Abstract
Main conclusion
A combination of unique EPSPS structure and increased gene copy number and expression contribute to natural glyphosate tolerance in three lilyturf species.
A few plants are naturally tolerant to glyphosate, the most widely used non-selective herbicide worldwide. Here, the basis for natural tolerance to glyphosate in three lilyturf species, Ophiopogon japonicus (OJ), Liriope spicata (LS), and Liriope platyphylla (LP), is characterized. These species tolerate glyphosate at about five times the commercially recommended field dose. They share three unique amino acids in their 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) that affect glyphosate binding. These correspond to Asp71Met, Ala112Ile, and Val201Met amino acid variations compared to 231 other published plant EPSPS amino acid sequences. There was also a common deletion at 91 of a highly conserved glutamic acid. Glyphosate-treated lilyturf plants accumulated little shikimic acid but had significantly higher levels of EPSPS mRNA than initially expressed in the control. The IC50 of LsEPSPS was 14.0 µM compared to the 5.1 µM of Arabidopsis thaliana. The higher K m and K i values of LsEPSPS kinetics showed that LsEPSPS had lower substrate binding affinity to glyphosate. Overexpression of LsEPSPS in the recombinant E. coli BL21 (DE3) strain enhanced its tolerance to glyphosate. Both OJ and LS had two copies of the EPSPS gene, while LP had three copies. Therefore, a combination of unique EPSPS structure and increased gene copy number and expression contribute to natural glyphosate tolerance in the three lilyturf species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glyphosate (N-(phosphonomethyl) glycine), a non-selective, systemic postemergence herbicide, has been recognized as a unique weed killer because of its intrinsic properties such as broad-spectrum, low toxicity, and lack of soil residual activity (Duke and Powles 2008). Since its commercial introduction in the early 1970s, glyphosate has increased its market share to become the most widely used herbicide (Woodburn 2000), with a current market value at USD 5.46 billion in 2012 and expected to reach USD 8.79 billion by 2019 (Albany 2014). However, with widespread planting of glyphosate-resistant (GR) crops and extensive reliance on glyphosate, resistant weed biotypes have evolved worldwide (Heap 2015).
Although glyphosate is regarded as non-selective, a naturally glyphosate-tolerant biotype of field bindweed (Convolvulus arvensis) was first reported with no history of glyphosate selection (DeGennaro and Weller 1984). Subsequently, numerous cases of glyphosate-tolerant species have been recognized including birdsfoot trefoil (Lotus corniculatus) (Boerboom et al. 1990), tropical spiderwort (Commelina benghalensis) (Culpepper et al. 2004), Asiatic dayflower (Commelina communis) (Culpepper 2006), and Chinese foldwing (Dicliptera chinensis) (Yuan et al. 2002). Some of these weed species became more problematic as they occupied newly available ecological niches in fields of GR crops, thus reducing the agronomic effectiveness of glyphosate (Nandula et al. 2005). Floristic changes with prevalence of glyphosate-tolerant species and evolved resistant biotypes of otherwise susceptible weeds are a major threat to glyphosate-dependent agricultural production.
Mechanisms of tolerance or evolved resistance to herbicides can be grouped into two types: target site and non-target site. In the case of glyphosate, the former is caused by mutation of the target enzyme (5-enolpyruvylshikimate-3-phosphate synthase, EPSPS, E.C.2.5.1.19) that decreases its binding affinity for the herbicide, or by target enzyme overexpression, that allows the plant to maintain the synthesis of aromatic amino acids that otherwise would be blocked by the inhibition of the shikimic acid pathway by glyphosate. Mutations conferring glyphosate resistance resulting in serine (P106S), alanine (P106A), threonine (P106T), or leucine (P106L) substitutions at proline position 106 of the EPSPS proteins have been reported in Eleusine indica (Baerson et al. 2002; Kaundun et al. 2008; Ng et al. 2003), Lolium rigidum (Kaundun et al. 2011; Wakelin and Preston 2006; Yu et al. 2007), Lolium perenne ssp. Multiflorum (Jasieniuk et al. 2008; Perez-Jones et al. 2007), Echinochloa colona (Alarcón-Reverte et al. 2013, 2014), and Amaranthus tuberculatus (Nandula et al. 2013). Double T102I-P106S mutations confer a high level of glyphosate resistance to E. indica (Chen et al. 2015; Yu et al. 2015). Additionally, mutations or differences in non-coding portions of the gene, i.e., promotor, may affect expression levels and enzyme efficiency, which could modify the plant’s response to glyphosate (Huang et al. 2015).
Target enzyme overexpression can be driven by gene amplification or mutation in a gene promoter. Increased gene copy number was first documented in GR Amaranthus palmeri from Georgia (Gaines et al. 2010) and later identified as a possible mechanism in biotypes from North Carolina and Mississippi (Chandi et al. 2012; Ribeiro et al. 2014) and GR L. perenne ssp. multiflorum from Arkansas (Salas et al. 2012). Amplification of the EPSPS gene has also been associated with glyphosate resistance in A. tuberculatus, Kochia scoparia, Bromus diandrus, and E. indica populations (Chen et al. 2015; Malone et al. 2015; Wiersma et al. 2015; Zhang et al. 2015; Jugulam et al. 2014; Tranel et al. 2011).
Major non-target mechanisms reported in glyphosate-resistant plants include reduced glyphosate translocation, vacuolar sequestration, and rapid leaf necrosis and cell death (Sammons and Gaines 2014; Vila-Aiub et al. 2012). Sorghum halepense resists glyphosate by reducing its foliar uptake and translocation to meristems (Powles and Yu 2010; Vila-Aiub et al. 2012). The same mechanism was identified in L. rigidum populations (Yu et al. 2009; Lorraine-Colwill et al. 2002). Rapid vacuolar sequestration conferred glyphosate resistance in horseweed (Conyza canadensis) (Ge et al. 2010, 2011). In addition, a biotype of Ambrosia trifida rapidly develops necrosis of glyphosate-treated leaves thus reducing herbicide translocation (Robertson 2010).
Ophiopogon japonicus (OJ), Liriope spicata (LS), and Liriope platyphylla, known as lily turfs or liriopogons, are perennial polyploid herbs (Lattier et al. 2014) of the Ruscaceae (formerly assigned to Liliaceae) (Kim et al. 2010), which are widely grown as ornamental and medicinal plants. They sometimes become pernicious weeds in gardens, orchards, and public green lands. The three lilyturf species were found to be naturally tolerant to glyphosate after being unintentionally exposed to the herbicide in private gardens (Qiang, personal observation). Some container-grown lily turfs have also been shown to withstand repeated applications of glyphosate (Van Hoogmoed et al. 2013). The objectives of this study were to assess their glyphosate tolerance level and elucidate tolerance mechanisms through biological (dose–response), physiological (shikimic acid accumulation and target enzyme inhibition), and molecular (cloning of EPSPS, overexpression of the cloned gene, southern-blot analysis, and determination of EPSPS gene mRNA level) experiments.
