Abstract
The inheritance of glyphosate resistance in two Amaranthus palmeri populations (R1 and R2) was examined in reciprocal crosses (RC) and second reciprocal crosses (2RC) between glyphosate-resistant (R) and -susceptible (S) parents of this dioecious species. R populations and Female-R × Male-S crosses contain higher 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene copy numbers than the S population. EPSPS expression, EPSPS enzyme activity, EPSPS protein quantity, and level of resistance to glyphosate correlated positively with genomic EPSPS relative copy number. Transfer of resistance was more influenced by the female than the male parent in spite of the fact that the multiple copies of EPSPS are amplified in the nuclear genome. This led us to hypothesize that this perplexing pattern of inheritance may result from apomictic seed production in A. palmeri. We confirmed that reproductively isolated R and S female plants produced seeds, indicating that A. palmeri can produce seeds both sexually and apomictically (facultative apomixis). This apomictic trait accounts for the low copy number inheritance in the Female-S × Male-R offsprings. Apomixis may also enhance the stability of the glyphosate resistance trait in the R populations in the absence of reproductive partners.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The non-selective herbicide glyphosate (N-(phosphonomethyl)glycine) has become the leading herbicide worldwide, largely because of its use with transgenic, glyphosate-resistant (GR) crops (Duke and Powles 2008). Glyphosate inhibits 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), an enzyme of the shikimate pathway, thereby, preventing the biosynthesis of the aromatic amino acids phenylalanine, tyrosine and tryptophan (Siehl et al. 1997; Steinrücken and Amrhein 1980). GR crop technology was utilized on ca. 128 million ha in 2012 worldwide (James 2012), boosting farm income globally by $32.2 billion (Brookes and Barfoot 2013) and significantly reducing environmental impact of weed management (Bonny 2011; Gardner and Nelson 2008; Kleter and Kuiper 2003).
Although strong arguments were made against the likelihood of weeds evolving resistance to glyphosate (Bradshaw et al. 1997), the first case of a GR weed was reported in Lolium rigidum (Gaud) about 20 years following the introduction of the herbicide glyphosate (Heap 2013; Powles et al. 1998; Pratley et al. 1999). Resistance to glyphosate has now been confirmed in 23 other weed species (Heap 2013). Evolved glyphosate resistance has recently emerged in Amaranthus palmeri (S. Wats.) populations (Culpepper et al. 2006).
Amaranthus palmeri is a tall, upright, and dioecious summer annual species that is native to the Mexican states of Sonora and Baja California, as well as parts of southern Arizona and California, USA (Ehleringer 1983). A. palmeri was first described from specimens collected from San Diego County, CA, and along the banks of the Rio Grande River (Watson 1877). The species has since spread into the southeastern United States, where it has become a common and competitive weed in row crop production. Interference from A. palmeri affects the growth and yield of several crops that are now mostly GR crops in the USA: maize (Zea mays L.) (Massinga et al. 2001), soybean [Glycine max (L.) Merr.] (Bensch et al. 2003), and cotton (Gossypium hirsutum L.) (Morgan et al. 2001). Currently, GR A. palmeri infests more than two million ha in 17 US states (Heap 2013). GR A. palmeri has become the single greatest threat to the economic sustainability of cotton production in infested fields, largely due to the lack of adequate control provided by available post emergence herbicides (Mayo et al. 1995).
The steady increase in the number of weed species with evolved resistance to glyphosate has been exacerbated with the introduction and widespread adoption of GR crops (Duke and Powles 2009; James 2012). In evolved resistance, single base pair mutations in EPSPS have not provided a high level of resistance (Baerson et al. 2002; Ng et al. 2003; Perez-Jones and Mallory-Smith 2010). Alteration of more than one codon, as with site-directed mutagenesis to produce the GA21 version of maize EPSPS, has resulted in commercial GR maize with a much higher level of resistance (Green 2009). GA21-type mutations resulting from the double Thr to Ile101 and Pro to Ser 106 (TIPS) mutation was recently reported in a Malaysian goosegrass biotype (Jalaludin et al. 2013). The most recently evolved GR weed populations have other mechanisms of resistance based on amplification of the EPSPS gene(s) (Gaines et al. 2010) or sequestration of glyphosate in the vacuole (Ge et al. 2010, 2011, 2012).
Gene amplification of target site genes is a known resistance mechanism for insecticides (Bass and Field 2011) and fungicides (Selmecki et al. 2008). In the laboratory, step-wise increases in glyphosate concentrations of plant cell cultures has been selected for gene amplification of EPSPS (Pline-Srnic 2006). Amplification of the EPSPS gene is a mechanism for field-evolved glyphosate resistance in A. palmeri (Gaines et al. 2010), Lolium perenne L. ssp. multiflorum (Lam.) Husnot (Salas et al. 2012) and Kochia scoparia (L.) Schrad. (Wiersma 2012). Glyphosate is the only herbicide for which there is field-evolved amplification of a target gene known to confer resistance.
In the case of A. palmeri, glyphosate resistance correlates with increased copy number of EPSPS genes, transcripts, protein content, and enzymatic activity (Gaines et al. 2011). These correlations indicate that between 30 and 50 EPSPS copies endow complete resistance to the normal field rate of glyphosate (0.5–1 kg ha−1). These extra copies are found throughout the genome, on every chromosome (Gaines et al. 2010). The mechanism leading to the multiplication of gene copies has recently been linked to sequences with homology to miniature inverted-repeat transposable elements (MITEs) flanking the EPSPS gene copies in R individuals (Gaines et al. 2013). Other genetic elements such as a putative activator (Ac) transposase and a repetitive sequence region were also reported, though the precise mechanism of gene amplification is yet to be fully determined. Studies by Gaines and co-workers were all conducted on an A. palmeri population that evolved in the southeastern coastal plain of the USA. GR populations of this species were later found in 17 other US states extending throughout the south and as far west as California and as far north as Michigan (Heap 2013). This GR trait can be transferred via pollen to at least three other Amaranthus species (Gaines et al. 2012).
Very little is understood about the inheritance of this novel mode of resistance to glyphosate. The objectives of this research were to investigate the molecular mechanism of resistance to glyphosate in A. palmeri populations from Mississippi and investigate the mode of inheritance of this novel mechanism of resistance in this GR weed.
Materials and methods
Plant material and general experimental conditions
Seeds of suspected GR (R) populations of A. palmeri were collected from 35 GPS site coordinates across the northwestern region of Mississippi (MS), USA, in 2007 (Nandula et al. 2012). Each location corresponded to a population, with each population defined as a group of seed collected from 10 to 20 randomly selected female plants. A preliminary screening with 0.84-kg glyphosate ha−1 was conducted on these 35 populations. The C1B1 and T4B1 accessions were selected for subsequent research because they had the greatest number of surviving individuals. A known susceptible (S) population, collected in Washington County, MS, was included for comparison in all experiments. Seeds were stored at 10 °C until further use.
Experiments were carried out as described by Nandula et al. (2012). Seeds were planted at 1-cm depth in 50-cm by 20-cm by 6-cm plastic trays with holes containing a commercial potting mix (Metro-Mix 360®, Sun Gro Horticulture, Bellevue, WA, USA). Two weeks after emergence, A. palmeri plants were transplanted into 6-cm by 6-cm by 6-cm pots containing potting mix. Plants were greenhouse grown under 30/20 °C day/night temperature with a 14-h photoperiod, sub-irrigated as needed and fertilized with a nutrient solution (All Purpose Miracle-Gro®, The Scotts Company, Marysville, OH, USA) containing 200 mg L−1 each of N, P2O5, and K2O 1 week after transplanting and then once per month. Plants were sprayed at the four- to six-leaf stage with glyphosate (Roundup WeatherMAX®, Monsanto Company, St. Louis, MO, USA) using an 8002E nozzle and overhead compressed-air sprayer calibrated to deliver 140 L ha−1 at 280 kPa.
