene in the 5′ of eGFP gene in pBSKII-eGFP vector to express GsERF6-eGFP fusion protein in plant cells. The resultant pBSKII-GsERF6-eGFP or the empty plasmid were transformed into onion epidermis cells separately by the particle bombardment method as described (Luo et al. 2013). The expression and localization of fluorescent proteins in the epidermis cells were imaged using a confocal laser-scanning microscope (SP5, Leica, Wetzlar, Germany).
In vitro transcriptional activation assay
GsERF6 and AtDREB1A were inserted into pGBKT7-BD vector. The resulted pGBKT7-GsERF6, pGBKT7-AtDREB1A or the empty plasmid was transformed into yeast strain AH109, respectively. The transformants were selected on solid SD/-Trp medium at 30 °C for 3 days. The grown cells from independent colonies were re-streaked on SD/-Trp medium for another 2 days, and then were collected for measuring β-galactosidase activity by colony-lift filter assay. At the same time, the independent colonies were picked and grown on SD/-Trp-His medium for another 3 days to evaluate the expression of HIS3 reporter gene. Transcriptional activation was analyzed according to the methods described on Yeast Protocols Handbook (Clontech).
Histochemical GUS assay
To investigate GsERF6 gene expression pattern in planta, approximately 2.5 kb of its promoter (−2572 to −14 upstream of the translation initiation codon) was amplified by PCR from wild soybean genomic DNA. The PCR product was inserted into the pCAMBIA 3301 vector between EcoRI and BglII sites in the upstream of GUS gene. The resulted construct or the empty vector was transformed into Agrobacterium tumefaciens strain LBA4404 and then into Arabidopsis thaliana by floral dip method as previously described (Clough and Bent 1998). Fifteen glyphosate-resistant transgenic (T1) plants were obtained and four independent T3 lines were analyzed.
For histologic analysis, the Arabidopsis seedlings were grown on 1/2 MS plates and soil separately. On the 2nd and 8th day after germination, three to four seedlings from each line on 1/2 MS plates were sampled and applied to GUS staining. On the other hand, the flowers and seed pods from 45-day-old plants grown on soil were sampled, and then subjected to GUS staining. GUS histochemical staining was performed using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) as substrate (Thomine et al. 2003).
To determine the GUS expression pattern in the transgenic Arabidopsis plants in response to alkali stress, the seeds from four independent transgenic lines were germinated on 1/2 MS medium for 7 days, and then transferred to filter papers soaked with 1/2 MS liquid medium containing 50 mM NaHCO3 for 6 or 12 h before GUS staining. Plants or tissues were then visualized using a stereo microscope (Olympus, Tokyo, Japan).
Vector construction and generation of transgenic Arabidopsis thaliana
The full coding region of GsERF6 was amplified from pEASY-GsERF6 with SmaI and XbaI linker primers 5′-TGACCCGGGATGGCTAACGCTGCTG-3′ and 5′-TGCTCTAGATCACACAGCCACGAGCGGT-3′, and then was cloned into the plant expression binary vector pCAMBIA2300 to generate pCAMBIA2300-GsERF6. The recombinant vector was transformed to Agrobaterium tumefaciens strain LBA4404 and then the targeted gene was transferred into Arabidopsis plants by floral dip method. The transgenic seedlings were selected on 1/2 MS medium plate containing 50 mg L−1 kanamycin. The positive plants were transferred into soil and the mature seeds were individually harvested. The T2 seeds were selected again on kanamycin-containing 1/2 MS medium and the homozygous plants were determined and identified by genotyping the T2 seedlings on selective medium. The gene expression levels were confirmed by RT-PCR analysis. The primers used for RT-PCR were listed in Supplementary Table S2.
Phenotypic analysis of transgenic Arabidopsis plants
The Arabidopsis seeds were firstly sterilized in 5 % NaClO solution for 5 min and washed with sterilized water for 5 times, and then were transferred onto wet filter paper and stored at 4 °C in the dark for 3–7 days. The plants were grown in a tissue culture room or a chamber under controlled environmental conditions of 21–23 °C, 100 μmol photons m−2 s−1, 60 % relative humidity and 16/8 h day-night cycles.
To analyze germination rates, the seeds of WT and GsERF6 OX lines were sown on 1/2 MS agar plates supplemented with 0, 6 or 8 mM NaHCO3, or 6 or 7 mM KHCO3, or 1 M KOH to adjust the medium pH at 7.5–8.2. The germination rates were recorded for 6 consecutive days after sowing. Pictures were taken to show the growth performance of each line, followed by measurement of the opening/greening of the leaves. For each experiment, ninety seeds were used and all experiments were generated at least three replications.
To characterize stress tolerance at the early seedling stage, WT and OX Arabidopsis seeds were germinated and grown on 1/2 MS medium for 7 days, and then were transferred to plates with vertical position containing 1/2 MS agar in the absence or presence of 6 mM NaHCO3, or 5–7 mM KHCO3, or at pH 7.5–8.2 (pH was adjusted with 1 M KOH) for 7–10 days before photographed. To analyze stress tolerance at adult stage, the seedlings were grown in pots filled with the mixture of vermiculite:peat moss:garden soil (volume ratio, 1:1:1). The plants were irrigated with 100 mM NaHCO3 solution every 4 days for a total of 16 days. The photos were taken at different stages of growth and development.
Statistical analysis
All experiments with each group were performed at least in triplicate. Data were reported as mean ± SD. Data were analyzed statistically by Duncan’s multiple range tests or Student’s t test. Results were considered statistically significant when P < 0.05.
Results
Isolation and sequence analysis of GsERF6
AP2/ERF TFs demonstrate to be related to many regulation processes when plants respond to various environmental stresses. In this study, GsERF6 was isolated from G. soja by homology-based cloning method according to the cDNA sequence of Glyma05g05130 from G. max. GsERF6 contains a complete open reading frame (ORF) of 837 bp which is 99 % identical to Glyma05g05130 based on the genome database from Phytozome.
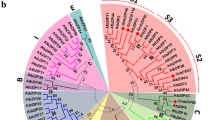
AP2/ERF family TFs are specifically evolved in plants and the evolutionary process of AP2/ERF proteins can be traced back to the lowest plant green alga Chlamydomonas reinhardtii (Shigyo et al. 2006). To know the phylogenetic relationship and classification of GsERF6, phylogenetic analysis was performed using MEGA 5.0 software based on GsERF6 sequence and other AP2/ERF proteins from Physcomitrella patens, Selaginella moellendorffii and Arabidopsis which represent mosses, lycophytes and angiosperms, respectively. GsERF6 is clustered to the B-3 subgroup of ERF subfamily and is closely related to AT5G47230 (AtERF5) in the phylogenetic tree as shown in Supplementary Fig. S1.
