Abstract
The Arabidopsis thaliana KNAT7 (KNOX family) and MYB75 (MYB family) transcription factors were each shown earlier to interact in yeast two-hybrid assays, and to modulate secondary cell wall formation in inflorescence stems. We demonstrate here that their interaction also occurs in vivo, and that specific domains of each protein mediate this process. The participation of these interacting transcription factors in secondary cell wall formation was then extended to the developing seed coat through the use of targeted transcript analysis and SEM in single loss-of-function mutants. Novel genetic and protein–protein interactions of MYB75 and KNAT7 with other transcription factors known to be involved in seed coat regulation were also identified. We propose that a MYB75-associated protein complex is likely to be involved in modulating secondary cell wall biosynthesis in both the Arabidopsis inflorescence stem and seed coat, and that at least some parts of the transcriptional regulatory network in the two tissues are functionally conserved.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Secondary cell walls in plants are composed of cellulose, hemicellulose, and lignin polymers, which together provide mechanical support and protection. These cell walls are found in diverse organs and tissues, including inflorescence stems, trichomes, the vasculature of leaves and roots, and the seed coat. The role of different transcription factors in regulating secondary cell wall deposition has been examined in numerous studies (for reviews, see Zhong and Ye 2007; Demura and Ye 2010; Zhong et al. 2010). These analyses have identified roles for individual members of various transcription factor families, including those of the NAC, MYB, and KNOX homeodomain classes. However, most transcription factors are unlikely to act in isolation, but rather as components of dynamic, multi-protein complexes. In addition, most transcription factor proteins have a modular structure that typically includes one or more DNA-binding or protein-binding domains together with effector domains. Together, these domains define multiple contact interfaces important for the formation of multi-component complexes that can stabilize interaction with particular DNA sequence motifs in their target promoters and thereby contribute to the specificity of transcriptional regulation (Wolberger 1999).
Several transcription factors involved in secondary cell wall formation have been shown to have physical interactions with other proteins, including members of the MYB and homeodomain families (Kumar 2006; Zimmermann et al. 2004; Li et al. 2011) and NAC domain proteins (Yamaguchi et al. 2010), but most of the transcription factors thought to be involved in cell wall deposition or re-modeling have not been characterized for protein–protein interactions.
MYB75 (PAP1 production of anthocyanin pigment1) regulates many metabolic pathways in plants, including anthocyanin accumulation (Gonzalez et al. 2008; Borevitz et al. 2000), proanthocyanidins (Matsui et al. 2004), and secondary cell wall formation in the Arabidopsis inflorescence stem (Bhargava et al. 2010). MYB transcription factors are known to form complexes with basic helix-loop-helix (bHLH) transcription factors and WD40 repeat proteins to regulate a range of developmental processes (Schellmann and Hulskamp 2005; Serna and Martin 2006; Serna 2005; Guimil and Dunand 2006; Martin and Glover 2007) and metabolic pathways in plants (Haughn and Chaudhury 2005; Broun 2005; Koes et al. 2005; Ramsay and Glover 2005; Western 2006), and physical interactions between MYB75 and the bHLH TFs, TT8, and bHLH012 (MYC1) have been reported earlier (Zimmermann et al. 2004).
Plant MYB proteins are characterized by a conserved MYB domain, which consists of one or two imperfect repeats referred to as R2 and R3, each of which forms a helix-turn-helix structure. The R3 domain has been specifically implicated in protein–protein interactions of MYB75 with bHLH proteins (Frampton et al. 1991; Kanei-Ishii et al. 1990, 1997; Zimmermann et al. 2004). In particular, it was shown earlier that anthocyanin accumulation in vegetative tissues is regulated by a putative transcriptional complex involving MYB75, MYB90, and three bHLH transcription factors, GL3, EGL3, and TRANSPARENT TESTA8 (TT8) (Borevitz et al. 2000; Zhang et al. 2003), while proanthocyanidin accumulation in seed coats is regulated by an R2R3 MYB gene, TRANSPARENT TESTA2 (TT2) and a bHLH gene, TT8 (Nesi et al. 2000, 2001). Seed coat mucilage production, on the other hand, is regulated by the activity of an R2R3 MYB gene, MYB61, and two bHLH genes, EGL3, and TT8 (Penfield et al. 2001; Zhang et al. 2003).
