Abstract
Main conclusion
Arabidopsis was engineered to produce 21.2 % punicic acid in the seed oil. Possible molecular factors limiting further accumulation of the conjugated fatty acid were investigated.
Punicic acid (18:3Δ9cis,11trans,13cis) is a conjugated linolenic acid isomer and is a main component of Punica granatum (pomegranate) seed oil. Medical studies have shown that punicic acid is a nutraceutical with anti-cancer and anti-obesity properties. It has been previously demonstrated that the conjugated double bonds in punicic acid are produced via the catalytic action of fatty acid conjugase (FADX), which is a homolog of the oleate desaturase. This enzyme catalyzes the conversion of the Δ12-double bond of linoleic acid (18:2Δ9cis,12cis) into conjugated Δ11trans and Δ13cis-double bonds. Previous attempts to produce punicic acid in transgenic Arabidopsis thaliana seeds overexpressing P. granatum FADX resulted in a limited accumulation of punicic acid of up to 4.4 %, accompanied by increased accumulation of oleic acid (18:1∆9cis), suggesting that production of punicic acid in some way inhibits the activity of oleate desaturase (Iwabuchi et al. 2003). In the current study, we applied a new strategy to enhance the production of punicic acid in a high linoleic acid A. thaliana fad3/fae1 mutant background using the combined expression of P. granatum FADX and FAD2. This approach led to the accumulation of punicic acid at the level of 21 % of total fatty acids and restored the natural proportion of oleic acid observed in the A. thaliana fad3/fae1 mutant. In addition, we provide new insights into the high oleate phenotype and describe factors limiting the production of punicic acid in genetically engineered plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most commercial oilseeds contain saturated and unsaturated fatty acids, such as palmitic (16:0), stearic (18:0), oleic (18:1Δ9cis), linoleic (18:2Δ9cis,12cis) and α-linolenic acid (18:3Δ9cis,12cis,15cis) (Li et al. 2010). Typically, the double bonds of polyunsaturated fatty acids are separated by one or more methylene groups (−CH2−). In contrast, seeds of a limited number of plant species contain oil enriched in fatty acids with conjugated non-methylene interrupted double bonds (Cahoon et al. 1999). Conjugated linolenic acids (CLNAs) are found in the seed oils of plant species from several families including Curcubitaceae, Punicaceae, Bignoniaceae, Rosaceae, Chrysobalanaceae, Lythraceae, Balasaminaceae and Euphorbiaceae (Smith 1971; Badami and Patil 1980; Rawat et al. 2012).
Oils enriched in CLNAs are valuable for nutraceutical and industrial applications (Cahoon et al. 1999; Rawat et al. 2012). CLNAs such as α-eleostearic acid (18:3Δ9cis,11trans,13trans), present in tung oil, are commonly used in formulations of inks, dyes, coatings and resins (Sonntag 1979; Hornung et al. 2004; Cahoon et al. 2006). There is also growing evidence showing that dietary supplementation with CLNAs has cytotoxic effects on tumor cells and alters lipid metabolism in animals (Igarashi and Miyazawa 2005; Vroegrijk et al. 2011). In particular, it has been shown that Punica granatum (pomegranate) seed oil, containing punicic acid (18:3Δ9cis,11trans,13cis) and polyphenols, retards oxidation and prostaglandin synthesis, inhibits breast and colon cancer cell proliferation and invasion, and promotes breast cancer cell apoptosis (Kim et al. 2002; Kohno et al. 2004). Also, in human clinical trials, it has been shown that consumption of pomegranate seed oil has a positive effect on cardiovascular health (Mirmiran et al. 2010). Therefore, there is a growing interest in transferring the genes responsible for the biosynthesis of punicic acid into crop plants to obtain significant amounts of it in the seed oil (Mietkiewska et al. 2014).
It has been previously demonstrated that the conjugated double bonds in punicic acid are synthesized by a divergent form of the Δ12-oleic acid desaturase (FAD2). This enzyme, which has been designated ‘fatty acid conjugase, or FADX’, catalyzes the conversion of the Δ12-double bond of linoleic acid into two conjugated double bonds at positions Δ11trans and Δ13cis from the carboxyl end of the fatty acid (Hornung et al. 2002; Iwabuchi et al. 2003). Fatty acid conjugase involved in the biosynthesis of punicic acid in P. granatum (PgFADX, GenBank# AY178446) and Trichosanthes kirilowii (TkFADX, GenBank# AY178444) was previously isolated by PCR-based cloning. Earlier attempts to produce punicic acid in transgenic plants resulted in the limited accumulation of the conjugated fatty acid. Arabidopsis thaliana seeds overexpressing PgFADX or TkFADX showed accumulation of punicic acid up to 4.4 % or 10 %, respectively (Iwabuchi et al. 2003). In addition, TkFADX Brassica napus was developed showing production of punicic acid up to 2.5 % of total fatty acids in transgenic seeds (Koba et al. 2007). These levels of punicic acid were considerably lower compared to those observed in P. granatum (up to 80 % w/w) or T. kirilowii (40 % w/w).