Materials and methods
Plant materials and growing conditions
Plants of the three lilyturf species were transplanted from cultivated lilyturf gardens at Nanjing Agricultural University campus and neighboring Xiamafang garden into pots (15 cm in diameter, 15 cm deep), individually, containing commercial potting media in a glasshouse under natural illumination at 28–35 °C. Plants were watered by drip irrigation and fertilized as needed, and allowed to grow for 2 months until they developed complete new leaves for dose–response and shikimic acid assay experiments. For EPSPS gene cloning, southern blot, and gene expression experiments, new plants were cultivated directly from May to June 2012 from garden collected seeds of each of the three species. Emerged seedlings were thinned to one per pot and allowed to reach the 4- to 5-leaf growth stage. Growth conditions were the same as those for transplanted seedlings. A. thaliana seeds were germinated on MS medium and emerged seedlings were individually transplanted into pots with vermiculite:peat moss:perlite (9:3:1) in a glasshouse at constant 23 °C.
Glyphosate dose–response experiments
To evaluate the response of the lilyturf species to increasing doses of glyphosate, 10- to 12-leaved plants individually transplanted into pots were sprayed with glyphosate (Roundup, 41 % glyphosate isopropylammonium, Monsanto) at 0, 0.375, 0.75, 1.5, 3.0, 6.0, 12.0, and 24.0 kg ae ha−1 using a compressed air tower sprayer (PT-1, China Agricultural University) calibrated to deliver 234 L ha−1 at 0.2 MPa. In the absence of a susceptible lilyturf accession, A. thaliana was used as control species. Four-week-old seedlings were sprayed with glyphosate at 0, 40, 120, 240, 400, and 600 g ha−1 (here and thereafter expressed as acid equivalent). Four pots, each containing one seedling, were used for each treatment. Symptom development was assessed visually 3 weeks after treatment (Table 1). Percent injury was estimated based on discrete herbicide injury severity (HIS) values (Song et al. 2011):
All experiments were repeated three times. Injury data were subjected to ANOVA (SPSS 17.0, SPSS Institute Inc.) test and, after conversion to percent, fitted with a three-parameter log-logistic equation since it was not possible to attain a lower limit even with the highest dose applied (Burgos et al. 2013; Ritz and Streibig 2005). Data and estimated curves were plotted with Origin 8.0 (Origin Lab. Co.).
Shikimic acid assay
Selected 10- to 12-leaved plants of the lilyturf species were treated with a critical dose of 3000 g ha−1 glyphosate based on the results of the dose–response experiments. Leaves were collected every 2 days after treatment (DAT), snap frozen in liquid nitrogen, and stored at −20 °C until use. Six seedlings (one per pot) of each species were used. Shikimic acid was extracted following the modified method of Singh and Shaner (1998), and the shikimic acid level was measured spectrophotometrically by the method of Cromartie and Polge (2000). Plant leaf tissue (0.5 g) was ground to fine powder in liquid nitrogen with a mortar and pestle and then transferred to a 10-mL tube containing 1 mL 0.25 mol L−1 HCl. The tube was incubated on a shaker for 1.5 h at room temperature followed by centrifugation at 12,000g for 15 min. The supernatant was then briefly re-centrifuged, and the new supernatant was used for the subsequent steps. The extract (200 µL) was mixed with 2.0 mL 1 % periodic acid. The mixture was incubated at room temperature for 3 h for shikimic acid oxidation. The solution was then mixed with 2.0 mL 1.0 mol L−1 NaOH and 1.2 mL 0.1 mol L−1 glycine. The absorbance at 380 nm was immediately measured spectrophotometrically. Shikimic acid concentration was calculated from a standard curve. The experiment was conducted three times with independent biological replicates.
Cloning of full-length EPSPS cDNA
Young leaves (c. 0.1 g) of the three lilyturf species were ground to a fine powder in liquid nitrogen, and the total RNA was extracted with Trizol reagent according to the manufacturer’s protocol (TANGEN Code: DP405). First-strand cDNA was synthesized with SMART scribe™ reverse transcriptase (Clontech catalog No: 639535). Total RNA (1.0 ng–1.0 µg) was incubated with Oligo(dT)18, primer and SMART scribe™ reverse transcriptase at 42 °C for 60 min according to the manufacturer’s instructions. The primers EPSP-CF and EPSP-CR (Supp. Table S2) were designed from EPSPS gene sequences in NCBI. Reverse transcription-polymerase chain reactions (RT-PCR) were carried out in 0.2-mL tubes (25 µl per tube) in a reaction buffer containing Ex-Taq polymerase, dNTPs, MgCl2, Ex-Taq buffer, 0.2 µM primers, and template cDNA. PCR was performed in a thermal cycler as follows: 5 min at 94 °C (1 cycle); 30 s at 94 °C; 30 s at 52 °C; 1 min at 72 °C (30 cycles); and 7 min at 72 °C (1 cycle). The obtained fragments were cloned into the PMD-19T vector. The nucleotide sequences were determined by Sangon Biotech (Shanghai) Co., Ltd. and compared using the NCBI-Basic Local Alignment Search Tool (BLAST) program (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
The total RNA of young lilyturf leaves was prepared as previously described. In OJ and LS, first-strand cDNA synthesis was performed using 1 g of poly (A) RNA as the template, according to recommendations in the 3′ or 5′ SMART RACE kit (Clontech catalog No. 634914). The sequence information obtained from the conserved region was used to design the 5′-end and the 3′-end primers (Supp. Table S2). PCR RACE was performed at the appropriate annealing temperature (Supp. Table S2). The complete nucleotide sequence was amplified (Supp. Table S2), determined, and confirmed as described before. Because of their high homology, the full length of EPSPS gene in LP was amplified according to that in LS. Additionally, sequences of plant EPSPS genes were obtained from GenBank, pooled, and redundancy was removed as in Garg et al. (2014). Phylogenetic trees were created by MEGA software (http://www.megasoftware.net/) for homology analysis.
EPSP synthase activity
Protein extraction and EPSPS assay were conducted following the procedures of Dayan et al. (2015). A leaf tissue sample (5 g) was ground to fine powder in a chilled mortar. The powder was transferred to tubes containing 100 mL of cold extraction buffer (100 mM of MOPS, 5 mM of EDTA, 10 % glycerol, 50 mM of KCl and 0.5 mM of benzamidine) with 1 % polyvinylpolypyrrolidone (PVPP) and 70 µL of fresh β-mercaptoethanol. Samples were homogenized for about 5 min with constant stirring to minimize foaming and then centrifuged for 40 min at 18,000×g at 4 °C. The supernatant was decanted into a cold beaker. Powdered ammonium sulfate was slowly added to the supernatant to make 45 % w v−1 concentration, stirred continuously for 30 min and centrifuged at 30,000×g for 30 min at 4 °C. Protein extracts were precipitated out of solution by the gradual addition of ammonium sulfate to a concentration of 80 % (w v−1) with gentle stirring and then centrifuged at 30,000×g for 30 min at 4 °C. Pellets were dissolved in about 3 mL of extraction buffer and dialyzed overnight in 2 L of dialysis buffer using a 30 mm, 10,000-MWC dialysis tubing at 4 °C on a stir plate.