Production of genetic populations
The first controlled crosses (Female-R × Male-R, R/R) were produced by first treating the C1B1 or T4B1 accessions (Nandula et al. 2012) with 0.84-kg glyphosate ha−1, as previously described. At least 100 surviving individuals of each gender were selected and grown together in isolation from other populations in different greenhouses to ensure genetic purity of each controlled cross. Pollen from the male plants was spread on the female plants every morning over a period of 2 months. A. palmeri inflorescence spikes were hand-harvested where the majority of the seed-possessed coats were black in color (seed maturity). Seeds were air-dried in a greenhouse (25/20 °C day/night, 12-h photoperiod under natural sunlight conditions) for 7 days, cleaned, and stored at 10 °C until further use. Resistance was confirmed by treating a subset of whole plant first R/R controlled cross progeny with 0.84-kg glyphosate ha−1 before proceeding with the second controlled crosses (data not shown). Second controlled crosses (R/R) were developed using at least 100 first R/R controlled crosses surviving individuals that were treated with 0.84-kg glyphosate ha−1, as described for the development of the first R/R controlled crosses. A subset of second R/R controlled cross plants was grown and their resistance to glyphosate was confirmed following the previously described procedure (data not shown). Seeds from the C1B1 or T4B1 accessions having been through the first and second controlled crosses were, respectively, defined as resistant parents R1 and R2 accessions to generate subsequent genetic populations.
Reciprocal crosses (Female-S × Male-R, S/R, and Female-R × Male-S, R/S) were made by growing S females in isolation with each R male (R1 and R2), and vice versa in separated greenhouses with the aim of generating four reciprocal crossed (RC) populations, hereafter referred to as S/R1, S/R2, R1/S, and R2/S. Plants were pollinated as previously described and resistance confirmed by the application of glyphosate at 0.42 kg ae ha−1 to a subset of the RC generations. The majority (≥85 %) of S/R1 and S/R2 cross plants treated with glyphosate at the described rate were susceptible (data not shown) indicating homozygous susceptible individuals.
Second reciprocal crosses (Female-S/R × Male-S/R, S/R//S/R, and Female-R/S × Male-R/S, R/S//R/S) were made by growing each RC female in isolation with each RC male in separated greenhouses with the aim of creating four second reciprocal crossed (2RC) populations, hereafter referred to as S/R1//S/R1, S/R2//S/R2, R1/S//R1/S, and R2/S//R2/S. Plants were pollinated as previously described and resistance confirmed by the application of glyphosate at the low rate of 0.42 kg ae ha−1 to a subset of the 2RC generations. The majority (≥70 %) of plants of S/R1//S/R1 and S/R2//S/R2 populations were susceptible, some plants of all four 2RC populations had an intermediate level of injury, and the majority (≥80 %) of plants of R1/S//R1/S and R2/S//R2/S were resistant (data not shown).
These experiments were designed to determine the inheritance of glyphosate resistance at a population level, rather than at the level of individuals. Therefore, crosses were performed with groups of plants with predetermined sensitivity or resistance to glyphosate. For these experiments, we assumed that little or no reproductive fitness penalty resulted in generating these crosses.
DNA, RNA isolation and cDNA synthesis
Total DNA and RNA were extracted from frozen 2- to 4-leaf stage tissue of S, R1, R2, RC, and 2RC plants. Genomic DNA (gDNA) was extracted using DNeasy Plant Mini kit (Qiagen, Valencia, CA, USA). Total RNA was isolated with the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions and treated with the RNase-Free DNase (Qiagen). The quality and quantity of prepared genomic DNA and total RNA were assessed according to the MIQE Guidelines (Bustin et al. 2009). The DNA and RNA were quantified using a NanoDrop (ND-1000) spectrophotometer (Thermo Scientific, Wilmington, DE 19810) and checked for quality and integrity by TAE agarose gel electrophoresis. For quantitative real-time PCR (qPCR), cDNA was synthesized from 2 μg of total RNA in a 20 μL reaction volume using a proprietary blend of oligo(dT) and random hexamer primers according to the manufacturer’s instructions (iScript cDNA Synthesis Kit, Bio-Rad Laboratories, Hercules, CA, USA).
EPSPS cDNA sequencing
Total RNAs for cDNA cloning were isolated from S, R1 and R2 populations as described above. First strand cDNA synthesis was then performed using 1 μg total RNAs and M-MuLV reverse transcriptase in a final volume of 20 μL using oligo(dT)23VN according to the manufacturer’s instructions (New England BioLabs, Ipswich, MA, USA). A pair of primers (sense: 5′-TGGCTCAAGCTACTACCATCAAC-3′; antisense: 5′-ATATAGCTACTCAATGCTTGGCGAAC-3′) was designed based on the EPSPS coding sequence from A. palmeri (GenBank accession number FJ861242) (Gaines et al. 2010). PCR reaction contained 1 μL cDNA; 0.1 mM each of forward and reverse primers; 0.2 mM each of dATP, dCTP, dGTP, and dTTP; 2 mM MgCl2; and 1 U of proof reading PfuUltra high-fidelity DNA polymerase (Stratagene, La Jolla, CA, USA) with a 1× concentration of supplied buffer in a final volume of 50 μL. The cycling conditions were 2 min at 95 °C followed by 30 cycles of 20 s at 95 °C, 20 s at 55 °C, and 1 min at 72 °C, with a final extension of 3 min at 72 °C. PCR products were ligated into pCR Blunt TOPO vector using Zero Blunt TOPO PCR Cloning Kit (Invitrogen, Carlsbad, CA, USA), transformed into Escherichia coli TOP10 cells and grown on selection media. Single colonies of six transformants of R1 and R2, and 11 transformants from S were cultured overnight in liquid LB media for plasmid extraction. Plasmid DNAs were sequenced and assembled using Lasergene version 10.0 SeqMan (DNASTAR, Madison, WI, USA). Multiple DNA sequence alignments of EPSPS, including sequences from both R and S A. palmeri populations (Gaines et al. 2010), GenBank accession numbers FJ861242 and FJ861243, were constructed using ClustalW version 10.0 (DNASTAR). RNA extraction and amplification of the EPSPS gene was performed on pooled leaf samples from S, R1 and R2 populations.
Quantitative PCR
Quantitative real-time PCR (qPCR) was used to measure EPSPS genomic copy number relative to the gene for acetolactate synthase (ALS) and cDNA expression level of EPSPS relative to ALS in S, R1, R2, RC, and 2RC A. palmeri plants according to previously described procedures (Gaines et al. 2010). The ALS gene was used as a low-copy control gene with known monogenic inheritance in other Amaranthus species (Trucco et al. 2005). The first experiment measured EPSPS genomic copy number and expression level in the population (i.e., combined leaf material from at least ten individuals per population studied). The second experiment measured EPSPS genomic copy number by individuals of each population (i.e., 30 individuals per population were analyzed separately).
Protein extraction and EPSPS enzyme activity assay
Ten grams of fresh leaf tissue was frozen in a −80 °C refrigerator prior to being ground to fine powder in a chilled mortar. Total soluble protein (TSP) was extracted from 2- to 4-leaf stage tissue of S, R1, R2, RC, and 2RC plants. Protein extraction and EPSPS assay were conducted following published protocols (Sammons et al. 2007; Webb 1992) and described in detail in Dayan et al. (2013).