Similar problems with low accumulation of other conjugated fatty acids were reported earlier in transgenic plants overexpressing cDNAs encoding other divergent FAD2 isoforms including the conjugase from Momordica charantia, Vernica fordii, Impatients balsamina and Calendula officinalis (Cahoon et al. 1999, 2001; Cahoon and Kinney 2004; Cahoon et al. 2006). This indicates that although the biosynthesis of conjugated fatty acids might be supported by a single gene trait (FADX), the accumulation of these unusual fatty acids in the storage lipids requires a network of additional genes/enzymes. In all of these cases, transgenic expression of FAD2 divergent genes resulted in low accumulation of modified fatty acids and was also accompanied by a significant increase in the content of oleic acid (Napier 2007; Carlsson et al. 2011).
Here, we describe a new strategy developed to increase the accumulation of punicic acid to higher than previously reported levels (Iwabuchi et al. 2003). First, overexpression of PgFADX in high oleic A. thaliana double mutant fad3/fae1 led to 2.6-fold higher amounts of punicic acid compared to that previously reported in the wild-type A. thaliana background. Secondly, combined overexpression of PgFADX with PgFAD2 in a fad3/fae1 mutant background brought the production of punicic acid to 21.2 % of total fatty acids. These investigations reveal metabolic constraints limiting the accumulation of punicic acid in developing transgenic seeds. Furthermore, the current data also provide new insights into the phenomenon of reduced endogenous FAD2 desaturase activity observed in plants engineered to produce punicic acid.
Materials and methods
RNA isolation and cDNA synthesis from P. granatum seeds
Punica granatum L. seeds were obtained from fruits purchased at the local market. Total RNA was isolated using Spectrum Plant Total RNA Kit (Sigma-Aldrich, Oakville, ON, Canada) followed by additional cleaning and concentration with RNeasy MinElute™ Cleanup Kit (Qiagen, Mississauga, ON, Canada). Single-stranded cDNA was synthesized at 42 °C from 1 µg of seed total RNA with Superscript II (Invitrogen, Burlington, ON, Canada) and used as a template for PCR amplification.
Preparation of plant expression constructs and transformation
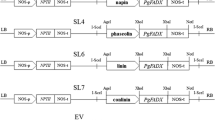
The napin promoter (Josefsson et al. 1987) was amplified by PCR with the primers:
F1: 5′-ATAGAATTCAAGCTTTCTTCATCGGTGAT-3′ (EcoRI site is underlined) and R1: 5′-ATACCCGGGGTCCGTGTATGTTTTTAATC-3′ (SmaI site is underlined). A P. granatum FADX ORF (GenBank# AY178446) was amplified with the primers: F2: 5′-TATCCCGGGATGGGAGCTGATGGAACA-3′ (SmaI site is underlined) and R2: 5′-CGCGCGGCCGCTCAGAACTTGCTCTTGAAC-3′ (NotI site is underlined).
The NOS terminator (Bevan 1983) was generated by PCR with the primers:
F3: 5′-CGCCGGCGGCCGCGATCGTTCAAACATTTGGCA-3′ (NotI site is underlined) and R3: 5′-TATGGTACCCGATCTAGTAACATAGATGAC-3′ (KpnI site is underlined).
Subsequently, napin, PgFADX and NOS PCR fragments digested with the appropriate endonucleases were ligated into EcoRI and KpnI sites of pRD400 (Datla et al. 1992) and that resulting in the NCJ construct.
Then, the napin promoter was amplified with the primers:
F4: 5′-ATAGGTACCAAGCTTTCTTCATCGGTGAT (KpnI site is underlined) and R4: 5′-ATACTCGAGGTCCGTGTATGTTTTTAATCT-3′ (XhoI site is underlined). A P. granatum FAD2 ORF (GenBank# AY178447) was generated by PCR with the primers: F5: 5′-TAACTCGAGATGGGAGCCGGTGGAAG-3′ (XhoI site is underlined) and R5: 5′-TATTCTAGATCAGAGGTTCTTCTTGTAC-3′ (XbaI site is underlined).
The NOS terminator was amplified with the primers:
F6: 5′-TATTCTAGAGATCGTTCAAACATTTGGCAA-3′ (XbaI site is underlined) and R6: 5′-ATAGTCGACCGATCTAGTAACATAGATGAC-3′ (SalI site is underlined). Subsequently, napin, PgFAD2 and NOS PCR fragments digested with the appropriate endonucleases were cloned into KpnI and SalI sites of NCJ resulting in the NCJD construct.
The binary vectors: NCJ or NCJD were electroporated into Agrobacterium tumefaciens cell strain GV3101 (Koncz and Schell 1986) and transformed into A. thaliana fad3/fae1 (Smith et al. 2003) mutant background using the floral dip method as described earlier by Clough and Bent (1998). Transgenic plants were selected and analyzed as described by Mietkiewska et al. (2007). Arabidopsis thaliana plants were grown in a growth chamber at 22 °C with a photoperiod of 18 h light and 250 µmol m−2 s−1 light intensity.
Gene expression analysis
Total RNA was extracted from T4 transgenic A. thaliana seed lines and wild type at the beginning of the late developmental stage (14 days after flowering) using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). First-strand cDNA synthesis was performed using 1 µg of total RNA as template and the QuantiTect reverse transcription kit (Qiagen) according to the manufacturer’s instructions. Developing seeds were harvested at 14 DAF for gene expression analysis, since it has been shown earlier that the accumulation of polyunsaturated fatty acids starts to increase significantly from 14 to 18 DAF (Baud and Lepiniec 2009).