EPSPS activity was determined at 28 °C for 20 min in 100 µL reaction mixtures (50 mM MOPS, 0.5 mM MgCl2, 5 % glycerol (v v−1), 1 mM sodium molybdate, 100 mM NaF, 10 µL purified enzyme, and different doses of substrate and glyphosate) by measuring the amount of inorganic phosphate produced in the reaction using the malachite green dye assay method (Lanzetta and Alvarez 1979). After reaction, 1600 µL of malachite green ammonium molybdate colorimetric solution was added, and color development was stopped after 1 min by the addition of 400 µL of a 34 % sodium citrate solution. 30 min later, the mixture was measured for absorbance at 660 nm. Reaction mixtures without substrate served as controls.
The K m values for PEP, the K i values for glyphosate, and the IC50 value for glyphosate were determined as described by Tian et al. (2010).
Appropriate dilutions of 10 mM KH2PO4 were used as standards, and the color reaction measured as described above. Determination of the soluble protein in plants was with reference to total protein quantitative assay kit (Nanjing Jiancheng Bioengineering Institute, Code: A045-2). Units of EPSPS specific activity were defined as µmoles of increased phosphate per minute per milligram of protein. The data represent the averages ± SD of three independent experiments.
Overexpression and identification of glyphosate tolerance
EPSPS gene of LS was selected for overexpression, after amplifying it by PCR using the primers EPSP1 (5′-CCATGGATGGAGCAAGCGATCATGGCTAA-3′) and EPSP2 (5′-CTCGAGGTGCGTTGCGAACCTCTGCAAT-3′). The PCR product was digested with NcoI and XhoI, cloned into the corresponding restriction sites of pET-28a (Novagen, Inc.) and confirmed by DNA sequencing. The plasmid was then transformed into E. coli BL21 (DE3) (Novagen, Inc.). The expressed protein was detected by discontinuous vertical SDS-PAGE electrophoresis.
The transgenic bacteria were inoculated into liquid LB medium containing 1.0 mmol L−1 IPTG and glyphosate at increasing concentrations (0, 1500, 3000, 4500, 6000, 7500, 9000, 10,500, and 12,000 mg L−1) and shaken at 37 °C for 12 h. A strain only transformed with plasmid PET-28a was used as a negative control. Cell concentrations were calculated by optical density (OD600) measurements with a spectrophotometer.
Southern-blot analysis
Total DNA (10 μg) from young leaves of the three lilyturf species was extracted by DNeasy Plant Mini Kit (QIAGEN) and digested with Hind III. The prepared samples were electrophoresed in 7 g L−1 agarose gels and transferred onto nylon membranes (Hybond-N+) using standard blotting techniques described in DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche). The membranes were pre-hybridized for 30 min at 60 °C and hybridized with the gene-specific cDNA fragments that were labeled with DIG-High Primer for 20 h at 60 °C in a rapid hybridization buffer (Roche). The membranes were then washed twice at room temperature in 2 × saline sodium citrate (SSC) buffer with 1 g L−1 SDS and subsequently washed twice in 2 × SSC with 1 g L−1 SDS for 15 min at 68 °C. The immunological identification was recorded according to the operation manual of DIG-High Prime DNA Labeling and Detection Start Kit I (Roche).
Quantitative real-time PCR
After treatment with glyphosate at 1.5 kg ha−1 (328 mg L−1 for A. thaliana), leaf samples of the lilyturfs and A. thaliana were harvested from four plants of each replicate at 0, 3, 6, 9, and 12 DAT, respectively. The experiments were independently repeated three times. Total RNA was isolated from leaf samples according to TANGEN Reagent’s recommendations (TANGEN Code: DP405). Recombinant DNase I (TaKaRa Code: D2270A) was used to degrade the DNA. First-strand cDNA was synthesized with Reverse Transcriptase M-MLV (TaKaRa Code: D2640S). Total RNA (1.0 ng–1.0 µg) was incubated with Oligo (dT)18 at 70 °C for 10 min. Primer and M-MLV reverse transcriptase were added into the mixture and incubated at 42 °C for 1 h according to the manufacturer’s instructions.
Primers 25SrRNA-F and 25SrRNA-R (Supp. Table S3) specific for the constitutively expressed 25SrRNA gene were used in real-time PCR to normalize the different samples for differences in the amounts of plant RNA. The LEPSPS-F and LEPSPS-R primers were used for the amplification of the lilyturf EPSPS genes, while the ATEPSPS-F and ATEPSPS-R primers (designed based on CAA29828.1 from NCBI) were used for the AT EPSPS gene. PCR reactions were according to SYBR Premix Ex-Taq™ (TaKaRa Code: DRR041A). The reactions were carried out in 0.2-mL tubes (25 µl per tube) in a reaction buffer containing 1 × SYBR Green I mix (Taq polymerase, dNTPs and SYBR Green dye), 0.2 µM primers EPSPS-F and EPSPS-R (Supp. Table S3), and template DNA. PCR conditions were 95 °C for 15 s, 60 °C for 15 s, and 68 °C for 20 s for 40 cycles on an Eppendorf Sequence Detection System. PCR efficiency for each target mRNA was obtained via the slope of the standard curve. The absence of primer–dimer formation that could interfere with specific amplification was verified in controls (without template DNA). Each time point was determined as an average from triplicate data. Relative gene expression was obtained with the formula fold induction = 2−∆∆CT, where ∆∆C T = [C TEPSPS (unknown sample) − C T25SrRNA (unknown sample)] − [C TEPSPS (calibrator sample) − C T25SrRNA (calibrator sample)]. The C T (threshold cycle) value represents the PCR cycle at which the copy number passes the fixed threshold and can be first detected. EPSPS is the gene of interest, and 25SrRNA is the gene used as internal control. The calibrator sample is the sample chosen to represent 1 × expression of the gene of interest (non-treated leaves). Non-treated leaves of OJ were used for calibration.
Results
Effect of glyphosate on three lilyturf species
The three lilyturf species and A. thaliana differed in their response to glyphosate. The log-logistic model provided a good description of the relationship between glyphosate dose and injury level (Fig. 1a). A. thaliana was substantially more susceptible than the lilyturf species, with an ED50 value of 61 g ha−1. The calculated ED50 values for OJ, LS, and LP were 7000, 7360, and 8230 g ha−1 glyphosate, respectively. These correspond to 4.7, 4.9, and 5.5 times the commercially recommended field rate (c. 1500 g ha−1 in China), and 114, 120, and 134 fold the ED50 value of the reference species (A. thaliana), respectively (Fig. 1). Based on their ED50 values, LP was significantly (p < 0.05) more tolerant to glyphosate than OJ (their confidence intervals did not overlap), but LS tolerance to the herbicide was similar to that of both LP and OJ.