Similarly, homeodomain (HD) transcription factors (e.g. KNOX family members) also show protein–protein interactions with other transcription factors belonging to different families (Viola and Gonzalez 2006; Bellaoui et al. 2001; Li et al. 2011), mediated by the KNOX domain. KNOX proteins typically possess a canonical homeodomain, an ELK domain, and KNOX domains that mediate physical interactions with other proteins. KNAT7, a KNOX family protein that has been shown to regulate secondary cell wall biosynthesis in Arabidopsis (Li et al. 2012; Romano et al. 2012), also shows protein–protein interactions with ovate family protein (OFP) transcriptional co-regulators and bell-like homeodomain (BLH) family transcription factors (Li et al. 2011; Li 2009; Hackbusch et al. 2005).
The KNOX domain is composed of two sub-domains (KNOX1 and KNOX2) that are thought to specifically interact with other transcription factors, such as BLH proteins (Burglin 1997; Kumar 2006). MYB75 and KNAT7 were recently shown to physically interact in vitro (Bhargava et al. 2010), but the interaction determinants were not defined.
Some interactions (physical or genetic) between different proteins have been shown to be conserved in cells found in different tissue types (Gonzalez et al. 2009; Nesi et al. 2000, 2001; Baudry et al. 2004), but conservation of transcription factor protein–protein interactions across different cell types specifically undergoing secondary cell wall formation has not previously been examined. Here, we identify the protein domains required for KNAT7–MYB75 interaction both in vitro and in vivo, and show that these two interacting transcription factors help regulate secondary cell wall formation in two very different tissues—the Arabidopsis inflorescence stem vasculature and the seed coat. We have also been able to position KNAT7 and MYB75 within the known genetic interaction matrix involved in regulating the mucilage/seed coat wall biosynthesis pathway(s), and propose a model for the role of KNAT7-MYB75 interactions in modulating secondary cell wall formation in distinct tissue types in Arabidopsis.
Materials and methods
Plant material
The Arabidopsis loss-of-function allele of MYB75 (myb75-1) described by Bhargava et al. (2010) was used. The loss-of-function mutant of KNAT7 (knat7-1) employed in this work was described earlier (Li 2009; Li et al. 2012). Homozygous plants of each genotype were used for all experiments and their phenotype was always compared to the phenotype of the relevant WT ecotype, WT-Col, or WT-Nos (myb75-1 is in the Nossen and knat7-1 in the Columbia background).
Seeds were surface-sterilized using 20 % commercial bleach, cold treated at 4 °C in the dark for 2 days and plated on ½ MS agar medium [2.16 g/l MS salts, 1 % sucrose, 1 % Bacto-agar pH 6.0 (Murashige and Skoog 1962)]. Ten-day-old seedlings were grown in 5 × 5 cm pots containing a moistened Sunshine Mix #1 (Sun Gro Horticulture Canada Ltd), with a 16/8 h (light/dark) photoperiod at approximately 120 μmol photons m−2 s−1 at 23°C, unless specified otherwise. For protoplast isolation, leaves from 3- or 4-week-old plants were used for protoplast isolation.
For the seed coat phenotype studies, the loss-of-function myb75-1 mutant seeds were compared to wild-type (WT) Nossen seeds. The knat7-1 mutant is in the Columbia background and so was compared to WT Columbia seeds in this work. Homozygous seeds of TF mutants (tt8-1, ap2-1, gl2-1, ttg1-1, and ttg2-1) were obtained from the Arabidopsis Biological Resource Center (ABRC). For seed coat isolation at specific development stages of the seed, WT (Columbia and Nossen) and mutant seeds (tt8-1, ap2-1, gl2-1, ttg1-1, ttg2-1, myb7-1, and knat7-1) were grown in soil as described above for protoplast isolation. Plants were monitored for their silique development, and stage-specific seed coat tissue was collected as described below.