Quantification of AtFAD2 transcripts in various transgenic lines was performed using 6 μL of a 1/10 dilution of cDNA as template. The reactions were performed as described by Chen et al. (2011) with minor modifications. In brief, the 25 μL reaction contained 12.5 μL of 2x SYBR-Green Master Mix (Molecular Biology Facility, University of Alberta, Canada), 1.2 μL of forward primer (10 μM; final concentration 48 nM) and 1.2 μL of reverse primer (10 μM; final concentration 48 nM). PCR was performed in a 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). Gene expression analyses were carried out using three biological replicates. Three technical replicate reactions were performed with each cDNA sample and individual primer pairs. The Arabidopsis 18S small subunit nuclear ribosomal RNA gene was used as the internal reference gene (GenBank# NC001284) and the relative transcript abundance of the target gene in the individual seed sample was calculated using the comparative Ct method (Livak and Schmittgen 2001). The primers for the three prime untranslated regions (3′ UTR) of AtFAD2 (GenBank# L26296) were: F7: 5′-ATGATGGTGAAGAAATTGTCG-3′ and R7: 5′-GTCATAACACAACAAAATGGAC-3′. The primers for 18S rRNA were F8: 5′-CAAAACGGCTCCGAAACAA-3′ and R8: 5′-ACTGGCAGTCCCTCGTGAGT-3′.
Lipid analysis
Analysis of fatty composition of plant material was performed by homogenizing 4−5 mg of A. thaliana seeds in 1 mL of a mixture of chloroform and isopropanol (2:1, v/v). The homogenate was then dried under N2 and transmethylated with 1 mL of 5 % sodium methoxide (NaOCH3) in methanol at room temperature for 30 min. The FAMEs were extracted twice with hexanes and after drying under N2 resuspended in 1 mL of iso-octane with internal standard (21:0, methyl heneicosanoin, 0.1 mg mL−1).
For separation of individual lipid classes by thin-layer chromatography (TLC), lipids were extracted from 100 mg of A. thaliana seeds as described earlier (Bligh and Dyer 1959). Lipid extracts were separated by one-dimensional TLC on silica gel plates (SIL G25, 0.25 mm, Macherey-Nagel, Düren, Germany) using the two solvent systems. The TLC plates were placed in the chamber with chloroform/methanol/acetic acid/formic acid/water (70:30:12:4:2, by vol.) for running until the first solvent reached halfway up the plate. Then, the plate was moved into the second solvent, hexane/diethyl ether/glacial acetic acid (70:30:1, by vol) and developed until the solvent was 1 cm from the top. Lipid classes were visualized under UV after spraying with 0.05 % primuline solution. Spots corresponding to TAG and PC were scraped out and transmethylated with 5 % sodium methoxide in methanol at RT for 30 min. The FAMEs were extracted with hexanes and dried under N2. Finally, FAMEs were resuspended in 1 mL of iso-octane with an internal standard (21:0, methyl heneicosanoin, 0.1 mg mL−1). The FAMEs were analyzed as described by Mietkiewska et al. (2011). ODP values were calculated as described earlier (Singh et al. 2001).
Results
Overexpression of PgFADX in A. thaliana
As described before, seed-specific overexpression of P. granatum conjugase (PgFADX) in the wild-type A. thaliana background led to the limited accumulation of punicic acid of up to 4.4 % (Iwabuchi et al. 2003). The level of linoleic acid which serves as a precursor for punicic acid synthesis is limited in the wild-type seeds of A. thaliana background to 27 % (Zhou et al. 2006). Therefore, to increase the production of punicic acid, we decided to overexpress PgFADX in a A. thaliana fad3/fae1 mutant background which has an increased content of linoleic acid in the seed oil of up to 51 %, due to inhibition of Δ15-desaturase (FAD3) and fatty acid elongase (FAE1) activity (Smith et al. 2003). We developed a binary vector NCJ carrying PgFADX under the control of a seed-specific napin promoter and expressed in A. thaliana fad3/fae1. Twenty eight kanamycin resistant plants were grown to seed maturity and seeds were collected individually from each plant. The fatty acid composition of T2 segregating seeds collected from all plants is shown in Table 1. Significant changes in fatty acid composition in comparison to the control lines (nt-fad3/fae1) were found. Seed-specific expression of PgFADX resulted in an increased proportion of punicic acid from 0 % in the control lines up to an average of 6.43 % of total fatty acids. In the highest transgenic line NCJ-11, punicic acid accounted for as much as 11.26 % of total fatty acids in T2 segregating seeds. The increased proportion of punicic acid was correlated with a concomitant reduction in the proportion of its corresponding precursor 18:2 (reduced on average by 33 %). The production of punicic acid in A. thaliana seeds was also accompanied on average by a 34 % increase in oleic acid content. This indicates that overexpression of PgFADX led to the inhibition of the native FAD2 desaturase activity. Decreases in the proportion of palmitic acid were also found in A. thaliana seeds engineered to produce punicic acid. Seeds from the T2 lines with the highest proportion of punicic acid were grown to obtain the T3 seed generation. As shown in Fig. 1, a consistent level of punicic acid production was observed in T3 seeds. The proportion of punicic acid increased from 0 % in wild-type and null segregant controls to as high as 11.2 and 11.5 % in the highest homozygous transgenic lines NCJ: 19-2 and 11-4, respectively. As in T2 segregating seeds, the production of punicic acid in the T3 generation was accompanied by a significant increase in oleic acid to levels of 50 % of the total fatty acids. Furthermore, seedlings of NCJ transgenic lines germinated as well as the corresponding controls (data not shown). The 100-seed weight averaged across eight T3 lines was 2.169 ± 0.049 mg (average ± standard error), compared to 2.226 ± 0.098 mg for plants of the non-transformed control. The average total fatty acid content of eight transgenic lines was 0.224 ± 0.004 mg/mg of seeds, compared with 0.211 ± 0.006 mg/mg for non-transformed control lines. Statistical analysis (t test) showed that the difference of 100-seed weight as well as total fatty acid content was not significant (P < 0.05) between the controls and the NCJ transgenic lines. Taken together, these data indicate that the accumulation of punicic acid has no detrimental effect on the size or total fatty acid accumulation in comparison with non-transformed controls.