The specific activity of EPSPS from A. palmeri plants was determined using EnzCheck phosphate assay kit (Invitrogen) with a UV-3101 spectrophotometer (Shimadzu North America, Columbia, MD, USA) as described in detail elsewhere (Dayan et al. 2013). The effect of glyphosate was tested in triplicates at concentrations ranging from 0.3 to 1,000 μM in half-log intervals. Two experiments were conducted with three replications per population. The collected samples consisted of combined leaf material from at least ten individuals per population studied.
Western blot analysis for the detection of EPSPS protein
Total cellular protein was isolated from 2- to 4-leaf stage tissue of S, R1, R2, RC, and 2RC plants. A quantity of 0.25 g of fresh leaf tissue was ground to fine powder in a chilled mortar as described above. Powdered tissues were homogenized in 500 μL cold extraction buffer (100 mM MOPS, 5 mM EDTA, 10 % glycerin, and 50 mM KCl, pH 7.0) with freshly added protease inhibitor (1 tablet per 10 mL of buffer) (Roche Applied Science, Indianapolis, IN, USA). The samples were thawed at room temperature and vortexed for 10 s. The mixtures were kept cold and placed on a Geno/Grinder mechanical shaker (SPEX SamplePrep, Metuchen, NJ, USA) for 1 min at 1,750 strokes per min. This step was repeated three times. Subsequently, the extract was centrifuged for 5 min at 18,000g. The supernatant was transferred to a new vial and it was centrifuged for 5 min at 18,400g. This step was repeated one more time and extract was stored at −80 °C. Protein concentrations were determined using a Bradford assay (Bradford 1976).
Western blot analysis for the detection of EPSPS protein was performed diluting soluble protein in 950-μL Laemmli premixed protein sample buffer (Bio-Rad Laboratories) and 0.71 M β-mercaptoethanol to reach a final concentration of 15 μg μL−1. The sample was transferred to a heating block at 90 °C for 4 min and allowed to cool to room temperature. The soluble protein was separated on a 12 % Tris–HCl precast polyacrylamide gel (Bio-Rad Laboratories). After separation, the proteins were blotted onto 0.45 μM Immobilon-FL PVDF membranes (EMD Chemicals, San Diego, CA, USA) and the membrane was equilibrated with transfer buffer [0.2 % methanol and Tris/glycine buffer (2.5 mM Tris, 19.2 mM glycine, pH 8.3)]. The membrane was washed with Tris-buffered saline (TBS) (20 mM Tris, pH 7.5 and 500 mM NaCl) and incubated overnight at 4 °C with 3 % gelatin from cold water fish skin. Western blot was probed with an EPSPS specific antibody (2 mg mL−1) developed against recombinant maize EPSPS (Monsanto Company) at dilution 1:2,000 and re-probed against Alexa Fluor 635 goat anti-rabbit (Invitrogen) at dilution 1:2,000. Within 20 min of the final wash with TBS buffer and Tween-20, the array was scanned and analyzed on a Pharos FX Plus Molecular Imager system equipped with an external 635-nm laser and quantified by densitometric analysis using Quantity One analysis software (Bio-Rad Laboratories) and represented as CNT (counts mm−2). The collected samples consisted of combined leaf material from at least ten individuals per population studied with three replications per population.
Glyphosate dose–response bioassay
Plants from parental lines (R1 and R2) and from the first and second reciprocal crosses were submitted to a dose–response experiment by applying glyphosate at 0, 0.052, 0.105, 0.21, 0.42, 0.84, 1.68, 3.36, and 6.72 kg ae ha−1 at the 3- to 4-leaf growth stage. Two weeks after treatment (WAT) shoots were clipped at soil level and fresh weight was determined. Biomass data are reported as percentage of the non-treated control. The experiment was conducted two times and arranged in a completely randomized design with triplicates.
Shikimate bioassay
Shikimate content of tissues of A. palmeri populations was determined spectrophotometrically in microtiter plates according to Shaner et al. (2005). Glyphosate concentrations ranged from 10 to 333 μM. The absorbance at 380 nm was measured within 30 min in a PowerWave XS microplate reader (Biotek, Winooski, VT, USA). Shikimate in μg mL−1 was determined based on a standard curve. Two experiments were conducted with four replications per population.
Testing for facultative apomixis
To test for apomixis in A. palmeri parent (S, R1 and R2) populations, pollination bags were placed over main inflorescences prior to emergence and were examined daily to determine their sex. The male plants were discarded and the female plants were isolated in different geographically located greenhouses to ensure repeatability of the apomixis test and exclude external sources of pollen contamination. The inflorescences were enclosed in pollination bags for the whole duration of the experiments. A total of 44 S individuals were grown, Spring/Summer, 2011, in Oxford (MS), 36 R1 individuals in Starkville (MS), and 38 R2 individuals in Abbeville (MS). A. palmeri inflorescence spikes were hand-harvested where the majority of the seed-possessed coats were black in color (seed maturity). A. palmeri seeds were air-dried, cleaned, and stored at 10 °C as previously described. The sex segregation ratio of the offsprings was determined by germinating the apomictically produced seeds and growing the plants until their sex could be determined by observing their flowers. A similar experiment was carried out during the winter, when no A. palmeri plants remained alive outside the greenhouse, to insure that no pollen could accidentally fertilize our reproductively isolated female plants. The inflorescence were also enclosed in pollination bags to further prevent pollination.
Intraspecific genetic diversity and relationships
Genetic markers that might confirm apomixis in A. palmeri were selected based on information from available literature (Chan and Sun 1997; Chandi et al. 2013; Lanoue et al. 1996; Lee et al. 2009; Ma et al. 2008; Popa et al. 2010; Wassom and Tranel 2005; Wetzel et al. 1999; Xu et al. 2011). RAPD (random-amplified polymorphic DNA) and ISSR (inter-simple sequence repeat) methods were used because they can provide up to a population level of identification of intraspecific variation.
DNA extraction was carried out as previously described from three different plants of the R1 population (n = 3) and diluted to 2 ng μL−1. RAPD analyses were performed using 12 decamer primers (Online Resource 1) randomly selected from a list of 100 primers (NAPS Unit, University of British Columbia, Biotechnology Laboratory, Vancouver, BC, Canada). ISSR analyses were performed using 15 primers (Online Resource 1) randomly selected from a list of 100 primers (NAPS Unit, University of British Columbia) and five provided by Natascha Techen (National Center for Natural Products, University of Mississippi, Oxford, USA). PCR reaction contained 10 ng DNA; 0.1 μM each of forward and reverse primers; 200 μM each of dATP, dCTP, dGTP, and dTTP (Promega, Madison, WI, USA); 1.5 mM MgCl2; and 1 unit of Platinum Taq DNA polymerase (Invitrogen) with a 1× concentration of supplied buffer in a final volume of 13.6 μL. The cycling conditions included 3 min at 94 °C followed by 45 cycles of 30 s at 94 °C, 30 s at 50 °C, and 3 min at 72 °C, with a final extension of 7 min at 72 °C in a thermal cycler (PTC-225, MJ Research, Waterton, MA, USA). PCR products were analyzed by electrophoresis on a 2 % TAE agarose gel stained with ethidium bromide and visualized under UV light using Quantity One (Bio-Rad, version 4.3.0). The sizes of the PCR products were compared to the molecular size standard 1 kb plus DNA ladder (Invitrogen).