Protoplast isolation, transfection, and GUS activity assay
Leaves from Columbia WT plants approximately 3–4-weeks-old were used for protoplast isolation, and subsequent transfection and GUS activity assays, as described previously (Wang et al. 2007). The plasmid DNAs for reporter and effector genes were isolated using Endofree Plasmid Maxi Kits (Qiagen, Mississauga, Ontario, Canada). Both effector plasmid (10 μg) and reporter plasmid (10 μg) were co-transfected and all assays were performed in triplicate. 35S::CAT plasmid DNA was used to equalize the amount of DNA in transfection.
Stage-specific seed coat separation, RNA isolation, and qRT-PCR
For timing of developmental stage-specific seed coat isolation (days post-anthesis; DPA), pollination was defined as the time at which the flower was beginning to open. Flowers at anthesis were marked using water-soluble paint, and a different color was used for each day of marking. Siliques were collected, pooled together and then used for RNA isolation. Total RNA was extracted from 3-, 7-, and 11-DPA WT or mutant seed coats using the RNAqueous kit (Ambion) and treated with RNAse-free DNAse I according to the manufacturer’s instructions (Qiagen). Total RNA (100 ng) was reverse transcribed using the SuperScript® VILO™ cDNA synthesis kit (Invitrogen) according to the manufacturer’s instructions, and 1.5 μl cDNA was used in each reaction in a 20 μl reaction volume. PCR amplification was performed with KNAT7- or MYB75-specific primers, using ACTIN8 as a normalization control. (An et al. 1996). The cDNA was amplified using the PerfeCta™ qPCR FastMix (Quanta Biosciences) on the DNA Engine Opticon® 2 (Bio-Rad). Relative values are arbitrary units and were calculated as described previously (Gutierrez et al. 2008).
Yeast two-hybrid (Y2H) assays
The ProQuest Y2H system (Invitrogen) was used with either full length or partial sequences (for domain interaction studies) of transcription factors in the pDEST32 (bait) or pDEST-22 (prey) vectors, and introduced into the yeast strain MaV203 in different combinations. Positive clones were isolated on the basis of three selectable markers: HIS3, URA3, and LacZ. Positive interactions were indicated by activation of HIS3 or URA3, according to the manufacturer’s instructions. The different domains were amplified from full-length cDNAs for each TF, using specific primers (Table S1). These amplified fragments were then cloned in the Gateway entry vector pCR8/GW/TOPO, and after sequence verification, were directionally recombined into the Gateway destination vectors, pDEST32, and pDEST22.
Bimolecular fluorescence complementation (BiFC) assays
Gateway entry vectors carrying either full length MYB75 or KNAT7, or different domains, were used. For N-terminal YFP-tagged constructs, the appropriate entry clone insert was transferred into the BiFC expression vector pSAT4-DEST-nEYFP1–174-C1 (pE3136) or pCL112 (pBATL) to produce nYFP constructs. The same procedure was used for C-terminal YFP-tagged constructs, using pSAT5-DEST-cEYFP175–end-C1 (B) (pE3130) or pCL113 (pBATL) to produce cYFP constructs. The resulting plasmids were co-transfected into freshly prepared Arabidopsis leaf mesophyll protoplasts and incubated for 20–22 h (Wang et al. 2007). Transformed protoplasts were examined for YFP fluorescence and photographed using a Leica DM-6000B upright fluorescence microscope with phase and differential interference contrast (DIC) equipped with a Leica FW4000 digital image acquisition and processing system (Leica Microsystems).