Fatty acid composition of mature seed of non-transformed A. thaliana fad3/fae1 mutant (nt-fad3fae1), null segregant (null segr) and T3 seeds of transgenic A. thaliana lines (NCJ) overexpressing the PgFADX gene under the control of the napin promoter. The values are the average of three determinations ± SD
Overexpression of PgFADX and PgFAD2 in A. thaliana
Since overexpression of PgFADX affected the activity of the native A. thaliana FAD2 desaturase, we decided to study if combined overexpression of PgFAD2 with PgFADX could reduce the negative effect on AtFAD2 desaturase activity and increase the accumulation of punicic acid. We developed a tandem construct (NCJD) carrying both genes under the control of napin promoters and introduced into the A. thaliana fad3/fae1 mutant. As shown in Table 1, the level of punicic acid found in lines overexpressing both PgFADX and PgFAD2 was higher compared to the lines overexpressing only PgFADX. On average the level of punicic acid was 9.2 % of total fatty acid in the seed oil, which was 43 % higher compared to the average level found in NCJ transgenic lines. The average level of oleic acid (33.5 %) found in the NCJD lines was lower compared with the level observed in lines overexpressing only PgFADX (NCJ) and was equivalent to the proportion observed in non-transformed A. thaliana fad3/fae1 seeds. These results indicate that overexpression of PgFAD2 could compensate for the negative effect of PgFADX on AtFAD2 desaturase activity observed in NCJ lines. Seeds from the best T2 transgenic lines with punicic acid content over 12 % were selected and grown to the T3 seed generation. As shown in Fig. 2, punicic acid accounted for as much as 21.2 % of total fatty acids in seeds of NCJD-30-2 and 34-3 transgenic lines representing a 61 % and 43 % increase, respectively, compared to the T2 parental lines. Increased production of punicic acid in these two NCJD lines correlated with decreased levels of oleic acid compared to non-transformed seed controls. This may indicate that T3 NCJD seed lines overexpressing PgFAD2 exhibited increased Δ12-desaturase activity (Fig. 2).
Effect of PgFADX on oleate desaturation levels in transgenic seeds
As shown in Table 1, production of punicic acid in the A. thaliana fad3/fae1 mutant was accompanied by increased accumulation of oleic acid. Therefore, to assess the cumulative effects of Δ12-desaturase activity, we applied an indirect method developed by Singh et al. (2001). Oleic acid desaturation proportion (ODP) values were calculated for all T2 NCJ and NCJD lines and controls (Fig. 3). Oleate desaturase is highly active in the developing seeds of wild-type A. thaliana, with 73 % of 18:1 being converted to 18:2 and 18:3, for an ODP value of 0.73 (Stoutjesdijk et al. 2002). In the A. thaliana fad3/fae1 mutant background with affected Δ15-desaturase activity, only 60 % of 18:1 is being further desaturated (ODP of 0.6, Fig. 3). NCJ transgenic lines overexpressing PgFADX exhibited a reduced level of Δ12-desaturase activity (ODP of 0.44−0.57). Regression analysis revealed a significant inverse correlation (R 2 = 0.761) between the amount of punicic acid and ODP value; higher accumulation of punicic acid was associated with lower ODP values (Fig. 3a). For instance, in NCJ-11 line with the highest content of punicic acid (11.26 %), ODP was only 0.44 indicating that the amount of Δ12-desaturated fatty acids was reduced by 27 % due to the overexpression of PgFADX compared to the non-transformed control. The linear regression analysis for NCJD lines exhibited a lower R 2 value (0.209), and there was also a change in the slope of the fitted line (Fig. 3b). The inverse relationship observed in lines overexpressing only PgFADX was found to be changed dramatically in the lines overexpressing both PgFADX and PgFAD2. The ODP levels for NCJD lines varied regardless of the level of punicic acid accumulation.