Statistical analysis
Data from EPSPS genomic copy number and gene expression, EPSPS protein quantity and shikimate levels were analyzed by ANOVA via the PROC GLM statement using SAS software (version 9.3, SAS Institute, Cary, NC, USA) to determine the main effects and interactions of the factors at P < 0.05. No significant experiment effect was observed in repeated experiments; therefore, data from those experiments were pooled.
Where ANOVA indicated significant differences between treatments, means separation were performed using Fisher’s protected least significant difference (LSD) test at P = 0.05 using SAS software. The data from EPSPS relative copy number were regressed against EPSPS cDNA relative expression level and against EPSPS enzyme activity.
The whole plant glyphosate dose–responses were analyzed using a four parameters log-logistic model (Seefeldt et al. 1995), whereas the in vitro dose–responses were analyzed with a three parameters log-logistic model using R software (version 2.15.2, R Foundation for Statistical Computing, Vienna, Austria) using package drc, drm and modelFit function. The graphs were generated with Sigma Plot (version 11, Systat Software, San Jose, CA, USA). Subsequently, the Student t test at P < 0.05 was used to determine any differences in potency between populations and the S population at the respective GR50 or IC50 effect level. The null hypothesis, H0: GR50 or IC50 populations/GR50 or IC50 S population = 1, was tested against the alternative hypothesis, Ha: GR50 or IC50 populations/GR50 or IC50 S population ≠ 1. This test was performed using the open-source R software using package drc, drm function, and the comparisons were given by means of the selectivity index (SI) function.
Results
EPSPS cDNA sequencing
There was little difference among EPSPS sequences from the R1 (six clones) and R2 (six clones) populations. Consequently, the consensus sequence represents residues common to all GR sequences (R_consensus) (Online Resource 2). Alignment of consensus sequences from R and S individuals had several polymorphisms in all S sequences in the alignment contig when compared with the reference S sequence (FJ861242, Gaines et al. 2010). This may be the result of combining seeds from several plants at the time of initial collection of this population.
The cDNA sequence analysis of the EPSPS gene in both GR (R1 and R2) populations revealed several nucleotide substitutions resulting in silent mutations when compared with the S population sequence. The S and R sequences had 99 and 100 % homology to the A. palmeri reference sequence FJ861242 (S) of Gaines et al. (2010), respectively (online resource 2). Only one of the three single nucleotide polymorphisms (SNP) between S and R clones (contig position 866) resulted in a non-synonymous mutation (arginine to lysine amino acid substitution) at position 215, based on the maize mature EPSPS numbering system (Fig. 1), in the GS A. palmeri (Lebrun et al. 1997).
Partial and deduced amino acid sequence alignment of the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene of glyphosate-susceptible (S) and glyphosate-resistant (R) A. palmeri populations. The boxed codon shows the substitutions of arginine (R) to lysine (K) at amino acid 215 and no substitution at proline (P) 106, when using the maize mature EPSPS numbering system. aFJ861243, glyphosate-resistant A. palmeri from Georgia (Gaines et al. 2010) sequence reference, sequence information can be found at http://www.ncbi.nlm.nih.gov/nuccore/ bFJ861242, glyphosate-susceptible A. palmeri from Georgia (Gaines et al. 2010) sequence reference, sequence information can be found at http://www.ncbi.nlm.nih.gov/nuccore/
EPSPS gene copy number correlates with EPSPS gene expression
Analysis of the genomic DNA extracted from pooled samples of several individuals for each population showed that on average, R1 and R2 populations contained 59 and 33 copies of the EPSPS gene, whereas the S population contained a single copy (Fig. 2b). Quantitative RT-PCR on cDNA revealed that the single copy of EPSPS gene was proportionally expressed, with a 1:1 correlation to EPSPS gene copy:EPSPS transcript richness (Figs. 2, 3). The EPSPS copy number was positively correlated with the gene expression level (r = 0.94, P < 0.0001, excluding R2 data, Fig. 3), and a pattern was observed where the reciprocal crosses generated by Female-S × Male-R (S/R) and by Female-S/R × Male-S/R (S/R//S/R) had lower EPSPS expression level than the reciprocal crosses generated by Female-R × Male-S (R/S) and by Female-R/S × Male-R/S (R/S//R/S). While the R2 population is shown in the graph (Fig. 3), it was excluded from the data used for the regression because it did not have a strong correlation between its relatively high number of gene copies and unexpectedly low number of mRNA transcript.
Glyphosate effect (a), genomic copy number of EPSPS (b), and relative transcription level of EPSPS (c) in glyphosate-susceptible (S), -resistant (R1 and R2), and first (R/S and S/R) and second (R/S//R/S and S/R//S/R) reciprocal crosses of A. palmeri populations. Vertical bars represent ± standard error of the mean (n = 20). Letters represent statistical difference based on Fisher’s LSD at P = 0.05
Positive correlation between increase in 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) relative to acetolactate synthase (ALS) genomic copy number with increase in EPSPS: ALS cDNA expression levels in glyphosate-susceptible (S), -resistant (R1 and R2), and first reciprocal crosses (R/S and S/R) of A. palmeri populations. Regression of the entire dataset indicated a good relationship between mRNA levels and copy numbers (solid line, r = 0.87, P < 0.0001). The R2 population did not fit the model as well as the other populations. Excluding this population from the dataset improved the strength of that relationship (dotted line, r = 0.94, P < 0.0001)
When individuals from each population were analyzed separately, there was a strong (P < 0.0001) relationship between EPSPS copy number and the populations (Fig. 4), with S, S/R1 and S/R2 with the fewest copy numbers. In fact, all of the S and 73 % of S/R1 and 70 % of S/R2 individuals analyzed had a single copy of the EPSPS gene. R1 and R2 populations had the highest copy numbers, and R1/S and R2/S had intermediate numbers (Fig. 4).
Genomic copy number of 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) relative to acetolactate synthase (ALS) genomic copy of 30 (n = 30) sampled individuals per A. palmeri population (dark circle), glyphosate-susceptible (S), -resistant (R1 and R2), and first (R/S and S/R) reciprocal crosses. Means of EPSPS relative gene copy number followed by the same letter are not significantly different by LSD test at 0.05. The EPSPS copy number segregation pattern observed in Fig. 2b, when combined sample population, is observed when averaging sampled individuals (dark gray square)
EPSPS gene copy number correlates with EPSPS enzyme activity and protein quantity
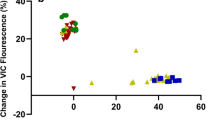
EPSPS specific activity in each A. palmeri population was inhibited by glyphosate concentrations (Online Resource 3, Fig. 5). The amounts of glyphosate needed to reduce the EPSPS activity by 50 % (IC50) ranged from 5.5 to 55 μM glyphosate (Online Resource 3), but there was no clear relationship between the IC50 values at the enzymatic level and in vivo sensitivity to glyphosate (i.e., the sensitive biotype had the highest IC50 value). Consequently, the R/S ratios for all populations were smaller than 1, suggesting that EPSPS from the GR plants was more sensitive to glyphosate than in the susceptible population.