GsERF6 contains one AP2 domain which is approximately 60 amino acids from 129 to 187 aa. Its AP2 domain is highly conserved compared with the other plant ERF proteins (Supplementary Fig. S2). Further structural analysis of GsERF6 protein showed that the conserved AP2 domain contains a three-strand β-sheet which is very important to recognize the target sequence and a C-terminal α-helix, which is similar with the structure of Arabidopsis ERF1 protein (Allen et al. 1998). This suggests that GsERF6 has ability to bind with ERE-like (ethylene response element–like) DNA sequences.
GsERF6 is a nuclear protein with transactivation activity
Transcription factors are a kind of proteins functioning as activators or repressors to regulate gene expression or transcription. To investigate the transcription activity of GsERF6 protein, we need firstly determine its subcellular localization. GsERF6 coding sequence without stop codon was fused at the 5′ end of eGFP (Fig. 1a) and the resultant construct was transformed into plant cells. The results of confocal imaging showed that GsERF6-eGFP fusion protein was localized in the nuclei of onion epidermal cells by a transient expression assay (Fig. 1b). As a control, eGFP protein alone was found in both the nucleus and cytoplasm. The nuclear localization of GsERF6 suggested that this protein may play a role as a transcription factor.
Subcellular localization of the GsERF6 protein. a Schematic representation of constructs used for subcellular localization analysis of GsERF6. b Images of onion epidermis cells expressing eGFP only (upper lane) and GsERF6-eGFP fusion protein (bottom lane). The subcellular localization was examined by confocal microscopy under fluorescent-field illumination, bright-field illumination and an overlay of bright and fluorescent illumination, respectively
The transcriptional activation activity of GsERF6 was investigated in yeast cells. First GsERF6 was inserted into pGBKT7-BD vector to fuse with GAL4 DNA-binding domain coding sequence (Fig. 2a) and then transformed to yeast AH109 strain with positive and negative controls. The colonies carrying the relevant plasmids were selected on SD/-Trp plates. The randomly selected colonies were used to measure the transcription of LacZ reporter gene using the colony-lift filter assay to test activation ability of GsERF6. As shown in Fig. 2b, the positive control exhibited strong β-galactosidase activity but the negative control did not. The transformants carrying pGBKT7-GsERF6 showed a little lower β-galactosidase activity than the positive but much stronger than the negative (Fig. 2b), indicating that GsERF6 may have transcriptional activation ability in yeast cells. Furthermore, we dropped a serial of diluted yeast cell suspensions of representative colonies from SD/-Trp medium onto SD/-Trp-His medium to see if GsERF6 can activate HIS3 reporter gene. As shown in Fig. 2c, all yeast cell lines grew similarly on SD/-Trp medium, but differently on SD/-Trp-His medium. The cells carrying GsERF6 grew almost as well as the positive control cells but the negative cells had no growth (Fig. 2c), suggesting GsERF6 can also activate HIS3 reporter gene as well. All of these data indicated that GsERF6 may be a putative transcription factor possessing transcriptional activation activity.
Transcription activation activity of GsERF6 in yeast. a Schematic representation of constructs used for transactivation activity analysis of GsERF6. b, c Transcriptional activation analysis of GsERF6 in yeast cells. pGBKT7-AtDREB1A and pGBKT7 were used as positive and negative controls, respectively
Spatial expression pattern of GsERF6
To investigate the spatial expression of endogenous GsERF6 in G. soja 07256, qRT-PCR was performed to analyze GsERF6 expression in different tissues including young leaves, old leaves, stems, root tips, hypocotyl and seed coats from mature G. soja 07256 seedlings. The qRT-PCR data showed GsERF6 was widely expressed in all tissues, but highly expressed in hypocotyls and roots with more than twofold in leaves (Fig. 3a).
Spatial expression pattern analysis of GsERF6. a Tissue-specific expression patterns of GsERF6 were examined in young leaf, old leaf, stem, flower, seed coat, hypocotyl and root from G. soja. Data were analyzed by Duncan’s multiple range tests. The mean value from three fully independent biological repeats and three technical repeats is shown. b–l Histochemical analyses for GUS expression in Arabidopsis thaliana transgenic plants under the control of GsERF6 promoter. b Control Arabidopsis with six leaves. c, d Young plants (2 and 10 days after germination). e Inflorescence. f Sepal. g Petal. h Cauline leaf below flowers. i Stamen. j Immature siliques. k–l Funiculus and micropilar endosperm. Scale bars 2 mm
To further determine GsERF6 promoter activity, we imaged representative GsERF6pro-driven GUS spatial expression patterns in transgenic Arabidopsis plants. The GUS activity was observed in all tissues, including root, leaf, stem, flowers and immature siliques (Fig. 3c–l). In most tissues, strong GUS expression was monitored in vasculatures, especially in young tissues. While, in young seedlings, the expression was mainly in the vascular bundles of leaves, cotyledons, hypocotyls and in the central cylinder of primary and secondary roots, but not found in root tips (Fig. 3c, d). In flowers, the staining was mainly observed in vascular bundles of sepal, petal, stigma and filament (Fig. 3e–i). Young siliques had more GUS expression than mature ones. In mature siliques (Fig. 3k, l), GUS activity decreased significantly but majorly expressed at hilum, where the only nutrition transport corridor from vegetative organs to seeds is. The tissue localization results indicated that GsERF6 may play important roles during the process of nutrient and mineral acquisition, absorption and transportation in the plant.
Expression of GsERF6 under NaHCO3 treatment
Our high throughput RNA seq data indicated that GsERF6 was highly induced in the roots of alkali-stressed G. soja (DuanMu et al. 2015). To determine the expression pattern of GsERF6 in response to alkali stimuli, we extracted total RNA from G. soja 07256 roots and used RT-PCR to measure GsERF6 expression levels from different treatments. Under alkali stress, GsERF6 transcription level was first reduced slightly at the 1 h point after treatment, and then increased rapidly and peaked at 6 h with a fivefold increase, and finally, the expression of GsERF6 came down to the normal level at 24 h (Fig. 4a). Nevertheless, the temporal GsERF6 promoter-driven GUS activity in Arabidopsis also showed dynamic change within the period of the treatment (Fig. 4b). Collectively, GsERF6 might be an important regulatory gene in alkaline-response pathway.