Relationship between oleate desaturation proportion (ODP) value and level of punicic acid in transgenic A. thaliana fad3/fae1 mutant in T2 segregating seeds. a Data for transgenic NCJ lines overexpressing PgFADX under the control of the napin seed-specific promoter. b Data for the transgenic NCJD lines overexpressing PgFADX and PgFAD2. In both panels, filled circle shows non-transformed A. thaliana fad3fae1 mutant
To further investigate the nature of FAD2 inhibition observed in PgFADX A. thaliana transgenic lines, we checked AtFAD2 transcript levels in three independent transgenic T4 homozygous seed lines at 14 DAF. For real-time PCR analysis, primers targeted to the 3′ UTR region of AtFAD2 were used since PgFADX and AtFAD2 exhibit high sequence identify (>65 %) in the coding region. As shown in Fig. 4, the relative AtFAD2 expression level was significantly reduced (up to 99 %) in the A. thaliana seeds overexpressing PgFADX only (NCJ lines) or in combination with PgFAD2 (NCJD line) compared to the non-transformed fad3/fae1 mutant line background. These data may indicate that punicic acid that accumulated in A. thaliana seeds overexpressing PgFADX may act as a transcriptional repressor causing significant reduction of AtFAD2 transcript level. Considering the high sequence identity (>65 %) of PgFADX and AtFAD2, the occurrence of post-transcriptional genes silencing in PgFADX transgenic lines cannot be completely excluded.
Relative expression of A. thaliana FAD2 gene in transgenic lines overexpressing PgFADX alone (NCJ lines) or in combination with PgFAD2 (NCJD line) and non-transformed control (nt-fad3fae1). Total RNA was obtained from T4 A. thaliana at 14 DAF. Equal amounts of total RNA were used for cDNA synthesis and serial dilutions of the resulting reaction were used for quantitative RT-PCR. Each bar represents the mean from three determinations ± SD with the Arabidopsis 18S small subunit nuclear ribosomal RNA gene as the internal reference gene
Relative content of fatty acid in phosphatidylcholine (PC) and triacylglycerol (TAG) from A. thaliana engineered to produce punicic acid
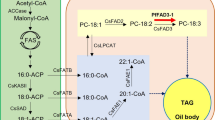
To investigate factors limiting the production of punicic acid in genetically engineered A. thaliana plants, we decided to check fatty acid composition of the selected lipid classes. Examination of the fatty acid content of the selected lipid classes was performed for the A. thaliana line overexpressing PgFADX+PgFAD2 (NCJD-30-2) with the highest content of punicic acid (21.2 %) in the T3 seeds (Fig. 2) and P. granatum seeds. In transgenic NCJD-30-2 A. thaliana seeds, the punicic acid content of PC was 12.5 %, which was higher than that observed in TAG (6.6 %). On the contrary, in P. granatum seeds, punicic acid accounted for 60 % of the fatty acids in TAG and only 0.8 % of fatty acids in PC (Fig. 5). Taken together, the above results indicate that in P. granatum an efficient mechanism of trafficking of punicic acid from PC to TAG has evolved and that mechanism is missing in developing A. thaliana seeds.
Discussion
Metabolic engineering of oilseed crops to produce large amounts of conjugated and other industrial fatty acids has proven to be very difficult (Mietkiewska et al. 2014). Over the years of intensive studies, enzymes involved in the biosynthesis of many unusual fatty acids have been identified and corresponding genes have been cloned. Conjugated fatty acids were shown to be synthesized by a divergent form of Δ12-desaturase (designated as FADX), either from oleic or linoleic acid precursors esterified to sn-2 position of PC (Cahoon et al. 2007; Vanhercke et al. 2013). Transgenic expression of FADX genes in A. thaliana or soybean (Glycine max) resulted in limited accumulation of the desired fatty acid product compared with the levels in seeds from plants of the source species.
As described earlier, punicic acid was synthesized from linoleic substrate on sn-2 position of PC by fatty acid conjugase (PgFADX) in P. granatum seeds. As with other unusual fatty acids, overexpression of P. granatum FADX led to the limited accumulation of punicic acid in A. thaliana seeds (Iwabuchi et al. 2003). Since the level of linoleic acid substrate is limited to <27 % in wild-type A. thaliana seeds, we overexpressed PgFADX in a high 18:2 A. thaliana fad3/fae1 mutant background. This resulted in increased accumulation of punicic acid up to 11.3 % in T2 segregating A. thaliana seeds (Table 1, Fig. 1) compared with the 4.4 % reported earlier by Iwabuchi et al. (2003). Our results have shown that a high 18:2 A. thaliana mutant background is a better host for transgenic production of punicic acid compared with the wild type. A similar observation was reported by Cahoon et al. (2006) when producing α-eleostearic and calendic acid (18:3Δ8trans,10trans,12cis) in A. thaliana seeds. The amounts of these fatty acids accumulated in seeds of wild-type background were below 5 % compared with more than 10 % found in the A. thaliana fad3/fae1 mutant background overexpressing either the M. charantia or C. officinalis conjugase. Similar effects of increased vernolic acid (12,13-epoxy-18:1Δ9cis) production in response to the increased linoleic acid availability in the A. thaliana fad3/fae1 double mutant were reported by Zhou et al. (2006). In contrast to the above, lack of such a direct relationship between substrate level (18:1) and ricinoleic acid (12-OH 18:1Δ9cis) synthesis catalyzed by FAH12 (another type of FAD2 divergent enzyme) was reported earlier (Smith et al. 2003; Napier 2007).