Glyphosate dose response and shikimic acid accumulation experiments in three lilyturf species, Ophiopogon japonicus (OJ), Liriope spicata (LS), and L. platyphylla (LP). a Response of three lilyturf species and Arabidopsis thaliana to increasing doses of glyphosate three weeks after treatment. b Symptoms of OJ, LS, and LP 20 days after treatment with 3000 and 12,000 g ha−1 glyphosate. c Shikimic acid accumulation in leaves of three lilyturf species after 3000 g ha−1 glyphosate treatment. Data were subjected to ANOVA (SPSS) test. Data are mean ± SE with three replicates
Foliar accumulation of shikimic acid in vivo
Glyphosate applied at 3000 g ha−1 induced the accumulation of shikimic acid in leaves of the three species, with a general increase with time until it reached a peak between 12 and 16 DAT and thereafter decreased until experiment end at 20 DAT. At this time, the level of shikimic acid was similar to that determined in the absence of glyphosate, i.e., at 0 DAT (Fig. 1c). Shikimic acid accumulation in OJ (with a peak of 76.4 µg g−1 FW at 16 DAT) was always higher than that found in the other two species (with peaks of 51.5 and 55.1 µg g−1 FW at 12 DAT for LP and LS, respectively) (Fig. 1c). However, the three lilyturf species withstood glyphosate at 3000 g ha−1 (Fig. 1b).
Cloning and sequence analysis of the EPSPS gene
Using gene-specific primers, PCR products of c. 500 bp of the EPSPS gene were amplified from the leaves of the three lilyturf species and sub cloned into the pMDTM19-T vector (detailed information of DNA markers used is shown in Supp. Table S4). Three targeted fragments were isolated by sequence analysis of the cloned ones. The full-length sequences were determined by 5′ and 3′ RACE. The opening reading-frames (ORF) of EPSPS gene cDNA in the three lilyturf species had 1551, 1557, and 1557 nucleotides (GenBank sequence accession numbers are KP143746, KP143747, and KP143748, respectively), coding 516, 518, and 518 amino acids (Fig. 2a–c). Molecular sizes of the predicted protein were 54.41, 54.55, and 54.51kD with isoelectric points of 7.27, 7.34, and 7.34, respectively. Identity among the three ORFs was 95 %.
The EPSPS gene sequences of the three lily turf species revealed high homology with the EPSPS from other plant species (as determined with BLAST), indicating that EPSPS of the three lily turfs belonged to the plant EPSPS family; this was also confirmed by phylogenetic analysis. The EPSPS genes of the three lilyturf species were widely dissimilar to other 47 EPSPS plant genes available in GenBank. They had the highest homology of 78 % with Allium macrostemon and together with it formed a distinct cluster (Fig. 2d).
There are three unique amino acids in the three lilyturf species compared to other 231 plant EPSPS registered in NCBI, 71Met, 112Ile, and 201Met (numbered according to A. tuberculatus EPSPS in which these amino acids correspond to Asp, Ala, and Val, respectively) (Fig. 3). The three sites were conserved in the genes from the three lilyturf species. There was also a common deletion at position 91 of a highly conserved glutamic acid compared with other plant EPSPS (Fig. 3). Additionally, there were a Pro and a Glu deletion at sites 79 and 80 in OJ, respectively, but not in LS and LP (Fig. 3).
Amino acid sequence alignment of EPSPS from A. tuberculatus, OJ, LS, and LP. A. tuberculatus: FJ869880 from NCBI. Quadrangular two deleted amino acid in OJ compared with LS and LP, arrowhead EPSPS unique amino acid residues in the lilyturf species, pentastar the deletion of 91Glu. The important sites are boxed
The three-dimensional structure prediction of the lilyturf EPSPS might help to explain a likely contribution of the unique enzyme to glyphosate tolerance. EPSPS of LS (LsEPSPS) was selected for this analysis. The sequence of LsEPSPS was submitted to Swiss Model (http://swissmodel.expasy.org/) under the automatic modeling mode. The output was analyzed with Swiss-PdbViewer 4.01 and PyMOL 1.5.0.3 softwares. The pdb of the X-ray structure used was DOI:10.2210/pdb2aay/pdb. The spatial positions of 71Met, 112Ile, and 201Met are shown in Fig. 4a. 112Ile is at the opposite end of the third helix away from 101Gly, along with 106Pro (Fig. 4b). In A. tuberculatus, 112Ala does not come in contact with 109Ala that was directly linked to 106Pro (Fig. 4c). However, 112Ile in LS could spatially conflict with 109Ala (Fig. 4b), which could shift the helix resulting in an adjustment of 101Gly at the active site.
Analysis of the three-dimensional structure of LS EPSP synthase by SPDBV4.01. a The 3D structure of LsEPSPS and three unique amino acid residues (green 71Met, blue 112Ile, pink 201Met). b The location of 112Ile in the third helix (blue 112Ile, red 109Ala, yellow 106Pro, orange 101Gly, magenta the substrate glyphosate, cyans the substrate S3P) of LsEPSPS. c 112Ala in A. tuberculatus EPSPS shown pink. d The hydrogen bond distance between glyphosate and 101Gly in E. coli (green), A. tuberculatus (white) and LsEPSPS (yellow)
The deletion at 91Glu is at the other end of the 101Gly helix. It can well cooperate with the 112Ile as this beta sheet shortens the reach of the helix with the 101Gly H-bond (Fig. 5a), possibly further adjusting the position of 101Gly. The model calculated that the hydrogen bond distance between glyphosate and 101Gly was shortened to 2.77 Å in LsEPSPS from 2.86 Å in E. coli and A. tuberculatus (Fig. 4d).
Residue 201Met is actually on the surface side midway down this helix under the surface beta sheet. This helix has a loop that connects and adjusts the exact location of the next helix containing the active site SSQ180 motif (Fig. 5b). The 180Gln H-bond stabilizes the glyphosate phosphonate or the PEP phosphate. Therefore, this Val201Met mutation might be a way to slightly contact or shift the Gln toward the phosphate due to the large size of 201Met (Fig. 5b). The hydrogen bond distance between glyphosate and 180Gln was lengthened from 2.69 Å in EPSPS of A. tuberculatus to 2.76 Å in that of LS. The H-bond between S3P and 180Gln was also extended by 0.39 Å (Fig. 5b). Finally, 71Met is on the turning of the second and third helix in domain 3, well away from the active sites, with which it does not interact given its surface exposure (Fig. 4a).
EPSP synthase activity in vitro
The specific activity of EPSPS from LS to A. thaliana in absence of glyphosate was 2.13 and 1.07 U/mg protein, respectively (Fig. 6a). The IC50 was 14.0 µM for LsEPSPS, almost three times higher than the 5.1 µM for EPSPS in A. thaliana (AtEPSPS) (Fig. 6b).
EPSPS kinetic analyses (Fig. 7) demonstrated that the K m value of LsEPSPS (4.27 µM) was higher than that of AtEPSPS (2.58 µM). The K i value of LsEPSPS (1.91 µM) was also higher than that of AtEPSPS (1.08 µM). Higher K m and K i values indicated that LsEPSPS had weaker substrate binding affinity thus conferring higher tolerance to glyphosate compared to AtEPSPS.