Dose–response of glyphosate against EPSPS (5-enolpyruvylshikimate-3-phosphate synthase) enzyme activity of glyphosate-susceptible (S), -resistant (R1 and R2), and first reciprocal cross (R/S and S/R) of A. palmeri populations. Glyphosate inhibition assays were normalized for total soluble protein (TSP) quantity. S black cross, 1 relative EPSPS copy, IC50 (glyphosate concentration that reduced enzyme activity by 50 %) = 55 μM; R1 dark gray square, 59 relative copies, IC50 = 16 μM; R2 black triangle, 33 relative copies, IC50 = 21 μM; R1/S light gray square, 43 relative copies, IC50 = 15 μM; S/R1 open square, 19 relative copies, IC50 = 7 μM; R2/S light gray triangle, 30 relative copies, IC50 = 6 μM; S/R2 open triangle, 1 relative copy, IC50 = 26 μM. Pi inorganic phosphate. Vertical bars represent ± standard error of the mean (n = 6)
The specific activity of EPSPS varied greatly between populations in the absence of glyphosate. The S biotype had the lowest activity (0.02 μmol μg−1 protein min−1), whereas both R1 and R2 parent populations had high activity (0.19 to 0.12 μmol μg−1 protein min−1, respectively) (Fig. 6c). In the RC populations, the R1/S and R2/S had intermediate EPSPS activity and had consistently higher activity than the S/R1 and S/R2 crosses (r = 0.87, P < 0.0001, including and excluding R2 data) and EPSPS expression level relative to ALS (r = 0.97, P < 0.0001, excluding R2 data; r = 0.84, P < 0.0001, including R2 data) (Fig. 7). In the second reciprocal crosses (2RC), R1/S//R1/S had a level of EPSPS activity similar to R1. The other 2RC populations had intermediate levels of EPSPS specific activity, which was positively correlated with EPSPS relative copy number. Consistent with these observations, EPSPS protein level, as measured with immunoblotting, correlated strongly with other EPSPS-related parameters such as EPSPS gene copy number and enzymatic activity (Fig. 6).
Positive correlation among EPSPS (5-enolpyruvylshikimate-3-phosphate synthase) genomic copy number (a), EPSPS quantity (b), and specific activity of EPSPS enzyme (absence of inhibitor glyphosate) in glyphosate-susceptible (S), -resistant (R1 and R2), and first (R/S and S/R) and second (R/S//R/S and S/R//S/R) reciprocal crosses of A. palmeri populations. Pi inorganic phosphate; TSP total soluble protein. Vertical bars represent ± standard error of the mean (n = 6). Means of EPSPS activity with the same letter are not significantly different by Fisher’s LSD test at 0.05
Positive correlation between specific activity of EPSPS (5-enolpyruvylshikimate-3-phosphate synthase) enzyme and EPSPS relative to acetolactate synthase (ALS) genomic copy number (a) and EPSPS: ALS cDNA expression levels (b) in glyphosate-susceptible (S), -resistant (R1 and R2), and first reciprocal cross (R/S and S/R) of A. palmeri populations. a regression of the entire dataset indicated a strong relationship between enzyme activity and copy number (solid and dotted line, r = 0.87, P < 0.0001). b Regression of the entire dataset indicated a good relationship between mRNA levels and enzyme activity (line not shown r = 0.84, P < 0.0001). The R2 population did not fit the model as well as the other populations. Excluding this population from the dataset improved the strength of that relationship (solid line, r = 0.97, P < 0.0001)
EPSPS gene copy number correlates with level of glyphosate resistance
The shoot biomass of all A. palmeri population decreased in a dose-dependent manner with increasing glyphosate dose (Online Resource 4). Consistent with our preliminary data, the S population was the most sensitive to glyphosate, with a GR50 of 94.4 g ae ha−1 (Table 1). The R1 and R2 populations were 17 and 14.5 times more resistant, respectively, to glyphosate than S.
The S biotype had the highest shikimate accumulation at all glyphosate treatment concentrations, differing from the grouped R1 and R2 populations, which had no accumulation except for the R2 at 333-μM glyphosate (Fig. 8). Shikimate accumulation was highly dependent on glyphosate concentration and between populations (P < 0.005 for both).
Effect of glyphosate concentration on shikimate levels in excised leaf discs of glyphosate-resistant (R1 and R2) and -susceptible (S) A. palmeri populations. Means of shikimate level with the same letter are not significantly different by Fisher’s LSD test at 0.05. Vertical bars represent ± standard error of the mean (n = 8)
In all cases, the progeny of the RC and 2RC crosses had less resistance to glyphosate (lower GR50) than the resistant parent (Table 1). Interestingly, the GR50 for the RC were closer to their respective maternal parent than the midpoint (Table 1). The 2RC dose–response was intermediate between those of the R and S populations, containing both highly susceptible and highly resistant individuals and a range of intermediate phenotypes (Table 1). Consistent with the pattern observed with the number of gene copy numbers, mRNA levels and protein expression, the greenhouse data suggest that the mode of inheritance of GR trait was maternally affected.
Facultative apomixis and intraspecific genetic diversity
Involvement of apomixis in glyphosate resistance inheritance was tested by measuring seed production in reproductively isolated female plants. In that experiment, reproductively isolated female individuals (44 S, 36 R1 and 38 R2) were studied. In all cases, seeds were produced with the exception of one R1 plant. From 60 to 100 % (depending on the population) of individuals studied produced 1–1,000 seeds, and some S individuals produced as many as 6,000 seeds (Table 2). The female:male ratio of the apomictically produced offspring was close to 1:1.
Attempts to monitor apomixis using RAPD and ISSR genetic markers were unsuccessful (Online Resources 5 and 6). No significant RAPD and ISSR polymorphism was observed in a preliminary test experiment on a subset of three R1 individuals. Therefore, none of these markers would have been useful in differentiating apomictically produced seeds from seeds resulting from a fertilized egg through sexual reproduction (Chandrika et al. 2010; Lata et al. 2010, 2011).
Discussion
Analysis of the EPSPS sequences from the S and R populations of A. palmeri from Mississippi revealed several polymorphisms. Only one of these SNPs resulted in a non-synonymous mutation (arginine to lysine amino acid substitution) at position 215 (Fig. 1). However, this mutation was in the EPSPS sequence from the glyphosate sensitive wild-type and is therefore not linked to resistance. Importantly, all the sequences possessed the normal proline 106 residue.
These A. palmeri populations from Mississippi evolved resistance to glyphosate by gene amplification of EPSPS as first reported by Gaines et al. (2010) in Georgia and in other species by others (Salas et al. 2012; Wiersma 2012). The populations have as many as 59 copies of EPSPS (Fig. 2b), which is similar to that reported in other GR biotypes of A. palmeri. The biotypes from Georgia had individual plants with as many as 160 copies of the EPSPS gene (Gaines et al. 2010), and six individuals from the S × R cross had 1–39 copies (Gaines et al. 2011). A GR population from North Carolina possessed 22–63 copies of EPSPS gene (Chandi et al. 2012).
As with other cases where resistance is associated in amplification of multiple EPSPS gene copy numbers, the specific activity of EPSPS in our R1 and R2 populations was six- to tenfold higher than in S plants (Fig. 6c and Online Resource 3). Similar differences in specific enzyme activity were observed in GR and GS A. palmeri from Georgia (Gaines et al. 2010), L. perenne L. ssp. multiflorum from Arkansas (Salas et al. 2012) and K. scoparia from the US central Great Plains (Wiersma 2012). R1 and R2 plants had a nine- to sixfold increase, respectively, in EPSPS enzyme activity relative to that of the S plants. Other A. palmeri populations had a 16-fold increase in specific activity between GR and GS (Gaines et al. 2010) and there was an average sixfold higher basal enzyme activity in GR L. perenne compared to the GS biotype (Salas et al. 2012).