Temporal expression patterns of GsERF6 under alkaline stress. a Expression levels of GsERF6 were up-regulated by alkali stress in G. soja roots. Total RNA was extracted at the indicated time points from roots of 21-day-old G. soja seedlings whose roots were submerged in nutrient solution with 50 mM NaHCO3. Untreated plants were used as controls. Relative transcript levels were determined by qRT-PCR using GAPDH as an internal control. The mean value from three fully independent biological repeats and three technical repeats is shown. b GUS expression in transgenic Arabidopsis under 50 mM NaHCO3 treatment. The GUS activity is controlled by GsERF6 promoter
We then analyzed the upstream sequence of GsERF6 by a MEME search. The result revealed that some stress- and defense-related elements, such as HSEs (position −258 to −267 and −1592 to −1601), LTR (positions −1281 to −1286), MBSs (position −386 to −391), TC-rich repeats (position −412 to −421, −673 to −682 and −952 to 961) and WUN-motif (positions −600 to −609) were enriched, implicating that GsERF6 is a stress-related gene. However, no other well-known motif was found, suggesting that there may be unknown cis-element(s) in the promoter leading to alkali-inducible transcription of GsERF6.
Overexpression of GsERF6 enhanced plant NaHCO3 tolerance
To determine the regulatory role of GsERF6 under NaHCO3 treatment in planta, we generated transgenic Arabidopsis lines carrying CaMV35S-driven GsERF6 constitutive overexpression (OX) cassette. Two homozygous T3 OX Arabidopsis lines (#7, #19) with relatively high GsERF6 expression level were selected for further analysis. Compared with the untransformed wild type (WT) plants, the increased transcription levels of GsERF6 in the OX lines were confirmed by semi-quantitative RT-PCR analysis (Fig. 5a). Meanwhile, the transcription levels of GsERF6 in OX lines were higher than those of its homologs AtERF5 and AtERF6 measured by semi-quantitative RT-PCR (Supplementary Fig. S3), indicating that the expression of GsERF6 is high enough to have an effect on regulating genes in Arabidopsis.
Overexpression of GsERF6 in Arabidopsis enhanced the alkaline tolerance. a The transcript levels of GsERF6 in the WT and OX lines was analyzed by semi-quantitative RT-PCR, the Actin2 gene was used as an internal standard. b, c The growth and germination rate of WT and OX seedlings on 1/2 MS medium with 6 and 8 mM NaHCO3. Photographs were taken 12 days after germination. d Quantitative evaluation of leaf opening and greening rate at the seed germination stage. Experiments were performed at least three times. The bars represent standard errors. The data show means (±SE) of three replicates (each with 100 seeds for each line). *P < 0.05, **P < 0.01 by Student’s t test. e, f Phenotypes and measurements of primary root lengths of WT and OX seedlings under normal and alkali stress. Seven-day-old seedlings grown on 1/2 MS were transferred to fresh solid agar plates supplemented with or without 6 mM NaHCO3. Photographs were taken after 10 days growth on the supplemented media. All values are means (±SE) from three independent experiments (30 seedlings per experiment). *P < 0.05, **P < 0.01 by Student’s t test
In the absence of NaHCO3, GsERF6 OX plants displayed similar performance with the WT plants, showing no difference in germination and development under normal conditions (Fig. 5b). In the presence of 6 mM NaHCO3 (pH 7.25 ± 0.05) or 8 mM NaHCO3 (7.70 ± 0.05), both the WT and OX lines exhibited very different initial but similar final germination rates up to 100 % (Fig. 5b, c). However, 12 days after germination, the OX lines had much higher percentages of seedlings with open and green leaves than the WT plants under various concentrations of NaHCO3 (Fig. 5b, d). For example, in the presence of 6 mM NaHCO3, the WT seedlings had approximately 30 % individuals with fully open cotyledons while GsERF6 OX seedlings had approximately 75 %. When NaHCO3 concentration increased to 8 mM, the percentage of the WT seedlings with fully open cotyledons was <5 % compared with approximately 20 % for the GsERF6 OX seedlings (Fig. 5d). We further investigated the effect of alkaline stress on root growth. As shown in Fig. 5e, f, when grown on solid medium supplemented with 6 mM NaHCO3, the leaf growth and root elongation of the WT plants were inhibited more severely than those of OX plants.
During the late growing and developmental stages, 4-weeks old OX lines grown in soils irrigated with 100 mM NaHCO3 (pH 8.2) for 16 days showed greater development ability than the WT plants (Fig. 6a). Under normal conditions, the WT plants and OX lines showed similar growth, while OX lines showed significant alkali stress tolerance with higher survival rate and greater height than WT plants after NaHCO3 treatment (Fig. 6a–c). Then we measured the total chlorophyll contents in the plants growing in pots after 100 mM NaHCO3 treatment. The contents of total chlorophyll were decreased in both WT and transgenic plants, but GsERF6 OX plants decolor less than the WT (Fig. 6d). These results indicate that GsERF6 overexpression alleviates the effects of high-alkali stress on chlorophyll deformation and enhances the alkaline tolerance of transgenic plants. It is generally accepted that the level of malondialdehyde (MDA) produced during peroxidation of membrane lipids is often used as an indicator of oxidative damage (Weber et al. 2004; Kotchoni and Gachomo 2006). Therefore, we measured the MDA contents in the transgenic and WT plants under both alkali stress and normal conditions, and found that MDA level in WT plants was significantly higher than that in GsERF6 OX lines (Fig. 6e). Together, these data indicate that overexpression of GsERF6 enhanced NaHCO3-stress tolerance in Arabidopsis.
Enhanced tolerance of transgenic plants to alkaline stress at the adult stage. a Phenotypes of WT and OX plants in response to alkali stress. b The survival rate of WT and OX plants under alkali stress. c The plant height of WT and OX plants. d The total chlorophyll content of WT and OX plants. e The total malondialdehyde (MDA) content of WT and OX plants. For the alkaline tolerance at the adult stage, 4-week-old plants were irrigated with 100 mM NaHCO3 solution every 4 days for a total of 16 days. Photos were taken on the 16th day after initial alkali treatment. All values are means (±SE) from three independent experiments (40 seedlings per experiment). *P < 0.05, **P < 0.01 by Student’s t test
Overexpression of GsERF6 specifically increased bicarbonate tolerance but not high pH tolerance
As HCO3 − ion and high pH resulted from NaHCO3 solution are the main contributors to alkali stress, we next examined the tolerating function of GsERF6 in response to HCO3 − ion and/or pure high pH stress.