The production of punicic acid obtained by overexpression of PgFADX was accompanied by changes in the relative proportion of other fatty acids in the seed oil. The most striking change was found for oleic acid which increased by up to 43 % compared to the seeds from non-transformed plants (Table 1, Fig. 1). This phenotype of reduced endogenous FAD2-mediated desaturase activity has also been observed in soybean somatic embryos producing α-eleostearic acid and A. thaliana seeds (Cahoon et al. 2006). An increased accumulation of oleic acid was also documented in transgenic plants overexpressing cDNAs encoding hydroxylases (Broun and Somerville 1997; Smith et al. 2003), epoxygenases (Singh et al. 2001; Li et al. 2010) and acetylenases (Thomaeus et al. 2001). In the current study, the suppression of AtFAD2 desaturase activity as indicated by lower ODP values correlated with higher punicic acid levels (Fig. 3a). To investigate the nature of this high oleic acid phenotype, we checked the AtFAD2 transcript levels using a quantitative RT-PCR approach with primers targeted to the 3′ UTR of AtFAD2. The results clearly demonstrate a reduced level of AtFAD2 mRNA in transgenic seeds at 14 DAF, which may suggest that accumulation of punicic acid may have repressed AtFAD2 mRNA expression (Fig. 4). Similar to the current study, jacaric acid (18:3Δ8cis,10trans,12cis) and another isomer of CLNA (18:4Δ9cis,11trans,10trans,12cis) were shown to decrease the expression level of stearoyl-CoA desaturase mRNA in mouse (Mus musculus) liver (Lee et al. 1998; Shinohara et al. 2012). Also, the occurrence of post-transcriptional genes silencing in PgFADX transgenic lines cannot be completely excluded, since PgFADX and AtFAD2 exhibit high sequence identity (>65 %). However, the possibility of decreased AtFAD2 mRNA level was excluded before for A. thaliana overexpressing Ricinus communis (RcFAH12) or Crepis palestina epoxygenase (Cpal2) based on Northern blot analysis which showed unaltered AtFAD2 transcript levels (Broun and Somerville 1997; Singh et al. 2001). This discrepancy could be possibly explained by the two different approaches used to check AtFAD2 transcript level (i.e., Northern blot versus q-RT-PCR in our study). Secondly, there was a difference in the plant material used. The current study used late developing seeds as a source of transcript, whereas earlier studies used entire siliques.
There are also other factors that could explain high oleate levels and low levels of punicic acid accumulation in transgenic plants. Given that AtFAD2 and PgFADX utilize membrane-linked substrates, impaired synthesis of 18:2 might result from inhibition of AtFAD2 desaturase activity by punicic acid. As we have shown (Fig. 5), punicic acid accounted for 12.5 % of total fatty acids of PC in transgenic A. thaliana compared to only 0.8 % in P. granatum seeds and that may have resulted in either the inhibition of AtFAD2 desaturase activity or in the reduction of oleic acid flux available for Δ12-desaturation. This scenario has been discussed before by Napier (2007) in relation to other unusual fatty acids synthesized by divergent FAD2. As in the current study, higher relative amounts of unusual fatty acids in PC compared to TAG were previously reported for the transgenic production of acetylenic, epoxy, hydroxy and other conjugated fatty acids (Thomaeus et al. 2001; Cahoon et al. 2006).
We have shown here that combined overexpression of PgFADX with PgFAD2 resulted in a 1.9-fold further increase in accumulation of punicic acid compared to the lines overexpressing only PgFADX along with restoration of oleic acid to the level found in non-transformed A. thaliana fad3/fae1 (Figs. 2, 3b). Based on the data presented here, it should be noted that overexpression of an extra PgFAD2 could restore Δ12-desaturation to the level found in non-transformed plants even in the presence of punicic acid. As described previously for enhanced accumulation of vernolic acid in Cpal2 + Cpdes A. thaliana (Zhou et al. 2006), the effect of an extra PgFAD2 expression on the increased punicic acid production and restoration of Δ12-desaturation in plants carrying PgFADX+PgFAD2 is more likely related to the higher levels of FAD2 enzyme availability.
Comparison of fatty acid composition of PC and TAG revealed that in transgenic A. thaliana seeds, an efficient mechanism of trafficking punicic acid from its site of synthesis to the storage lipids is missing. By contrast, such a mechanism evolved in P. granatum seeds since punicic acid accounted for less than 0.8 % of total fatty acids in PC (Fig. 5). Similar to the studies described here, increased accumulation of other unusual fatty acids in PC was reported in A. thaliana engineered to produce hydroxy, epoxy, or conjugated fatty acids (Thomaeus et al. 2001; Cahoon et al. 2006; Napier 2007). As has also been shown for transgenic production of hydroxy and epoxy fatty acids (Burgal et al. 2008; Li et al. 2010), further increase in the transgenic production of punicic acid could be possibly achieved by combined overexpression of PgFADX and PgFAD2 with cDNAs encoding specialized diacylglycerol acyltransferases and/or phospholipid:diacylglycerol acyltransferase from P. granatum. In addition, enzymes such as phospholipases C or D, lysophosphatidylcholine acyltransferase, or phospholipid:diacylglycerol cholinephosphotransferase sourced from P. granatum and introduced into transgenic plants might be useful for efficient removal of punicic acid from PC via acyl editing and exchange mechanisms (Vanhercke et al. 2013).