In our study, the 2RC genomic copy number varied greatly and was an intermediate value between those of the R and S populations (R1/S//R1/S: 53, S/R1//S/R1: 5, R2/S//R2/S: 15, and S/R2//S/R2: 6), containing both highly susceptible and highly resistant individuals and a range of intermediate genotypes (Fig. 2b). The molecular mechanism leading to the insertion of multiple EPSPS copies throughout the genome of A. palmeri (Gaines et al. 2010) is not fully understood, but it is linked to the presence of MITEs flanking the EPSPS gene copies in R individuals (Gaines et al. 2013). Furthermore, this process is very dynamic, where one individual A. palmeri (S/R × S/R) had a higher relative copy number than the sum of their relative copy number from both parents (Gaines et al. 2010).
Reciprocal crosses generated by Female-R × Male-S (R/S) and by Female-R/S × Male-R/S (R/S//RS) of the populations tended to have higher EPSPS copy number than the other crosses (Fig. 2b). This pattern of inheritance where the direction of the cross affected the level of resistance suggests a maternal influence.
While the transfer of glyphosate resistance via pollen movement is evident in A. palmeri, the rate of transfer is inversely proportional to the distance from a R male parent (Sosnoskie et al. 2012). In fact, that study in which only GS female plants surrounded a central group of GR male plants, no resistance was transferred to the seeds of female plants located 600 m or more from the resistant male pollen donor. Nevertheless, GS seeds were produced by plants at all distances from the GR male plants (40 % at 5 m to 80 % at 250 m), and there was little difference in the number of seeds produced at distances of 50 m and farther from the pollen source. The authors proposed that these findings may be due to fertilization by autopollination or from pollen providers outside the boundary of the experiment. These authors discounted agamospermy (apomixis) because the proportion of male to female offsprings approximated 1:1. However, all but one of the reproductively isolated A. palmeri plants in our study underwent apomixis and produced seeds (Table 2) that yielded a similar ratio of male to female offspring.
Little is known about the inheritance of this incompletely dominant, nuclear encoded GR trait, but it might follow a polygenic additive pattern in populations of A. palmeri from Georgia and North Carolina (Chandi et al. 2012; Gaines 2009). However, both of these studies observed an unpredictable behavior in the inheritance of glyphosate resistance in some populations, which may be the result of facultative apomixis, even though the overall contribution of this process may be small at the population level under most conditions. Indeed, a relatively small number of seeds are produced by apomixis (Table 2) when compared with ‘normal’ seed production per female plant in the field reaching 200,000–600,000 seeds (Keeley et al. 1987). Nonetheless, production of apomictic seeds would be sufficient to maintain the resistant trait in a population.
Our observations that A. palmeri can produce seeds both sexually and apomictically (facultative apomixis) are consistent with some of the data by other groups (Chandi et al. 2012; Gaines 2009; Gaines et al. 2010, 2011; Sosnoskie et al. 2012). The occurrence of agamospermy in A. palmeri is also evident in several studies demonstrating that crosses between A. palmeri and A. rudis can yield non-hybrid progeny (Franssen et al. 2001; Steinau et al. 2003; Trucco et al. 2007; Wetzel et al. 1999). The ability of plants to reproduce apomictically is particularly advantageous because it “guarantees a protracted existence to the individual genotypes, over long periods and over wide areas” (Gustafsson 1947).
The current understanding of sex determination in dioecious species suggests that the change from unisexuality to bisexuality has been a short step in evolution (Chattopadhyay and Sharma 1991). Apomictic seeds derived from unreduced cells (apomeiosis) should be 100 % female. However, reproductively isolated A. palmeri individuals produced 1:1 female:male ratio of offspring apomictically. The production of both male and female apomictic offspring would require meiotic events and subsequent fusion (to restore the somatic number). Such events are known among apomictic species as synergid or antipodal apogamy. If the Y male sex determinant in A. palmeri were a non-heteromorphic chromosome, then it would segregate and assort during meiosis I with the other chromosomes of the genome. If gender of this dioecious species were determined by homozygous presence or absence of genes, then synergids or antipodals with W (conferring femaleness) or Y (conferring maleness) would produce genotypes; WW (a female) or YY (a male) in a 1:1 phenotypic ratio. While Westergaard (1958) reports the normal (sexual) male of Acnida spp. (an amaranth) to be heterozygous (XY); he also reports 8 of 18 plant families investigated showing possibility of the homozygous genotype (however, viability is unknown for this amaranth species). It is possible that the offsprings we are observing in this 1:1 ratio are homozygous (normal) females and homozygous males.
Alternatively, if gender were determined by an X:autosome balance as found in Rumex and Humulus (Parker and Clark 1991; Jacobsen 1957, respectively), a 1:1 ratio of male to female would be possible in apomictic offspring. In this system, the X:autosome ratio determines the sexual fate of floral primordial, and thus gender of the plant, rather than the presence or absence of the Y chromosome. In diploid Rumex, XX are female and XYY are male. In polyploidy species of the genus Rumex, an X:autosome ratio of 1.0 or higher is female; a ratio of 0.5 or lower is male (Parker and Clark 1991). In domestic hops (Humulus spp.) a XY system is known. XX is female while XY is male. Polyploid variants (multiple X) of hops show XXXX to be female and XXYY to be male. In the above synergid/antipodal apogamy system proposed, meiosis I could change the ratio by eliminating the X chromosome from (half) the megaspore mother cells being generated, thereby generating males (00:autosomes) and females (XX:autosomes) in a 1:1 ratio.
A third possibility is that the amplification of EPSPS copy number is altering gene sequences that determine gender. Since A. palmeri does not have a heteromorphic sex chromosome (Grant 1959a; Westergaard 1958), it is possible that glyphosate-induced EPSPS gene copy number amplification occurs concurrent with or adjacent to segments of the genome that cause female/maleness. Selection for high EPSPS copy number would alter the ratio of sex factor:autosome resulting in the production of male and female apomictic offspring, however, a fixed ratio of 1:1 is unlikely.
However, A. palmeri/A. rudis non-hybrid apomictic offsprings were all females (Trucco et al. 2007). A. palmeri does not have a heteromorphic sex chromosome (Grant 1959a; Westergaard 1958), and the factors that determine sex are unknown. The dioecious condition may have evolved in Amaranthus after the split of 32/34 chromosomes, and sex expression is likely under the control of one or more genes (Grant 1959b). It is possible that the sex determination factors are present in both female and male dioecious plants. Glyphosate could affect sex expression by effects on sex-modifying hormones. Environmental stress on female A. palmeri is known to modulate hormonal balance (abscisic acid levels) in seeds (Jha et al. 2010). Therefore, the strong selection pressure imposed by glyphosate combined with its ability to reduce of indole-3-acetic acid (IAA) levels (e.g., Lee 1984) may contribute to this phenomenon.
The level of R alleles in a population under the selection pressure of glyphosate is dependent on the mode of inheritance of the resistance trait. Facultative apomixis would maintain the presence of individuals with high levels of EPSPS gene copy number in A. palmeri populations with GR individuals, even in the presence of overwhelming amounts of GS pollen or absence of pollen donors. A deeper understanding of the mechanism of gene amplification (Gaines et al. 2013) and the regulation of apomixis and sex determination factors in A. palmeri populations are essential for understanding the stability of multiple EPSPS gene copies in populations.