To explore the role of GsERF6 on pure high pH stress, we analyzed plant tolerance under a range of pH from 5.8 to 8.2 at germinating and seedling stages. We grew Arabidopsis seedlings on the standard medium (pH 5.8) for 7 days and then transferred the seedlings to 1/2MS medium at pH 5.8 (control), or pH 7.5 or pH 8.2 (adjusted by KOH). We can see both GsERF6 OX and WT had 100 % seed germination rates and similar growing and developmental performances (Supplementary Fig. S4a). The results from vertical growing plants showed that the primary roots elongation of both OX and WT plants were obviously inhibited under high pH stress, but also no significant difference between GsERF6 OX and WT plants (Supplementary Fig. S4b, c). Our data indicated that GsERF6 had no contribution to the single high pH stress.
Then we further characterized the function of GsERF6 under HCO3 − stress. The existence of HCO3 − gives double injuries including bicarbonate ion and high pH stress caused by bicarbonate ion. For HCO3 − stress, here we used KHCO3 instead of NaCO3 because potassium ion (K+) is less toxic than sodium ion (Na+). As shown in Fig. 7, similar with the phenotype under NaHCO3 stress conditions, the OX plants had higher survival rate with more open and green leaves than the WT under 6 mM (pH 7.35 ± 0.05) and 7 mM KHCO3 (pH 7.65 ± 0.05) treatments (Fig. 7a, b), and the OX plants were more tolerant with better growth and longer root length at seedling stage (Fig. 7c, d).
Overexpression of GsERF6 in Arabidopsis enhanced KHCO3 tolerance. a The growth of WT and OX seedlings on 1/2 MS medium with 6 and 7 mM KHCO3. Photographs were taken 12 days after germination. b Quantitative evaluation of leaf opening and greening rate. Experiments were performed at least three times. The bars represent standard errors. The data show means (±SE) of three replicates (each with 100 seeds for each line). *P < 0.05, **P < 0.01 by Student’s t test. c, d Phenotypes and measurements of primary root length of WT and OX seedlings under normal and KHCO3 stress. Seven-day-old seedlings grown on 1/2 MS were transferred to new solid agar plates supplemented with and without 5, 6 and 7 mM KHCO3. Photographs were taken after 7 days growth on the supplemented media. All values are means (±SE) from three independent experiments (30 seedlings per experiment). *P < 0.05, **P < 0.01 by Student’s t test
GsERF6 overexpression altered expression patterns of stress-responsive genes
To explore the mechanism of the increased bicarbonate tolerance conferred by GsERF6 overexpression, the transcription levels of some alkaline stress-related genes were analyzed under 50 mM NaHCO3 treatment. Biochemically, NADP-ME, H+-Ppase and H+-ATase have been shown the ability to adjust the pH in the cytoplasm (Marrè and Ballarin-Denti 1985; Kurtz 1987). Therefore we selected NADP-ME, H +-Ppase and H +-ATase as marker genes under NaHCO3 stress. After qRT-PCR analysis, we found that the expression levels of these three genes were significantly induced by NaHCO3 stress at 6 and 12 h time points. Both WT and GsERF6 OX plants showed similar expression profiles (Fig. 8). However, the expression levels were significantly higher in transgenic plants than those in WT at 6 and 12 h (Fig. 8), suggesting that GsERF6 positively and directly or indirectly regulated the expression of NADP-ME, H +-Ppase and H +-ATPase genes. Furthermore, we examined the effects of GsERF6 on the transcription levels of several other stress-responsive marker genes including COR47, RD29A and KIN1. Real-time PCR results showed that in the presence of 50 mM NaHCO3 treatment, the expression of all these genes was up-regulated in WT and GsERF6 OX lines. However, their expression levels were significantly higher in OX lines than WT (Fig. 8). Therefore, we inferred that GsERF6 might promote the bicarbonate defense by regulating expression of the stress-inducible marker genes.
Expression patterns of stress-induced marker genes in WT and GsERF6 transgenic Arabidopsis seedlings in response to alkali stress. 2-week-old WT and OX seedlings were treated with 1/2 MS solution containing 50 mM NaHCO3 for 0, 6 and 12 h. The induction of stress-responsive genes NADP-ME, H +-ATPase, H +-Ppase, RD29A, COR47 and KIN1 were measured by qRT-PCR analysis. Expression of Actin2 was used as an internal control. Values represent the means of three biological replicates and three technology replicates for each. *P < 0.05, **P < 0.01 by Student’s t test
GsERF6 overexpression altered expression patterns of plant hormone synthesis and hormone-responsive genes
Previous studies have suggested that ERF transcription factors can affect the endogenous hormone levels and plant responses to the hormones such as abscisic acid (ABA), ET and JA (Cheng et al. 2013; Lee et al. 2015; Wang et al. 2015; Muller and Munne-Bosch 2015). These hormones are important for plants to adapt to the changeable environments and stress conditions. Therefore, in this study, we further examined the potential role of GsERF6 to affect the hormone levels and the signal responses by analyzing the expression patterns of some hormone synthesis and responsive marker genes. It is well known that the endogenous ABA levels can be indirectly evaluated by analyzing ABA biosynthesis and catabolism in plant cells (Nambara and Marion-Poll 2005). 9-cis-epoxycarotenoid dioxygenase (NCED) genes encode key enzymes for ABA biosynthesis (Dong et al. 2015). Here, we found that these two ABA biosynthesis-related genes, AtNCED3 and AtNCED5, were both up-regulated under NaHCO3-stress treatment, and their increasing extents in GsERF6 OX plants are much higher than those in WT plants (Fig. 9). In accordance with this observation, the expression levels of some ABA-responsive genes such as RAB18 and RD29B were also significantly higher in GsERF6 transgenic plants than in WT even at 12 h after treatment (Fig. 9). We also performed the analyses of ET-synthesis marker genes, AtACS1, AtACS2 and AtACS5, and found all were induced under NaHCO3 treatment and the increasing fold in GsERF6 OX plants was much higher than that in transgenic plants (Fig. 9). The analyses of ET and JA-inducible genes such as EIN3, ERF1 and PDF1.2 revealed that EIN3 and ERF1 were induced under NaHCO3 stress. Intriguingly, the expression level of EIN3 was higher in transgenic plants but the expression level of ERF1 was lower in transgenic plants than in the WT plants (Fig. 9). In contrast, the expression levels of ERF1-target genes such as P5CS and PDF1.2 were not significantly lower in transgenic plants. P5CS showed similar expression level compared with WT but PDF1.2 showed higher expression level in transgenic plants. The similar or higher expression of ERF1-target genes is possibly due to the highly induced levels of hormones such as ABA and JA because P5CS can also be induced by ABA and PDF1.2 can be induced by JA (Cheng et al. 2013). The expression levels of GsERF6 homologs, AtERF5 and AtERF6, were also examined under NaCO3 stress in WT and transgenic plants, and we found both of them were induced under the stress and had higher expression in transgenic plants. Taken together, alkaline (NaHCO3) stress could induce ABA and ET synthesis and strengthen the hormone responses during adaptation to alkaline (NaHCO3) stress. The varying expression levels of these marker genes are possibly due to the regulation of GsERF6 across the hormones transduction and feedback, and the further analyses are required to reveal its precise roles in the hormone signaling pathway.