Overexpression and identification of glyphosate tolerance
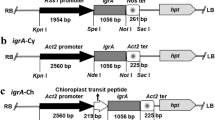
The construction of overexpression vectors with LS EPSPS genes is shown in Fig. 8a–c. The recombinant plasmids LsEPSPS-PET-28a effectively expressed in E. coli BL 21 (DE3). The protein was c. 47kD without the signal peptide as determined by discontinuous vertical SDS-PAGE electrophoresis (Fig. 8d).
Overexpression of EPSPS genes in L. spicata specie. a Recombinant vector LsEPSPS-PMD-19T. b Double enzymes digestion of LsEPSPS-PMD-19T and PET-28a. c Recombinant vectors LsEPSPS-PET-28a. d SDS-PAGE analysis of LsEPSPS-PET-28a [BL21 (DE3)] induced by IPTG for 8 h. e Dose–Response curve of transgenic BL21 (DE3) with PET and LsEPSPS-PET-28a in response to different doses of glyphosate 12 h after treatment
The recombinant strains (with LsEPSPS-PET-28a plasmid) and the control strain (with only PET-28a plasmid) were cultured in the presence of increasing concentrations of glyphosate. The response of the strains to increasing doses of glyphosate based on OD600 growth values was subjected to a log-logistic analysis. The calculated ED50 value for the recombinant strains was 9080 mg L−1, 1.3 times higher than that of the control strain that was 6840 mg L−1 (Fig. 8e). The LsEPSP was also transformed into A. thaliana. In the growth assay, most calli of wild-type A. thaliana showed severe yellowing at the lowest glyphosate dose of 50 mg L−1 glyphosate, whereas LS-transgenic callus grew well even when treated with 600 mg L−1 glyphosate (Supp. Fig. S1–S3). For whole plant assay, the total EPSPS gene expression was significantly higher in transgenic A. thaliana than in wild-type population (Supp. Fig. S5). The calculated ED50 value of transgenic A. thaliana was 220 g ha−1, c. 3.6-fold that of the wild-type (Supp. Fig. S1, S2, S4). Thus, the LsEPSPS gene conferred transformed E. coli and A. thaliana, the ability to withstand higher glyphosate concentrations than the wild types.
Southern-blot analysis
DNA extraction and digestion are illustrated in Fig. 9a–c. According to the Southern blot, both OJ and LS species had two copies of the EPSPS gene, while LP had three copies (Fig. 9d). The two restriction bands of OJ and LS digested by Hind III were at different locations (Fig. 9d), indicating that the EPSPS genes were in different positions of DNA genome. One of the EPSPS gene copies of LP is almost at the same position to that of OJ, another copy shares the same position with that of LS, and the third copy is at a distinct position. In addition, the chromosome number of OJ was determined as 2n = 4X = 72 and that of both LS and LP was 2n = 2X = 36 (Supp. Fig. S6).
EPSPS gene expression
To further examine the glyphosate tolerance mechanism in the three lilyturf species, the EPSPS mRNA level was determined by real-time PCR analysis in the young leaves of A. thaliana and the three species (Supp. Fig. S7). The relative levels of EPSPS mRNA in LS and LP species slightly increased at 3 DAT and then further increased to reach a peak at 9 DAT to c. 43-and 99-fold of the initial expression, respectively (Fig. 10). The EPSPS gene expression in OJ was lower and reached a peak at 6 DAT that was only c. 17-fold of the initial level. The expression tendency in A. thaliana was similar to OJ (Fig. 10). Its peak was significantly lower than that observed in the lilyturf species.
EPSPS mRNA level at different times after the treatment of glyphosate at 1500 g ha−1 by real-time RT-PCR in A. thaliana and three lilyturf species at the 4–5 leaf stage. Relative mRNA level was calculated using the ∆∆C T method from triplicate data. 25SrRNA gene was used as internal control to normalize the different samples with the same amount of plant RNA. Non-treated leaves of OJ were used as 1 × expression level. Data are mean ± SE with three replicates
Discussion
Several plants with different levels of natural glyphosate tolerance have been identified (Boerboom et al. 1990; Culpepper 2006; DeGennaro and Weller 1984; Huang et al. 2014; Owen and Zelaya 2005; Westhoven et al. 2008; Yuan et al. 2001). Among them, D. chinensis survived glyphosate at 3.3 kg ha−1. Its GR50 value was about 2–7-fold that of Ageratum houstonianum (a reference species) in a glasshouse dose–response study (Yuan et al. 2001). Recommended field doses of herbicides vary among countries and regions based on agronomic management of crops and product marketing. Glyphosate is commonly used at doses ranging from 900 to 1500 g ha−1. Thus, the three lilyturf species were tolerant (at GR50 level) to glyphosate applied at 4.7–5.5 times the highest commercial dose (Fig. 1a). Tolerance to glyphosate in the three lily turfs is substantially higher than in any of the recognized tolerant species so far and even surpasses the resistance level evolved in the field by many weeds selected by this herbicide. The mechanism of glyphosate tolerance in the three lilyturf species may be helpful to better understand the contribution of a low-sensitivity target site alone or in combination with other mechanisms conferring a modicum of tolerance as well as the molecular role of EPSPS isozymes.
Inhibition of EPSPS by glyphosate in susceptible plants leads to the accumulation of shikimic acid. Weeds that have evolved target-site resistance to this herbicide accumulate shikimate at levels much lower than those of the susceptible counterparts, making shikimic acid, an important biomarker to detect differential responses to glyphosate (Singh and Shaner 1998). Decreased shikimic acid accumulation has been observed in many tolerant and resistant plants after exposure to glyphosate including transgenic glyphosate-resistant crops (Singh and Shaner 1998), field-evolved resistant weeds (Yu et al. 2009), in vitro selected resistant mutants (Tong et al. 2009), and differentially or naturally tolerant crops and weeds (Henry et al. 2007; Zhang et al. 2011). The highest accumulation of shikimic acid in the leaves of LS, LP, and OJ lily turfs (between 12 and 15 DAT) was 55.1, 51.5, and 76.4 μg g−1 FW, respectively, after treatment with 3000 g ha−1 glyphosate (Fig. 1c). The low accumulation of shikimic acid in the foliage of OJ, LS, and LP may be an indication that the EPSPS has low sensitivity to glyphosate or that the herbicide is not reaching its target site.
Target-site mutations are now well established as one of the mechanisms of glyphosate resistance (Sammons and Gaines 2014). Several weeds evolving resistance have single mutations at their EPSPS, particularly at Pro106 position (Sammons and Gaines 2014; Powles and Yu 2010). Resistant biotypes of E. indica carrying two mutations in their EPSPS were recently reported (Chen et al. 2015, Yu et al. 2015). The ability to withstand glyphosate has also been conferred to plants by introducing point mutations in the EPSPS gene through site-directed mutagenesis, gene targeting, and DNA shuffling (He et al. 2003; Tian et al. 2013; Zhang et al. 2011). Naturally glyphosate-tolerant Allium macrostemon has an EPSPS that transformed and expressed into E. coli, and tobacco confers them resistance to the herbicide (Huang et al. 2009; Zhou et al. 2009). Similarly, the EPSPS of C. arvensis has a Phe96Ser (numbered according to A. tuberculatus EPSPS but reported in the original paper as Phe101Ser based on E. coli numbering) substitution that could contribute to its glyphosate tolerance (Zhang et al. 2011). Conversely, mutations of EPSPS residues Val133Ile and Pro382Leu in a glyphosate-resistant biotype of E. indica from China did not contribute to its resistance at the enzyme level (Zhang et al. 2015).