Abbreviations
- ALS:
-
Acetolactate synthase
- CNT:
-
Counts per mm2
- EPSPS:
-
5-Enolpyruvylshikimate-3-phosphate synthase
- GR:
-
Glyphosate-resistant
- GS:
-
Glyphosate-susceptible
References
Baerson SR, Rodriguez DJ, Tran M, Feng Y, Best NA, Dill GM (2002) Glyphosate-resistant goosegrass: identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Plant Physiol 129:1265–1275
Bass C, Field LM (2011) Gene amplification and insecticide resistance. Pest Manag Sci 67:886–890
Bensch CN, Horak MJ, Peterson D (2003) Interference of redroot pigweed (Amaranthus retroflexus), Palmer amaranth (A. palmeri), and common waterhemp (A. rudis) in soybean. Weed Sci 51:37–43
Bonny S (2011) Herbicide-tolerant transgenic soybean over 15 years of cultivation: pesticide use, weed resistance, and some economic issues. The case of the USA. Sustainability 3:1302–1322
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem 72:248–254
Bradshaw LD, Padgette SR, Kimball SL, Wells BH (1997) Perspectives on glyphosate resistance. Weed Technol 11:189–198
Brookes G, Barfoot P (2013) The global income and production effects of genetically modified (GM) crops 1996–2011. GM Crops Food 4:1–10
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Chan KF, Sun M (1997) Genetic diversity and relationships detected by isozyme and RAPD analysis of crop and wild species of Amaranthus. Theor Appl Genet 95:865–873
Chandi A, Milla-Lewis SR, Giacomini D, Westra P, Preston C, Jordan DL, York AC, Burton JD, Whitaker JR (2012) Inheritance of evolved glyphosate resistance in a North Carolina Palmer amaranth (Amaranthus palmeri) biotype. Int J Agron. doi:10.1155/2012/176108
Chandi A, Jordan DL, Milla-Lewis SR, Jordan DL, York AC, Burton JD, Zuleta CM, Whitaker JR, Culpepper AS (2013) Use of AFLP markers to assess genetic diversity in Palmer amaranth (Amaranthus palmeri) populations from North Carolina and Georgia. Weed Sci 61:136–145
Chandrika M, Rai VR, Thoyajaksha V (2010) ISSR marker based analysis of micropropagated plantlets of Nothapodytes foetida. Biol Plant 54:561–565
Chattopadhyay D, Sharma AK (1991) Sex determination in dioecious species of plants. Feddes Repertorium 102:29–55
Culpepper AS, Grey TL, Vencill WK, Kichler JM, Webster TM, Brown SM, York AC, Davis JW, Hanna WW (2006) Glyphosate-resistant Palmer amaranth (Amaranthus palmeri) confirmed in Georgia. Weed Sci 54:620–626
Dayan FE, Owens DK, Corniani N, Silva FML, Watson SB, Howell J, Shaner DL (2013) Biochemical markers and enzyme assays for herbicide mode of action and resistance studies. Weed Sci. doi:10.1614/WS-D-11-00194.1
Duke SO, Powles SB (2008) Glyphosate: a once in a century herbicide. Pest Manag Sci 64:319–325
Duke SO, Powles SB (2009) Glyphosate-resistant crops and weeds: now and in the future. AgBioForum 12:346–357
Ehleringer J (1983) Ecophysiology of Amaranthus palmeri, a sonoran desert summer annual. Oecologia 57:107–112
Franssen AS, Skinner DZ, Al-Khatib K, Horak MJ, Kulakow PA (2001) Interspecific hybridization and gene flow of ALS resistance in Amaranthus species. Weed Sci 49:598–606
Gaines TA (2009) Molecular genetics of glyphosate resistance in Palmer amaranth (Amaranthus palmeri L.). Ph.D. Dissertation, Colorado State University, Fort Collins
Gaines TA, Zhang W, Wang D, Bukun B, Chisholm ST, Shaner DL, Nissen SJ, Patzoldt WL, Tranel PJ, Culpepper AS, Grey TL, Webster TM, Vencill WK, Sammons RD, Jiang J, Preston C, Leach JE, Westra P (2010) Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci USA 107:1029–1034
Gaines TA, Shaner DL, Ward SM, Leach JE, Preston C, Westra P (2011) Mechanism of resistance of evolved glyphosate-resistant Palmer amaranth (Amaranthus palmeri). J Agric Food Chem 59:5886–5889
Gaines TA, Ward SM, Bukun B, Preston C, Leach JE, Westra P (2012) Interspecific hybridization transfers a previously unknown glyphosate resistance mechanism in Amaranthus species. Evol Appl 5:29–38
Gaines TA, Wright AA, Molin WT, Lorentz L, Riggins CW, Tranel PJ, Beffa R, Westra P, Powles SB (2013) Identification of genetic elements associated with EPSPS gene amplification. PLoS One 8:e65819
Gardner JG, Nelson GC (2008) Herbicides, glyphosate-resistance and acute mammalian toxicity: simulating an environmental effect of glyphosate-resistant weeds in the USA. Pest Manag Sci 64:470–478
Ge X, d’Avignon DA, Ackerman JJH, Sammons RD (2010) Rapid vacuolar sequestration: the horseweed glyphosate resistance mechanism. Pest Manag Sci 66:345–348
Ge X, d’Avignon DA, Ackerman JJH, Duncan B, Spaur MB, Sammons RD (2011) Glyphosate-resistant horseweed made sensitive to glyphosate: low-temperature suppression of glyphosate vacuolar sequestration revealed by 31P NMR. Pest Manag Sci 67:1215–1221
Ge X, d’Avignon DA, Ackerman JJH, Collavo A, Sattin M, Ostrander EL, Hall EL, Sammons RD (2012) Vacuolar glyphosate-sequestration correlates with glyphosate resistance in ryegrass (Lolium spp.) from Australia, South America, and Europe: A 31P NMR investigation. J Agric Food Chem 60:1243–1250
Grant WF (1959a) Cytogenetic studies in Amaranthus. III. Chromosome numbers and phylogenetic aspects. Can J Genet Cytol 1:313–328
Grant WF (1959b) Cytogenetic studies in Amaranthus. I. Cytological aspects of sex determination in dioecious species. Can J Bot 37:413–417
Green JM (2009) Evolution of glyphosate-resistant crop technology. Weed Sci 57:108–117
Gustafsson A (1947) Apomixis in higher plants. III. Biotype and species formation. Lunds Univ Arsskr 43:71–178
Heap I (2013) International Survey of Herbicide Resistant Weeds. Online. Internet. www.weedscience.org. Accessed 7 August 2013
Jacobsen P (1957) The sex chromosoes in Humulus L. Hereditas 43:357–370
Jalaludin A, Han H, Powles S (2013) Evolution in action: a double amino acid substitution in the EPSPS gene endows high-level glyphosate resistance. In: Proceedings of the global herbicide resistance challenge conference, Perth, p 35
James C (2012) 2012 ISAAA report on global status of Biotech/GM Crops. International Service for the Acquisition of Agri-Biotech Applications Brief 44-2012, Ithaca
Jha P, Norsworthy JK, Riley MB, Bridges W (2010) Shade and plant location effects on germination and hormone content of Palmer amaranth (Amaranthus palmeri) seed. Weed Sci 58:16–21
Keeley PE, Carter CH, Thullen RM (1987) Influence of planting date on growth of Palmer amaranth (Amaranthus palmeri). Weed Sci 35:199–204
Kleter GA, Kuiper HA (2003) Environmental fate and impact considerations related to the use of transgenic crops. In: Voss G, Ramos G (eds) Chemistry of plant protection. Wiley-VCH, Weinheim, pp 304–321
Lanoue KZ, Wolf PG, Browning S, Hood EE (1996) Phylogenetic analysis of restriction-site variation in wild and cultivated Amaranthus species (Amaranthaceae). Theor Appl Genet 93:722–732