In conclusion, by using an A. thaliana fad3/fae1 mutant host combined with seed-specific overexpression of P. granatum conjugase (FADX) with FAD2, the transgenic production of punicic acid was increased to 21.2 % of total fatty acids, the highest level reported to date. These results indicate that the transgenic production of very high levels of punicic acid will require identification and characterization of other specialized genes/enzymes involved in the metabolic network in plants naturally accumulating punicic acid.
Author contribution
EM and RJW designed the research project, analyzed the data and wrote the manuscript. EM, RM, AW, AFS, MSG and GC conducted experiments and helped with data analysis. All authors read and approved the manuscript.
Abbreviations
- CLNAs:
-
Conjugated linolenic acids
- FAD2:
-
Δ12-oleic acid desaturase
- FAD3:
-
Δ15-linoleic acid desaturase
- FADX:
-
Fatty acid conjugase
- ODP:
-
Oleic acid desaturation proportion
- PC:
-
Phosphatidylcholine
- 3′ UTR:
-
Three prime untranslated region
- TAG:
-
Triacylglycerol
References
Badami RC, Patil KB (1980) Structure and occurrence of unusual fatty acids in minor seed oils. Prog Lipid Res 19:119–153
Baud S, Lepiniec L (2009) Regulation of de novo fatty acid synthesis in maturing oil seeds of Arabidopsis. Plant Physiol Biochem 47:448–455
Bevan M (1983) Binary Agrobacterium vectors for plant transformation. Nucleic Acid Res 12:8711–8721
Bligh EG, Dyer WJ (1959) A rapid method for total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Broun P, Somerville C (1997) Accumulation of ricinoleic, lesquerolic and densipolic acids in seeds of transgenic Arabidopsis plants that express a fatty acyl hydroxylase cDNA from castor. Plant Physiol 113:933–942
Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, Browse J (2008) Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J 6:819–831
Cahoon E, Kinney A (2004) Dimorphecolic acid is synthesized by the coordinate activities of two divergent Δ12-oleic acid desaturases. J Biol Chem 279:12495–12502
Cahoon EB, Carlson TJ, Ripp KG, Schweiger BJ, Cook GA, Hall SE, Kinney AJ (1999) Biosynthetic origin of conjugated double bonds: production of fatty acid components of high-value drying oils in transgenic soybean embryos. Proc Natl Acad Sci USA 96:12935–12940
Cahoon E, Ripp K, Hall S, Kinney A (2001) Formation of conjugated Δ8, Δ10-double bonds by Δ12-oleic-acid desaturase-related enzymes. Biosynthetic origin of calendic acid. J Biol Chem 276:2637–2643
Cahoon EB, Dietrich CR, Meyer K, Damude HG, Dyer JM, Kinney AJ (2006) Conjugated fatty acids accumulate to high levels in phospholipids of metabolically engineered soybean and Arabidopsis seeds. Phytochemistry 67:1166–1176
Carlsson AS, Yilmaz JL, Green AG, Stymne S, Hofvander P (2011) Replacing fossil oil with fresh oil—with what and for what? Eur J Lipid Sci Technol 113:812–831
Chen X, Truksa M, Snyder CL, El-Mezawy A, Shah S, Weselake RJ (2011) Three homologous genes encoding sn-glycerol-3-phosphate acyltransferase 4 exhibit different expression patterns and functional divergence in Brassica napus. Plant Physiol 155:851–865
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Datla RSS, Hammerlindl JK, Panchuk B, Pelcher LE, Keller W (1992) Modified binary plant transformation vectors with the wild-type gene encoding NPTII. Gene 122:383–384
Hornung E, Pernstich C, Feussner I (2002) Formation of conjugated Δ11 Δ13-double bonds by Δ12-linoleic acid (1,4)-acyl-lipid-desaturase in pomegranate seeds. Eur J Biochem 269:4852–4859
Igarashi M, Miyazawa T (2005) Preparation and fractionation of conjugated trienes from alpa-linolenic acid and their growth-inhibitory effects on human tumor cells and fibroblasts. Lipids 40:109–113
Iwabuchi M, Kohno-Murase J, Imamura J (2003) Δ12-Oleate desaturase-related enzymes associated with formation of conjugated trans-Δ11, cis-Δ13 double bonds. J Biol Chem 278:4603–4610
Josefsson LG, Lenman M, Ericson ML, Rask L (1987) Structure of a gene encoding the 1.7 S storage protein, napin, from Brassica napus. J Biol Chem 262:12196–12201
Kim ND, Mehta R, Yu W, Neeman I, Livney T, Amichay A, Poirier D, Nicholls P, Kirby A, Jiang W, Mansel R, Ramachandran C, Rabi T, Kaplan B, Lansky E (2002) Chemopreventive and adjuvant therapeutic potential of pomegranate (Punica granatum) for human breast cancer. Breast Cancer Res Treat 71:203–217
Koba K, Imamura J, Akashoshi A, Kohno-Murase J, Nishizono S, Iwabuchi M, Tanaka K, Sugano M (2007) Genetically modified rapeseed oil containing cis-9, trans-11, cis-13-octadecatrienoic acid affects body fat mass and lipid metabolism in mice. J Agric Food Chem 55:3741–3748