Expression patterns of hormone synthesis and responsive marker genes in WT and GsERF6 transgenic Arabidopsis seedlings under alkali stress. 2-week-old WT and OX seedlings were treated with 1/2 MS solution containing 50 mM NaHCO3 for 0, 6 and 12 h. The expression levels of the marker genes were measured by qRT-PCR analysis. Expression of Actin2 was used as an internal control. Values represent the means of three biological replicates and three technology replicates for each. *P < 0.05, **P < 0.01 by Student’s t test
Discussion
In plants, tolerance or susceptibility to alkaline stress is a coordinated action involved in various genes such as calcium-binding proteins and other components in stress signaling pathways. These components may have cross talk with each other, however little is known on this aspect. Based on the transcriptome sequencing data of wild soybean G07256 roots, 320 TFs belonging to 32 different families were found to be induced during NaHCO3 treatment (DuanMu et al. 2015). Among them, the ERF TFs were shown to be involved in responses to environmental stresses and exhibited multiple, complex and flexible expression patterns (Xu et al. 2011). In this study, we focused on a novel AP2/ERF family gene GsERF6, which was identified as a key gene responding to alkaline stress based on the transcriptome sequencing data.
The AP2/ERF family is a large TF family specifically in plants and the members share a highly-conserved DNA-binding domain (Mizoi et al. 2012). Cluster analysis and phylogenic tree showed that GsERF6 was an ERF TF with one conserved AP2 domain and belonged to ERF superfamily B-3 subgroup (Supplementary Fig. S1). The AP2 domain of GsERF6 is located in the middle region and shows significant sequence homology with those of the other ERF proteins, but the sequences outside AP2 domain are quite different. Subcellular localization showed that GsERF6 was a nuclear protein (Fig. 1). Previous studies showed that ERFs usually function as activators or repressors of downstream genes during multiple signals pathways (Shinshi 2008). Our transactivation assays in yeast cells indicate that GsERF6 had transactivation activity to activated two reporter genes (Fig. 2). These studies suggested that GsERF6 can putatively function as a TF and have a role in the activation of downstream target genes in plants.
Various TFs show quite different responses to abiotic stresses in plants (Tran et al. 2004; Liu et al. 2007a). Here, we demonstrated that the expression of GsERF6 displays dynamic changes during NaHCO3 stress in wild soybean. Consistent with the RNA-seq data, an increase in GsERF6 expression in G. soja roots occurred when the plants were exposed to 50 mM NaHCO3 (Fig. 4a). This is consistent with the previous findings that other alkali-regulated genes in plants are also regulated at the transcriptional level (Zhu et al. 2011, 2012; Liu et al. 2015). The up-regulation of GsERF6 during early stages under NaHCO3 stress may induce downstream components to cope with the alkaline stress. In agreement with the data obtained from the qRT experiment, the GUS activity driven by the GsERF6 promoter was up-regulated under alkaline conditions in Arabidopsis (Fig. 4b). Thus, the GsERF6 promoter is sufficient to confer up-regulation of GsERF6/GUS activity under alkaline stress and there may be alkali stress-responsive cis-acting elements in the upstream region of GsERF6.
The AP2/ERF genes are also involved in plant growth and development besides stress response. The spatial expression analysis of GsERF6 in G. soja shows it expressed in both the roots and shoots but higher in the roots (Fig. 3a). In Arabidopsis, higher expression levels of GUS occur in the root stele and in the vascular bundles of leaves and stems (Fig. 3b–l). The tissue-specific expression pattern of GsERF6 in root stele supports the hypothesis that GsERF6 plays an important role in the primary uptake and transport of nutrients or ions from the soil. The location of GsERF6-GUS in stem and leaf vasculars bundles proposes a possible role of GsERF6 in long-distance transport of nutrients or ions between shoot and root tissues. Based on the specific tissue-location and basic gene expression, we assumed that GsERF6 may play a direct or indirect role to regulate plant growth and development or alkaline-response.
Some of ERF genes can be induced by the abiotic stresses such as salt, drought and cold, and overexpression of these ERF genes can improve the tolerance to these stresses correspondingly (Xu et al. 2008; Zhu et al. 2010). For example, overexpression of TaERF1 in Arabidopsis improved the salt and cold tolerance (Xu et al. 2007). Overexpression of RAP2.6L and DREB19 also enhanced tolerance to salt and drought stresses (Krishnaswamy et al. 2011). To evaluate the function of GsERF6 in response to alkaline stress, we generated transgenic Arabidopsis with overexpression of GsERF6 by Agrobacterium mediated transformation. As shown in Figs. 5 and 6, the GsERF6 OX lines demonstrated an obvious alkali tolerance with higher leaf opening and green leaves rates at the whole plant growing phases, and longer primary roots, higher total chlorophyll content and lower levels of malondialdehyde (MDA) than the wild type. Solaiman et al. (2007) found that canola genotypes maintained higher uptake of P and accumulated greater biomass compared to wheat genotypes on alkaline soils, and the better growth and higher P content in canola genotypes was mainly due to the fact that greater root length could exploit greater soil volume. Thus longer primary roots of GsERF6 transgenic plants at seedling stage may help the plants to resist against the alkaline stress better (Fig. 5).