Lata H, Chandra S, Techen N, Khan IA, ElSohly MA (2010) Assessment of the genetic stability of micropropagated plants of Cannabis sativa by ISSR markers. Planta Med 76:97–100
Lata H, Chandra S, Techen N, Khan IA, ElSohly MA (2011) Molecular analysis of genetic fidelity in Cannabis sativa L. plants grown from synthetic (encapsulated) seeds following in vitro storage. Biotech Lett 33:2503–2508
Lebrun M, Sailland A, Freyssinet G (1997) Mutated 5-enol pyruvylshikimate-3-phosphate synthase, gene coding for said protein and transformed plants containing said gene. World Patent Application No. WO9704103
Lee TT (1984) Release of lateral buds from apical dominance by glyphosate in soybean and pea seedlings. J Plant Growth Regul 3:227–235
Lee RM, Thimmapuram J, Thinglum KA, George G, Hernandez AG, Wright CL, Klim RW, Mikel MA, Tranel PJ (2009) Sampling the waterhemp (Amaranthus tuberculatus) genome using pyrosequencing technology. Weed Sci 57:463–469
Ma GG, Park YJ, Lee GA, Lee JR, Lee SY, Kwak JG, Kim TS, Cho EG (2008) SSR primer sets for identification of Amaranth species. Rep Korean Kongkae Taeho Kongbo Patent Oct 18, 2008, KR 2008092187, A20081015
Massinga RA, Currie RS, Horak MJ, Boyer J (2001) Interference of Palmer amaranth in corn. Weed Sci 49:202–208
Mayo CM, Horak MJ, Peterson DE, Boyer JE (1995) Differential control of four Amaranthus species by six postemergence herbicides in soybean (Glycine max). Weed Technol 9:141–147
Morgan GD, Baumann PA, Chandler JM (2001) Competitive impact of Palmer amaranth (Amaranthus palmeri) on cotton (Gossypium hirsutum) development and yield. Weed Technol 15:408–412
Nandula VK, Reddy KN, Koger CH, Poston DH, Rimando AM, Duke SO, Bond JA, Ribeiro DN (2012) Multiple resistance to glyphosate and pyrithiobac in Palmer amaranth (Amaranthus palmeri) from Mississippi and response to flumiclorac. Weed Sci 60:179–188
Ng CH, Wickneswary R, Salmijah S, Teng YT, Ismail BS (2003) Gene polymorphisms in glyphosate-resistant and -susceptible biotypes of Eleusine indica from Malaysia. Weed Res 43:108–115
Parker JS, Clark MS (1991) Dosage sex-chromosome systems in plants. Plant Sci 80:79–92
Perez-Jones A, Mallory-Smith C (2010) Biochemical mechanisms and molecular basis of evolved glyphosate resistance in weed species. In: Nandula VK (ed) Glyphosate resistance in crops and weeds. Wiley, Hoboken, pp 119–140
Pline-Srnic W (2006) Physiological mechanisms of glyphosate resistance. Weed Technol 20:290–300
Popa G, Cornea CP, Ciuca M, Babeanu N, Popa O, Marin D (2010) Studies on genetic diversity in Amaranthus species using the RAPD markers. Analele Universitătii din Oradea-Fascicula Biologie 17:280–285
Powles SB, Lorraine-Colwill DF, Delow JJ, Preston C (1998) Evolved resistance to glyphosate in rigid ryegrass (Lolium rigidum) in Australia. Weed Sci 46:604–607
Pratley J, Urwin N, Stanton R, Baines P, Broster J, Cullis K, Schafer D, Bohn J, Krueger R (1999) Resistance to glyphosate in Lolium rigidum. I. Bioevaluation. Weed Sci 47:405–411
Salas RA, Dayan FE, Pan Z, Watson SB, Dickson JW, Scott RC, Burgos NR (2012) EPSPS gene amplification in glyphosate-resistant Italian ryegrass (Lolium perenne ssp. multiflorum) from Arkansas. Pest Manag Sci 68:1223–1230
Sammons RD, Meyer J, Hall E, Ostrander E, Schrader S (2007) A simple continuous assay for EPSP synthase from plant tissue. http://www.cottoninc.com/2007-Glyphosate-Resistant-Palmer-Amaranth/11a-Industry-Sammons-NCWSS07-poster.pdf. Accessed 20 January 2012
Seefeldt SS, Jensen JE, Fuerst EP (1995) Log-logistic analysis of herbicide dose-response relationships. Weed Technol 9:218–227
Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J (2008) An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol 68:624–641
Shaner DL, Nadler-Hassar T, Henry WB, Koger CH (2005) A rapid in vivo shikimate accumulation assay with excised leaf discs. Weed Sci 53:769–774
Siehl DL, Subramaniam MV, Walters EW, Blanding JH, Niderman T, Weinmann C (1997) Evaluating anthranilate synthase as a herbicide target. Weed Sci 45:628–633
Sosnoskie LM, Webster MTM, Kichler JM, MacRae AW, Grey TL, Culpepper AS (2012) Pollen-mediated dispersal of glyphosate-resistance in Palmer amaranth under field conditions. Weed Sci 60:366–373
Steinau AN, Skinner DZ, Steinau M (2003) Mechanism of extreme genetic recombination in weedy Amaranthus hybrids. Weed Sci 51:696–701
Steinrücken H, Amrhein N (1980) The herbicide glyphosate is a potent inhibitor of 5-enolpyruvylshikimic acid-3-phosphate synthase. Biochem Biophys Res Com 94:1207–1212
Trucco F, Jeschke MR, Rayburn AL, Tranel PJ (2005) Promiscuity in weedy amaranths: high frequency of female tall waterhemp (Amaranthus tuberculatus) × smooth pigweed (A. hybridus) hybridization under field conditions. Weed Sci 53:46–54
Trucco F, Zheng D, Woodyard AJ, Walter JR, Tatum TC, Rayburn AL, Tranel PJ (2007) Nonhybrid progeny from crosses of dioecious Amaranths: implications for gene-flow research. Weed Sci 55:119–122
Wassom JJ, Tranel PJ (2005) Amplified fragment length polymorphism-based genetic relationships among weedy Amaranthus species. J Hered 96:410–416
Watson S (1877) Descriptions of new species of plants, with revisions of certain genera. Proc Am Acad Arts Sci 12:246–278
Webb MR (1992) A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci USA 89:4884–4887
Westergaard M (1958) The mechanism of sex determination in dioecious plants. Adv Genet 9:217–281
Wetzel DK, Horak MJ, Skinner DZ (1999) Use of PCR-based molecular markers to identify weedy Amaranthus species. Weed Sci 47:518–523
Wiersma A (2012) Regional whole plant and molecular response of Kochia scoparia to glyphosate. M.S. thesis, Colorado State University, p 55
Xu ZL, Ali Z, Yi JX, He XL, Zhang DY, Yu GJ, Khan AA, Khan IA, Ma HX (2011) Expressed sequence tag-simple sequence repeat-based molecular variance in two Salicornia (Amaranthaceae) populations. Genet Mol Res 10:1262–1276
Acknowledgments
This research was funded in part by Monsanto Company. We thank Dr. Natasha Techen for her advice with the genetic markers. Susan B. Watson, Marilyn Ruscoe and J’Lynn Howell provided excellent technical support. Dr. R. Douglas Sammons and Dr. Dafu Wang’s help with the Western blot analysis was greatly appreciated.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ribeiro, D.N., Pan, Z., Duke, S.O. et al. Involvement of facultative apomixis in inheritance of EPSPS gene amplification in glyphosate-resistant Amaranthus palmeri . Planta 239, 199–212 (2014). https://doi.org/10.1007/s00425-013-1972-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-013-1972-3