The unique structure of LsEPSPS may play an important role in glyphosate tolerance. In susceptible EPSPS, Pro106 adjusts the position of the 101Gly H-bond (Healy-Fried et al. 2007), and then it is likely that the deletion of 91Glu may also allow for a similar adjustment for 101Gly at the end of the helix (represented as an inflexible rod in Fig. 5a). At the opposite end of the third helix away from 101Gly, 112Ile has a conflict with 109Ala due to its large molecular volume (Fig. 4b). Thus, 112Ile may induce a change in the glyphosate-binding site by a shift in the 101Gly. The mentioned deletion at 91Glu can very well cooperate with the 112Ile (Fig. 5a). LsEPSPS had higher K m (PEP) and K i (glyphosate) values (Fig. 7), compared with the reference species AtEPSPS, which indicated that LsEPSPS had lower substrate binding affinity. This is a highly likely scenario since Gly101Ala confers very high resistance but substantially increases the K m for PEP and thus lowers dramatically the catalytic activity of the enzyme; this deleterious effect, however, is considerably compensated by a second mutation such as Ala192Thr (Eichholtz et al. 2001). Transgenic oil seed rape (Brassica napus) expressing the double mutant Gly101Ala + Ala192Thr is resistant to glyphosate (Kahrizi et al. 2007). In addition, the SSQ motif which stabilizes glyphosate could well be shifted “toward” the phosphonate due to the large size of 201Met in the adjacent helix connected by a loop (that stabilizes S3P binding) (Fig. 5b). Thus, it is possible for residues distantly located from the active site to exert indirect effects on glyphosate/PEP binding leading to improved glyphosate tolerance.
In the experiment of overexpression, the calculated ED50 value for the LsEPSPS transgenic strains was 1.3 times higher than that of the control E. coli strain. Similarly, expression of LsEPSPS in A. thaliana provided a 3.6-fold increase in its ED50 (Supp. Fig. S4). These results alone would not explain the high tolerance of the lilyturf species at the whole plant level. However, there was just one copy of LsEPSPS transformed into E. coli and A. thaliana. But it is clear that copy number and expression of EPSPS gene contribute to glyphosate tolerance in the three lilyturf species.
Increased expression of the EPSPS has been reported in several species including glyphosate-tolerant D. chinensis (Yuan et al. 2002) and glyphosate-resistant A. palmeri (Ribeiro et al. 2014; Chandi et al. 2012; Gaines et al. 2011) and C. canadensis (Dinelli et al. 2006). Overexpression of the target enzyme could be caused by gene amplification or mutation in a gene promoter. Gene amplification was found in GR A. palmeri (Gaines et al. 2011; Tranel et al. 2011) and L. perenne (Salas et al. 2012) species but not in glyphosate-tolerant D. chinensis (Yuan et al. 2002). The three species in this study contained more than one copy of the EPSPS gene (Fig. 9d). The OJ accession in this study is a tetraploid; LS and LP are diploids (Supp. Fig. S6). The additional EPSPS enzyme produced from the multiple gene copies would be expected to increase the ability of the three lilyturf species to withstand high glyphosate doses. The EPSPS expression level in LP was twofold higher than that of OJ and LS. However, the EPSPS transcript levels were not proportional to the gene copy numbers. Both OJ and LS had two copies of the EPSPS gene, but the maximum EPSPS expression in LS was much higher than that in OJ (Figs. 9d, 10). The higher EPSPS gene expression level in LS may be related to the different positions of its EPSPS genes in the genome compared to OJ (Fig. 9d). Further work should be carried out to provide additionally insight into the glyphosate tolerance mechanism in these species.
Conclusions
We have demonstrated that three lily turfs, OJ, LS, and LP, are naturally tolerant to glyphosate and accumulate little shikimic acid when treated with the herbicide at doses much higher than those commercially recommended. EPSPS of the three lilyturfs share three distinctive functional amino acid residues and a common 91Glu deletion that may alter the affinity of EPSPS for glyphosate. The EPSPS in L. spicata had higher K m (PEP), K i (glyphosate) and IC50 values, compared with the reference species, A. thaliana EPSPS. Their genomes have two (OJ and LS) or three (LP) copies of the EPSPS genes, and all have increased EPSPS gene expression after treatment with glyphosate. It would be relevant to determine in a future study the expression patterns of the EPSPS gene in different plant parts. Overall, unique EPSPS structure, multiple EPSPS copies, and higher expression level altogether contribute to natural tolerance of the three lilyturf species to glyphosate.
Author contribution statement
S. Qiang designed research; C.J. Mao, H.J. Xie, and S.G. Chen conducted experiments; C.J. Mao, B.E. Valverde, and S. Qiang analyzed and interpreted the data and wrote the paper.