Many bicarbonate-regulated genes are involved in metabolism, signal transduction and transcription (Alhendawi et al. 1997). NADP-MEI, NADP-MEII and V-H-PPase have been proven to be induced by bicarbonate stress (Fushimi et al. 1994; Liu et al. 2007b). These proteins play important roles with the function of regulating intracellular pH, helping plant cells to cope with the potential acidification in cytoplasm from environmental stresses (Davies 1986; Tang et al. 1993). In this study, transgenic Arabidopsis with GsERF6 showed increased tolerance to alkaline stress, especially bicarbonate stress, and the expression levels of these detected stress-inducible genes (NADP-ME, H +-Ppase, H +-ATPase, COR47, RD29A and KIN1) were significantly higher in GsERF6 OX lines than in WT plants. The up-regulated expression of other stress-related genes such as RD29A, COR47 and KIN1, whose transcripts can be strongly induced by cold, drought and ABA (Horvath et al. 1993; Wang et al. 1995; Kurkela and Franck 1990; Seki et al. 2003), may also contribute to the alkaline tolerance in OX lines, because the high accumulation of these proteins plays an important role in adjusting physiological conditions in plant cells (Puhakainen et al. 2004; Msanne et al. 2011). The ability of GsERF6 upregulating downstream genes suggests that it may be an important regulator in the alkali stress signaling transduction and alkali stress resistance.
Alkali stress, especially alkaline soils in nature, elicits four primary effects on plants: ionic toxicity, osmotic stress, high-pH and bicarbonate. We found that in the 1/2 MS medium, low concentration of NaHCO3 (8 mM) resulted in a lethal phenotype but high concentration of salt stress (NaCl, 100 mM) did not, indicating that injures from NaHCO3 are mainly due to the high pH and bicarbonate (HCO3 −) ion, but not to Na+. To elucidate the mechanisms of GsERF6 transgenic plants responsible for the enhanced alkaline stress tolerance, we analyzed the tolerance of GsERF6 OX plants under pure high pH stress and HCO3 − ion condition separately. Interestingly, the GsERF6 OX lines had a significantly increased tolerance to bicarbonate stress caused by NaHCO3 and KHCO3, but showed no tolerance to high pH (Figs. 4, 5, 7; Supplementary Fig. S4). We found that the plants grew better under single alkaline pH treatment than under a similar pH caused by HCO3 − ions (7 mM KHCO3 resulting in pH 7.65 ± 0.05 in 1/2 MS medium) (Supplementary Fig. S4; Fig. 7). Therefore, we speculate that alkali stress triggers injuries in plants largely through HCO3 − ions, rather than the pure pH, consistent with previous reports (Lee and Woolhouse 1969; Romera et al. 1992; Petersen 1996). The alkali tolerance conferred by GsERF6 appears to be mainly based on an improved bicarbonate ion tolerance rather than based on pure high pH tolerance. The up-regulated expression of H +-Ppase, NADP-ME and H +-ATPase induced by the pH change in the cytoplasm caused by bicarbonate may help to maintain the homeostasis of the internal environment. Taken together, the novel characteristic of GsERF6 implies complex roles of AP2/ERF TFs in alkali response; the specific role of GsERF6 in tolerance to bicarbonate stress provides a perception to elucidate the mechanisms underlying tolerance to alkaline stress.
Plant hormones like ABA, ET and JA are considered to be involved in many aspects of plant growth and development, and responses to environmental challenges including mutiple biotic and abiotic stresses (Danquah et al. 2014; Thao et al. 2015). Many ERF subfamily members participate in the stress signaling pathways and serve as connecting factors in the crosstalk of adversity and hormone signaling networks (Pandey et al. 2005; Lee et al. 2015; Wang et al. 2015; Muller and Munne-Bosch 2015). We investigated the effects of GsERF6 on the hormone synthesis and response in transgenic plants. By preforming the qRT-PCR we analyzed the expression pattern of some hormone synthesis and responsive genes in both transgenic and WT plants under NaHCO3 stress. The results indicated that NaHCO3 stress can increase the expression levels of both ABA and ET synthesis and responsive genes in plants, such as the ABA synthesis genes AtNCED3 and AtNCED5, and the ABA-responsive genes RAB18 and RD29B, ET-synthesis related genes AtACS1, AtACS2 and AtACS5, and the ET responsive genes EIN3, ERF1, ERF5, ERF6 and the JA responsive gene PDF1.2 (Fig. 9), suggesting that GsERF6 may regulate plant responses to NaHCO3 stress through regulating the synthesis, signaling transduction of the relevant endogenous hormones like ABA, ET and JA. Based on the above findings, we proposed that overexpression of GsERF6 increases bicarbonate stress tolerance might be due to induction of the plant hormones such as ABA and ET as signaling molecules to activated a number of hormone- and stress-responsive genes such as RAB18, RD29A, RD29B and COR47 genes, and some ERF like proteins. Consequently, these signaling molecules regulate the defense responses of plants through the synergistic or antagonistic effect. The mechanisms of GsERF6 and its role in bicarbonate signaling and signal transduction of other plant hormones need to be further analyzed in details.
In conclusion, we demonstrate here a novel TF gene from wild soybean, GsERF6, which can modify plant tolerane to alkali stress by specifically increasing tolerance to bicarbonate stress. It is significant to further investigate the precise function and the mechanism of GsERF6 in bicarbonate uptake, translocation and stress resistance. Further investigations include the relationship between bicarbonate stress and the ethylene signal transduction pathway, as well as the role of GsERF6 in the crosstalk of ethylene and bicarbonate-mediated signaling systems are also need to be conducted.
Author contribution statement
YY and YZ designed the research; YY, AL, XD, SW, CC, LC, JX and QL performed experiments; XS and HD analyzed data; YY wrote the manuscript; DZ and XD revised the manuscript. All authors contributed to editing and approving the final version of the manuscript.