Kohno H, Suzuki R, Yasui Y, Hosokawa M, Miyashita K, Tanaka T (2004) Pomegranate seed oil rich in conjugated linolenic acid suppresses chemically induced colon carcinogenesis in rats. Cancer Sci 95:481–486
Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes by a novel type of Agrobacterium binary vector. Mol Gen Genet 204:383–396
Lee KN, Pariza MW, Ntambi JM (1998) Conjugated linoleic acid decreases hepatic stearoyl-CoA desaturase mRNA expression. Biochem Biophys Res Commun 248:817–821
Li R, Yu K, Hatanaka T, Hildebrand DF (2010) Vernonia DGATs increase accumulation of epoxy fatty acids in oil. Plant Biotechnol J 8:184–195
Livak K, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods 25:402–408
Mietkiewska E, Brost JM, Giblin EM, Barton DL, Taylor DC (2007) Cloning and functional characterization of the Fatty Acid Elongase 1 (FAE1) gene from high erucic Crambe abyssinica cv. Prophet Plant Biotechnol J 5:636–645
Mietkiewska E, Siloto RM, Dewald J, Shah S, Brindley DN, Weselake RJ (2011) Lipins from plants are phosphatidate phosphatases that restore lipid synthesis in a pah1Δ mutant strain of Saccharomyces cerevisiae. FEBS J 278:764–775
Mietkiewska E, Lin Y, Weselake RJ (2014) Engineering production of C18 conjugated fatty acids in developing seeds of oil crops. Biocatal Agric Biotechnol 3:44–48
Mirmiran P, Fazeli MR, Asghari G, Shafiee A, Azizi F (2010) Effect of pomegranate seed oil on hyperlipidaemic subjects: a double-blind placebo-controlled clinical trial. Br J Nutr 104:402–406
Napier JA (2007) The production of unusual fatty acids in transgenic plants. Annu Rev Plant Biol 58:295–319
Rawat R, Yu X-H, Sweet M, Shanklin J (2012) Conjugated fatty acid synthesis: residues 111 and 115 influence product partitioning of Momordica charantia conjugase. J Biol Chem 287:16230–26237
Shinohara N, Ito J, Tsuduki T, Honma T, Kijima R, Sugawara S, Arai T, Yamasaki M, Ikezaki A, Yokoyama M, Nishiyama K, Nakagawa K, Miyazawa T, Ikeda I (2012) Jacaric acid, a linolenic acid isomer with a conjugated triene system, reduces stearoyl-CoA desaturase expression in liver of mice. J Oleo Sci 61:433–441
Singh S, Thomaeus S, Lee M, Stymne S, Green A (2001) Transgenic expression of a Δ12-epoxygenase gene in Arabidopsis seeds inhibits accumulation of linoleic acid. Planta 212:872–879
Smith CR Jr (1971) Occurrence of unusual fatty acids in plants. Prog Chem Fats Other Lipids 11:137–177
Smith MA, Moon H, Chowrira G, Kunst L (2003) Heterologous expression of a fatty acid hydroxylase gene in developing seeds of Arabidopsis thaliana. Planta 217:507–516
Sonntag NOV (1979) Composition and characteristics of individual fats and oils. In: Swern D (ed) Bailey’s industrial oil and fat products. Wiley, New York, pp 289–477
Stoutjesdijk PA, Singh SP, Liu Q, Hurlstone CJ, Waterhouse PA, Green AG (2002) hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol 129:1723–1731
Thomaeus S, Carlsson AS, Lee M, Stymne S (2001) Distribution of fatty acids in polar and neutral lipids during seed development in Arabidopsis thaliana genetically engineered to produce acetylenic, epoxy and hydroxy fatty acids. Plant Sci 161:997–1003
Vanhercke T, Wood CC, Stymne S, Singh SP, Green AG (2013) Metabolic engineering of plant oils and waxes for use as industrial feedstocks. Plant Biotechnol J 11:197–210
Vroegrijk IO, van Diepen JA, van den Berg S, Westbroek I, Keizer H, Gambelli L, Hontecillas R, Bassaganya-Riera J, Zondag GC, Romijn JA, Havekes LM, Voshol PJ (2011) Pomegranate seed oil, a rich source of punicic acid, prevents diet-induced obesity and insulin resistance in mice. Food Chem Toxicol 49:1426–1430
Zhou X-R, Singh S, Liu Q, Green A (2006) Combined transgenic expression of Δ12-desaturase and Δ12-epoxygenase in high linoleic acid seeds leads to increased accumulation of vernolic acid. Funct Plant Biol 333:585–592
Acknowledgments
We thank Drs. Ljerka Kunst (University of British Columbia, Vancouver, Canada) and Mark Smith (National Research Council of Canada) for providing seeds of A. thaliana fad3/fae1 mutant. We would like to thank Dr. Joseph Boothe for his critical assessment of the manuscript. We are grateful for the support provided by Alberta Innovates Bio Solutions, Alberta Enterprise and Advanced Education, the Canada Foundation for Innovation and the Canada Research Chairs Program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mietkiewska, E., Miles, R., Wickramarathna, A. et al. Combined transgenic expression of Punica granatum conjugase (FADX) and FAD2 desaturase in high linoleic acid Arabidopsis thaliana mutant leads to increased accumulation of punicic acid. Planta 240, 575–583 (2014). https://doi.org/10.1007/s00425-014-2109-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2109-z