References
Alarcón-Reverte R, García A, Urzua J, Fischer AJ (2013) Resistance to glyphosate in junglerice (Echinochloa colona) from California. Weed Sci 61:48–54
Alarcón-Reverte R, García A, Watson SB, Abdallah I, Sabate S, Hernández MJ, Dayan FE, Fischer AJ (2014) Concerted action of target-site mutations and high EPSPS activity in glyphosate-resistant junglerice (Echinochloa colona) from California. Pest Manag Sci 71:996–1007
Albany (2014) Global glyphosate market is expected to reach USD 8.79 Billion by 2019. Transparency Market Research. PRNewswire Publishers, New York, p 1
Baerson SR, Rodriguez DJ, Tran M, Feng YM, Biest NA, Dill GM (2002) Glyphosate-resistant goosegrass: identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Plant Physiol 129:1265–1275
Boerboom CM, Wyse DL, Somers DA (1990) Mechanism of glyphosate tolerance in birdsfoot trefoil (Lotus corniculatus). Weed Sci 38:463–467
Burgos NR, Tranel PJ, Streibig JC, Davis VM, Shaner DL, Norsworthy K, Ritz C (2013) Confirmation of resistance to herbicides and evaluation of resistance levels. Weed Sci 61:4–20
Chandi A, Milla-Lewis SR, Giacomni D, Westra P, Preston C, Jordan DL, York AC, Burton JD, Whitaker JR (2012) Inheritance of evolved glyphosate resistance in a North Carolina Palmer amaranth (Amaranthus palmeri) biotype. Int J Agron. doi:10.1155/176108
Chen J, Huang H, Zhang C, Wei S, Huang Z, Chen J, Wang X (2015) Mutations and amplification of EPSPS gene confer resistance to glyphosate in goosegrass (Eleusine indica). Planta. doi:10.1007/s00425-015-2324-2
Cromartie TH, Polge ND (2000) An improved assay for shikimic acid and its use as a monitor for the activity of sulfosate. Proc Weed Sci Soc Am 40:291
Culpepper AS (2006) Glyphosate-induced weed shifts. Weed Technol 20:277–281
Culpepper AS, Flanders JT, York AC, Webster TM (2004) Tropical spiderwort (Commelina benghalensis) control in glyphosate-resistant cotton. Weed Technol 18:432–436
Dayan FE, Owens DK, Corniani N, Silva FML, Watson SB, Howell JL, Shaner DL (2015) Biochemical markers and enzyme assays for herbicide mode of action and resistance studies. Weed Sci 63:23–63
DeGennaro FP, Weller SC (1984) Differential susceptibility of field bindweed (Convolvulus arvensis) biotypes to glyphosate. Weed Sci 32:472–476
Dinelli G, Marotti I, Bonetti A, Minelli M, Catizone P, Barnes J (2006) Physiological and molecular insight on the mechanisms of resistance to glyphosate in Conyza canadensis L. Cronq Biotypes Pestic Biochem Physiol 86:30–41
Duke SO, Powles SB (2008) Glyphosate: a once-in-a-century herbicide. Pest Manag Sci 64:319–325
Eichholtz DA, Alan D, Gasser CS, Scott C, Kishore GM, Murthy G (2001) Modified gene encoding glyphosate-tolerant 5-enolpruvyl-3-phosphoshikimate synthase. United States Patent No. 6,225,114
Gaines TA, Zhang WL, Wang DF, Bukun B, Chisholm ST, Shaner DL, Nissena SJ, Patzoldt WL, Tranel PJ, Culpepper AS, Grey TL, Webster TM, Vencill WK, Sammons RD, Jiang J, Preston C, Leach JE, Westra P (2010) Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci USA 107:1029–1034
Gaines TA, Shaner DL, Ward SM, Leach JE, Preston C, Westra P (2011) Mechanism of resistance of evolved glyphosate resistant Palmer amaranth (Amaranthus palmeri). J Agric Food Chem 59:5886–5889
Garg B, Vaid N, Tuteja N (2014) In-silico analysis and expression profiling implicate diverse role of EPSPS family genes in regulating developmental and metabolic processes. BMC Res Notes 7:58
Ge X, Avignon DA, Ackerman JJ, Sammons RD (2010) Rapid vacuolar sequestration: the horseweed glyphosate resistance mechanism. Pest Manag Sci 66:345–348
Ge X, Avignon DA, Ackerman JJ, Duncan B, Spaur MB, Sammons RD (2011) Glyphosate-resistant horseweed made sensitive to glyphosate: low-temperature suppression of glyphosate vacuolar sequestration revealed by 31P NMR. Pest Manag Sci 67:1215–1221
He M, Nie YF, Xu P (2003) A T42 M substitution in bacterial 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) generates enzymes with increased resistance to glyphosate. Biosci Biotechnol Biochem 67:1405–1409
Healy-Fried ML, Funke T, Priestman MA, Han H, Schönbrunn E (2007) Structural basis of glyphosate tolerance resulting from mutations of Pro101 in Escherichia coli 5-enolpyruvylshikimate-3-phosphate Synthase. J Biol Chem 282:32949–32955
Heap IM (2015) International Survey of Herbicide-Resistant Weeds. http://www.weedscience.org/. Accessed 10 April 2015
Henry WB, Shaner DL, Mark SE (2007) Shikimate accumulation in sunflower, wheat and prosomillet after glyphosate application. Weed Sci 55:1–5
Huang LH, Jiang X, Li B, Li YQ, Zhang XW (2009) Transformation of EPSP synthase gene from Allium macrostemon Bunge into tobacco and improvement of resistance in transgenic plants to glyphosate. Acta Agron Sin 35:855–860
Huang ZF, Zhang CX, Huang HJ, Wei SH, Liu Y, Cui HL, Chen JC, Yang L, Chen JY (2014) Molecular cloning and characterization of 5-enolpyruvylshikimate-3-phosphate synthase gene from Convolvulus arvensis L. Mol Biol Rep 41:2077–2084
Huang ZF, Wang GR, Huang HJ, Wei SH, Zhou XX, Chen JY, Chen JC, Zhang CX (2015) Isolation and functional analysis of Convolvulus arvensis EPSPS promoter. Plant Mol Biol Rep. doi:10.1007/s11105-015-0861-2
Jasieniuk M, Ahmad R, Sherwood AM, Firestone JL, Perez-Jones A, Lanini WT, Mallory-Smith C, Stednick Z (2008) Glyphosate-resistant Italian ryegrass (Lolium multiflorum) in California: distribution, response to glyphosate, and molecular evidence for an altered target enzyme. Weed Sci 56:496–502
Jugulam M, Niehues K, Godar AS, Koo DH, Danilova T, Friebe B, Sehgal S, Varanasi VK, Wiersma A, Westra P, Stahlman PW, Gill BS (2014) Tandem amplification of a chromosomal segment harboring 5-enolpyruvylshikimate-3-phosphate synthase locus confers glyphosate resistance in Kochia scoparia. Plant Physiol 166:1200–1207
Kahrizi D, Salmanian A, Afshari A, Moieni A, Mousavi A (2007) Simultaneous substitution of Gly96 to Ala and Ala183 to Thr in 5-enolpyruvylshikimate-3-phosphate synthase gene of E. coli (k12) and transformation of rapeseed (Brassica napus L.) in order to make tolerance to glyphosate. Plant Cell Rep 26:95–104
Kaundun SS, Zelaya IA, Dale RP, Lycett AJ, Carter P, Sharpies KR, McIndoe E (2008) Importance of the P106S target-site mutation in conferring resistance to glyphosate in a goosegrass (Eleusine indica) population from the Philippines. Weed Sci 56:637–646
Kaundun SS, Dale RP, Zelaya IA, Dinelli G, Marotti I, McIndoe E, Cairns A (2011) A novel P106L mutation in EPSPS and an unknown mechanism(s) act additively to confer resistance to glyphosate in a South African Lolium rigidum population. J Agric Food Chem 59:3227–3233
Kim J, Kim D, Forest F, Fay M, Chase M (2010) Molecular phylogenetics of Ruscaceae sensu lato and related families (Asparagales) based on plastid and nuclear DNA sequences. Ann Bot 106:775–790