Abbreviations
- ABA:
-
Abscisic acid
- ET:
-
Ethylene
- JA:
-
Jasmonate
- OX:
-
Overexpression
- TF:
-
Transcription factor
- WT:
-
Wild type
References
Alhendawi RA, Römheld V, Kirkby EA, Marschner H (1997) Influence of increasing bicarbonate concentrations on plant growth, organic acid accumulation in roots and iron uptake by barley, sorghum, and maize. J Plant Nutr 20:1731–1753
Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M (1998) A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J 17:5484–5496
Bui LT, Giuntoli B, Kosmacz M, Parlanti S, Licausi F (2015) Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Sci 236:37–43
Chaney RL, Coulombe BA, Bell PF, Angle JS (1992) Detailed method to screen dicot cultivars for resistance to Fe-chlorosis using FeDTPA and bicarbonate in nutrient solutions. J Plant Nutr 15:2063–2083
Chen W, Feng C, Guo W, Shi D, Yang C (2011) Comparative effects of osmotic-, salt- and alkali stress on growth, photosynthesis, and osmotic adjustment of cotton plants. Photosynthetica 49:417–425
Cheng MC, Liao PM, Kuo WW, Lin TP (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 162:1566–1582
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Danquah A, de Zelicourt A, Colcombet J, Hirt H (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv 32:40–52
Davies D (1986) The fine control of cytosolic pH. Physiol Plant 67:702–706
Dong T, Park Y, Hwang I (2015) Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem 58:29–48
DuanMu H, Wang Y, Bai X, Cheng S, Deyholos MK, Wong GK-S, Li D, Zhu D, Li R, Yu Y (2015) Wild soybean roots depend on specific transcription factors and oxidation reduction related genesin response to alkaline stress. Funct Integr Genomics 15:1–10
Fushimi T, Umeda M, Shimazaki T, Kato A, Toriyama K, Uchimiya H (1994) Nucleotide sequence of a rice cDNA similar to a maize NADP-dependent malic enzyme. Plant Mol Biol 24:965–967
Ge Y, Li Y, Zhu YM, Bai X, Lv DK, Guo DJ, Ji W, Cai H (2010) Global transcriptome profiling of wild soybean (Glycine soja) roots under NaHCO3 treatment. BMC Plant Biol 10:1–14
Guo R, Yang Z, Li F, Yan C, Zhong X, Liu Q, Xia X, Li H, Zhao L (2015) Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol 15:1–13
Hajiboland R, Yang XE, Romheld V, Neumann G (2005) Effect of bicarbonate on elongation and distribution of organic acids in root and root zone of Zn-efficient and Zn-inefficient rice (Oryza sativa L.) genotypes. Environ Exp Bot 54:163–173
Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R (2010) Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol 153:757–772
Horvath DP, McLarney BK, Thomashow MF (1993) Regulation of Arabidopsis thaliana L. (Heyn) cor78 in response to low temperature. Plant Physiol 103:1047–1053
Kotchoni SO, Gachomo EW (2006) The reactive oxygen species network pathways: an essential prerequisite for perception of pathogen attack and the acquired disease resistance in plants. J Biosci 31:389–404
Krishnaswamy S, Verma S, Rahman MH, Kav NN (2011) Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol Biol 75:107–127
Kurkela S, Franck M (1990) Cloning and characterization of a cold-and ABA-inducible Arabidopsis gene. Plant Mol Biol 15:137–144
Kurtz I (1987) Apical Na+/H+ antiporter and glycolysis-dependent H+-ATPase regulate intracellular pH in the rabbit S3 proximal tubule. J Clin Invest 80:928–935
Lee J, Woolhouse H (1969) A comparative study of bicarbonate inhibition of root growth in calcicole and calcifuge grasses. New Phytol 68:1–11
Lee SB, Lee SJ, Kim SY (2015) AtERF15 is a positive regulator of ABA response. Plant Cell Rep 34:71–81
Liu J-X, Srivastava R, Che P, Howell SH (2007a) An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19:4111–4119
Liu S, Cheng Y, Zhang X, Guan Q, Nishiuchi S, Hase K, Takano T (2007b) Expression of an NADP-malic enzyme gene in rice (Oryza sativa. L) is induced by environmental stresses; over-expression of the gene in Arabidopsis confers salt and osmotic stress tolerance. Plant Mol Biol 64:49–58
Liu A, Yu Y, Duan X, Sun X, Duanmu H, Zhu Y (2015) GsSKP21, a Glycine soja S-phase kinase-associated protein, mediates the regulation of plant alkaline tolerance and ABA sensitivity. Plant Mol Biol 87:111–124
Luo X, Bai X, Sun X, Zhu D, Liu B, Ji W, Cai H, Cao L, Wu J, Hu M (2013) Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J Exp Bot 64:2155–2169
Mahajan S, Pandey GK, Tuteja N (2008) Calcium- and salt-stress signaling in plants: shedding light on SOS pathway. Arch Biochem Biophys 471:146–158
Marrè E, Ballarin-Denti A (1985) The proton pumps of the plasmalemma and the tonoplast of higher plants. J Bioenerg Biomembr 17:1–21
Mazarei M, Elling AA, Maier TR, Puthoff DP, Baum TJ (2007) GmEREBP1 is a transcription factor activating defense genes in soybean and Arabidopsis. Mol Plant Microbe Interact 20:107–119
Misra A, Tyler G (1999) Influence of soil moisture on soil solution chemistry and concentrations of minerals in the calcicoles Phleum phleoides and Veronica spicata grown on a limestone soil. Ann Bot 84:401–410
Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) AP2/ERF family transcription factors in plant abiotic stress responses. BBA-Gene Regul Mech 1819:86–96
Msanne J, Lin J, Stone JM, Awada T (2011) Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234:97–107
Muller M, Munne-Bosch S (2015) Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol 169:32–41
Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci 10:339–346
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56:165–185
Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139:1185–1193
Petersen F (1996) Water testing and interpretation. Water media, and nutrition for greenhouse crops. Ball Publishing, Batavia, pp 31–49
Puhakainen T, Hess MW, Mäkelä P, Svensson J, Heino P, Palva ET (2004) Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol Biol 54:743–753
Romera F, Alcantara E, De La Guardia M (1992) Effects of bicarbonate, phosphate and high pH on the reducing capacity of Fe-deficient sunflower and cucumber plants. J Plant Nutr 15:1519–1530
Rouphael Y, Cardarelli M, Di Mattia E, Tullio M, Rea E, Colla G (2010) Enhancement of alkalinity tolerance in two cucumber genotypes inoculated with an arbuscular mycorrhizal biofertilizer containing Glomus intraradices. Bio Fert Soils 46:499–509
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Seki M, Kamei A, Yamaguchi-Shinozaki K, Shinozaki K (2003) Molecular responses to drought, salinity and frost: common and different paths for plant protection. Curr Opin Biotechnol 14:194–199
Shasha C (2012) Comparative effects of neutral salt and alkaline salt stress on seed germination, early seedling growth and physiological response of a halophyte species Chenopodium glaucum. Afr J Biotechnol 11:9572–9581
Shigyo M, Hasebe M, Ito M (2006) Molecular evolution of the AP2 subfamily. Gene 366:256–265
Shinshi H (2008) Ethylene-regulated transcription and crosstalk with jasmonic acid. Plant Sci 175:18–23
Solaiman Z, Marschner P, Wang D, Rengel Z (2007) Growth, P uptake and rhizosphere properties of wheat and canola genotypes in an alkaline soil with low P availability. Biol Fert Soils 44:143–153
Tang C, Robson A, Longnecker N, Greenway H (1993) Physiological responses of lupin roots to high pH. Plant Soil 155:509–512
Thao NP, Khan MI, Thu NB, Hoang XL, Asgher M, Khan NA, Tran LS (2015) Role of ethylene and its cross talk with other signaling molecules in plant responses to heavy metal stress. Plant Physiol 169:73–84
Thomine S, Lelièvre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H (2003) AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J 34:685–695
Tran L-SP, Nakashima K, Sakuma Y et al (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498
Valdez-Aguilar LA, Reed DW (2010) Growth and nutrition of young bean plants under high alkalinity as affected by mixtures of ammonium, potassium, and sodium. J Plant Nutr 33:1472–1488
Wang H, Datla R, Georges F, Loewen M, Cutler AJ (1995) Promoters from kin1 and cor6.6, two homologous Arabidopsis thaliana genes: transcriptional regulation and gene expression induced by low temperature, ABA, osmoticum and dehydration. Plant Mol Biol 28:605–617
Wang P, Du Y, Zhao X, Miao Y, Song C-P (2013) The MPK6-ERF6-ROS-responsive cis-acting Element7/GCC box complex modulates oxidative gene transcription and the oxidative response in Arabidopsis. Plant Physiol 161:1392–1408
Wang X, Liu S, Tian H, Wang S, Chen JG (2015) The small ethylene response factor ERF96 is involved in the regulation of the abscisic acid response in Arabidopsis. Front Plant Sci 6:1064
Weber H, Chételat A, Reymond P, Farmer EE (2004) Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J 37:877–888
Xu Z-S, Xia L-Q, Chen M, Cheng X-G, Zhang R-Y, Li L-C, Zhao Y-X, Lu Y, Ni Z-Y, Liu L (2007) Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol Biol 65:719–732
Xu Z-S, Chen M, Li L-C, Ma Y-Z (2008) Functions of the ERF transcription factor family in plants. Botany 86:969–977
Xu ZS, Chen M, Li LC, Ma YZ (2011) Functions and application of the AP2/ERF transcription factor family in crop improvement. J Integr Plant Biol 53:570–585
Yang CW, Chong JN, Li CY, Kim CM, Shi DC, Wang DL (2007) Osmotic adjustment and ion balance traits of an alkali resistant halophyte Kochia sieversiana during adaptation to salt and alkali conditions. Plant Soil 294:263–276
Yang CW, Wang P, Li CY, Shi DC, Wang DL (2008) Comparison of effects of salt and alkali stresses on the growth and photosynthesis of wheat. Photosynthetica 46:107–114
Yang CW, Xu HH, Wang LL, Liu J, Shi DC, Wang DL (2009) Comparative effects of salt-stress and alkali-stress on the growth, photosynthesis, solute accumulation, and ion balance of barley plants. Photosynthetica 47:79–86
Yang L, Ji W, Zhu Y, Gao P, Li Y, Cai H, Bai X, Guo D (2010) GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J Exp Bot 61:2519–2533
Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, Ma Y (2009) Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot 60:3781–3796
Zhang GY, Chen M, Chen XP, Xu ZS, Li LC, Guo JM, Ma YZ (2010) Isolation and characterization of a novel EAR-motif-containing gene GmERF4 from soybean (Glycine max L.). Mol Biol Rep 37:809–818
Zhu Q, Zhang J, Gao X, Tong J, Xiao L, Li W, Zhang H (2010) The Arabidopsis AP2/ERF transcription factor RAP2. 6 participates in ABA, salt and osmotic stress responses. Gene 457:1–12
Zhu D, Bai X, Chen C, Chen Q, Cai H, Li Y, Ji W, Zhai H, Lv DK, Luo X, Zhu YM (2011) GsTIFY10, a novel positive regulator of plant tolerance to bicarbonate stress and a repressor of jasmonate signaling. Plant Mol Biol 77:285–297
Zhu D, Cai H, Luo X, Bai X, Deyholos MK, Chen Q, Chen C, Ji W, Zhu Y (2012) Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem Biophys Res Commun 426:273–279
Acknowledgments
This work was supported by the “863” project (2008AA10Z153), the National Natural Science Foundation of China (31171578), Heilongjiang Provincial Higher School Science and Technology Innovation Team Building Program (2011TD005), the National Natural Science Foundation of China (31501331), Advanced Talents Foundation of QAU (6631115032).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2016_2532_MOESM1_ESM.tif
Phylogenetic tree of the AP2/ERF family transcription factors in green plants. The names of each subfamily and their subgroups are given according to (Sakuma et al. 2002). Classifications of small groups in the DREB and ERF subfamilies are given in parentheses according to (Nakano et al. 2006). A consensus tree (after 1000 bootstrap samplings) is shown, and support values are indicated on the sides of important nodes. The name of each branch indicates its locus identifier or transcript ID in Phytozome. The scale bar indicates the substitution rate per residue (TIFF 4729 kb)
425_2016_2532_MOESM2_ESM.tif
Multiple sequence alignment of amino acid sequences of GsERF6 with homologous AP2/ERFs from Arabidopsis. Sequences were aligned using ClustalX. The typical AP2 domain was underlined (TIFF 4223 kb)
425_2016_2532_MOESM3_ESM.tif
Transcript expression levels of GsERF6 and GsERF6 homologs in WT and transgenic Arabidopsis. The transcript expression levels were analyzed by semi-quantitative RT-PCR, the Actin2 gene was used as an internal standard (TIFF 158 kb)
425_2016_2532_MOESM4_ESM.tif
No effect of GsERF6 overexpression on plant tolerance of high pH stress. a The growth of WT and OX seedlings on 1/2 MS medium with pH 7.5 or 8.2. Photographs were taken 10 days after germination. Experiments were performed at least three times (each with 100 seeds for each line). b-c Phenotypes and measurements of primary root lengths of WT and OX seedlings under normal and high pH stress. Seven-day-old seedlings grown on 1/2 MS were transferred to new solid agar plates with pH 5.8, 7.5 and 8.2, respectively. Photographs were taken after 7d growth on the media. All values are means (±SE) from three independent experiments (30 seedlings per experiment). *P < 0.05, **P < 0.01 by Student’s t test (TIFF 9883 kb)
Rights and permissions
About this article
Cite this article
Yu, Y., Liu, A., Duan, X. et al. GsERF6, an ethylene-responsive factor from Glycine soja, mediates the regulation of plant bicarbonate tolerance in Arabidopsis . Planta 244, 681–698 (2016). https://doi.org/10.1007/s00425-016-2532-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2532-4