Lanzetta PA, Alvarez LJ (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem 100:95–97
Lattier J, Ranney T, Fantz PR, Avent T (2014) Identification, nomenclature, genome sizes, and ploidy levels of Liriope and Ophiopogon Taxa. HortSci 49:145–151
Lorraine-Colwill DF, Powles SB, Hawkes TR, Hollinshead PH, Warner SAJ, Preston C (2002) Investigations into the mechanism of glyphosate resistance in Lolium rigidum. Pestic Biochem Physiol 74:62–72
Malone JM, Morran S, Shirley N, Boutsalis P, Preston C (2015) EPSPS gene amplification in glyphosate-resistant Bromus diandrus. Pest Manag Sci. doi:10.1002/ps.4019
Nandula VK, Reddy KN, Duke SO, Poston DH (2005) Glyphosate-resistant weeds: current and future outlook. Outlook Pest Manag 16:183–187
Nandula VK, Ray JD, Ribeiro DN, Pan Z, Reddy KN (2013) Glyphosate resistance in tall waterhemp (Amaranthus tuberculatus) from Mississippi is due to both altered target-site and nontarget-site mechanisms. Weed Sci 61:374–383
Ng CH, Wickneswari R, Salmijah S, Teng YT, Ismail BS (2003) Gene polymorphisms in glyphosate-resistant and -susceptible biotypes of Eleusine indica from Malaysia. Weed Res 43:108–115
Owen MDK, Zelaya IA (2005) Herbicide-resistant crops and weed resistance to herbicides. Pest Manag Sci 61:301–311
Perez-Jones A, Park KW, Polge N, Colquhoun J, Mallory-Smith CA (2007) Investigating the mechanisms of glyphosate resistance in Lolium multiflorum. Planta 226:395–404
Powles SB, Yu Q (2010) Evolution in action: plants resistant to herbicide. Ann Rev Plant Biol 61:317–347
Ribeiro DN, Pan Z, Duke SO, Nandula VK, Baldwin BS, Shaw DR, Dayan FE (2014) Involvement of facultative apomixis in inheritance of EPSPS gene amplification in glyphosate-resistant Amaranthus palmeri. Planta 239:199–212
Ritz C, Streibig JC (2005) Bioassay analysis using R. J Stat Softw 12:1–22
Robertson RR (2010) Physiological and biochemical characterization of glyphosate resistant Ambrosia trifida L. MS Thesis, Purdue University
Salas RA, Dayan FE, Pan Z, Watson SB, Dickson JW, Scott RC, Burgos NR (2012) EPSPS gene amplification in glyphosate- resistant Italian ryegrass (Lolium perennessp. multiflorum) from Arkansas. Pest Manag Sci 68:1223–1230
Sammons RD, Gaines TA (2014) Glyphosate resistance: state of knowledge. Pest Manag Sci 70:1367–1377
Singh BK, Shaner DL (1998) Rapid determination of glyphosate injury to plants and identification of glyphosate-resistant plants. Weed Technol 12:527–530
Song XL, Wu JJ, Zhang HJ, Qiang S (2011) Occurrence of glyphosate-resistant horseweed (Conyza canadensis) population in China. Agric Sci China 10:1049–1055
Tian YS, Xiong AS, Xu J, Zhao W, Gao F, Fu XY, Xu H, Zheng JL, Peng RH, Yao QH (2010) Isolation from Ochrobactrum anthropi of a novel class II 5-enopyruvylshikimate-3-phosphate synthase with high tolerance to glyphosate. Appl Environ Microbiol 76:6001–6005
Tian YS, Xu J, Peng RH, Xiong AS, Xu H, Zhao W, Fu XX, Han HJ, Yao QH (2013) Mutation by DNA shuffling of 5-enolpyruvylshikimate-3-phosphate synthase from Malus domestica for improved glyphosate resistance. Plant Biotechnol J 11:829–838
Tong XH, Daud MK, Sun YQ, Zhu SJ (2009) Physiological and molecular mechanisms of glyphosate tolerance in an in vitro selected cotton mutant. Pestic Biochem Phys 94:100–106
Tranel PJ, Riggins CW, Bell MS, Hager AG (2011) Herbicide resistances in Amaranthus tuberculatus: a call for new options. J Agric Food Chem 59:5808–5812
Van Hoogmoed J, Gilliam CH, Wehtje GR, Knight PR, Foshee WG, Olive JW, Murphy AM (2013) Effects of repeated applications of Roundup Pro® over the top of container-grown nursery crops. J Environ Hort 31:234–240
Vila-Aiub MM, Balbi MC, Distéfano AJ, Fernández L, Hopp E, Yu Q, Powles SB (2012) Glyphosate resistance in perennial Sorghum halepense (Johnsongrass), endowed by reduced glyphosate translocation and leaf uptake. Pest Manag Sci 68:430–436
Wakelin AM, Preston C (2006) A target-site mutation is present in a glyphosate-resistant Lolium rigidum population. Weed Res 46:432–440
Westhoven AM, Kruger GR, Gerber CK, Stachler JM, Loux MM, Johnson WG (2008) Characterization of selected common lambsquarters (Chenopodium album) biotypes with tolerance to glyphosate. Weed Sci 56:685–691
Wiersma AT, Gaines TA, Preston C, Hamilton JP, Giacomini D, Robin Buell C, Leach JE, Westra P (2015) Gene amplification of 5-enol-pyruvylshikimate-3-phosphate synthase in glyphosate-resistant Kochia scoparia. Planta 241:463–474
Woodburn AT (2000) Glyphosate: production, pricing and use worldwide. Pest Manag Sci 56:309–312
Yu Q, Cairns A, Powles SB (2007) Glyphosate, paraquat, and ACCase multiple herbicide resistance evolved in a Lolium rigidum biotype. Planta 225:499–513
Yu Q, Abdallah I, Han HP, Owen M, Powles SB (2009) Distinct non-target site mechanisms endow resistance to glyphosate, ACCase and ALS-inhibiting herbicides in multiple herbicide-resistant Lolium rigidum. Planta 230:713–723
Yu Q, Jalaludin A, Han HP, Chen M, Sammons RD, Powles SB (2015) Evolution of a double amino acid substitution in the 5-enolpyruvylshikimate-3-phosphate synthase in Eleusine indica conferring high-level glyphosate resistance. Plant Physiol 167:1440–1447
Yuan CI, Chen YM, Chaing MY (2001) Responses of Dicliptera chinensis to glyphosate. Plant Prot Bull Taiwan 43:29–38
Yuan CI, Chaing MY, Chen YM (2002) Triple mechanisms of glyphosate-resistance in a naturally occurring glyphosate-resistant plant Dicliptera chinensis. Plant Sci 163:543–554
Zhang M, Liu Y, Zhang CX, Wei SH, Huang HJ (2011) Glyphosate-tolerant mechanisms in field bindweed Convolvulus arvensis. Acta Phytophylacica Sin 38:551–556
Zhang C, Feng L, He TT, Yang CH, Chen GQ, Tian XS (2015) Investigating the mechanisms of glyphosate resistance in goosegrass (Eleusine indica) population from South China. J Integr Agr 14:909–918
Zhou ZH, Huang LH, Jiang X, Li YQ, Zhang XW (2009) Cloning and prokaryotic expression of EPSP synthase gene cDNA of Allium macrostemon Bunge. Sci Agr Sin 42:2297–2304
Acknowledgments
This research was financially supported by China Transgenic Organism Research and Commercialization Project (2014ZX08011), National Science and Technology Pillar Program (2012BAD19B02), and the 111 project. The authors thank Vijay K. Nandula and Franck Dayan (USDA, USA) for the helpful comments and the improvement of the manuscript. The authors, however, accept full responsibility for the results and their interpretation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mao, C., Xie, H., Chen, S. et al. Multiple mechanism confers natural tolerance of three lilyturf species to glyphosate. Planta 243, 321–335 (2016). https://doi.org/10.1007/s00425-015-2408-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2408-z