Scanning electron microscopy
For scanning electron microscopy studies of Arabidopsis seeds (WT and mutants), the seeds were attached to standard EM stubs and sputter-coated with gold-palladium alloy using a Hummer VI sputtering system (Anatech, Union City, CA, USA). Specimens were visualized using a Hitachi Model S-800 scanning electron microscope (Hitachi, Norcross, GA, USA) and images were captured using an Evex Nano Analysis digital imaging system (Princeton, NJ, USA).
Resin embedding for bright field microscopy
Mature seeds were fixed in 3 % glutaraldehyde as described by Stork et al. (2010). All samples were slowly infiltrated with Spurr’s epoxy resin after transferring to propylene oxide solution. Dehydration, embedding, and sectioning were performed as described by Western et al. (2000). An Axioskop 2 microscope (Carl Zeiss) was used to photograph the sections. Height and width of the columella and radial wall were determined as described by Stork et al. (2010). The height of the radial walls was measured from the bottom of the mucilage pocket to the top of the wall in stained sections of the seed coat epidermis, using ImageJ software http://rsbweb.nih.gov/ij/. Similarly, width at the midpoint was measured for each radial wall. A total of at least ten cell walls were measured for each genotype, and Student’s t test was used to determine the significance of any differences in measured values.
Histochemical analyses
Staining with Ruthenium Red (Sigma, St. Louis, MO, USA) dissolved in water (0.2 % w/v) was used to monitor mucilage production from imbibed seeds (30 min at 25 °C), according to Beeckman et al. (2000). The stained seeds were photographed with an Axioskop 2 microscope image system (Carl Zeiss).
Statistical analysis of the results
Three biological replicates were used unless indicated. Pictures shown are the representative of the results. Two-sample analyses were done using Student’s t test. Error bars shown are either SEM or SD as indicated.
Accession number
GenBank database accession numbers for the genes investigated in this study and not indicated elsewhere are MYB75 (At1g56650), PAL1 (At2g37040), C4H (At2g30490), 4CL1 (At1g51680), HCT (At5g48930), C3H1 (At2g40890), CCoAOMT1 (At4g34050), CCR1 (At1g15950), F5H1 (At4g36220), COMT (At5g54160), CAD5 (At4g34230), CesA4 (At5g44030), CesA7 (At5g17420), CesA8 (At4g18780), IRX8 (At5g54690), IRX9 (At2g37090), CesA1 (At4g32410), CesA3 (At5g05170), CesA6 (At5g64740), FRA8 (At2g28110), MYB5 (At3g13540), TTG2 (At2g37260), TTG1 (At5g24520), AP2 (At4g36920), GL2 (At1g79840), and TT8 (At4g09820).
Results
Interaction of MYB75 with KNAT7 in vivo depends on the R2–R3 repeat of the MYB75 domain and the KNOX2 domain of KNAT7
The earlier observation that MYB75 and KNAT7 were able to physically interact in a Y2H assay (Bhargava et al. 2010), and that both transcription factors were involved in secondary cell wall deposition, suggested that these two phenomena might be functionally related. To better understand this relationship, we examined the interaction between MYB75 and KNAT7 more closely. In a protoplast transfection assay for transcription activation/repression activity, we tested the ability of effector plasmids carrying either KNAT7 or MYB75 to affect the expression of the co-transfected β-glucuronidase (GUS) reporter gene, which was under the control of the GAL4 DNA-binding site (GAL4::GUS) (Fig. 1a). When the protoplasts were transfected with an effector plasmid encoding only the yeast GAL4 DNA-binding domain polypeptide (GD), a modest level of the GUS reporter gene expression resulted (Fig. 1b), which represents the background level of GUS activity in this experiment. When the MYB75 open reading frame was fused with GD and used as an effector plasmid co-transfected with the GAL4::GUS reporter, a slight increase in GUS activity was seen, indicating that MYB75 might act as a weak transcriptional activator (Fig. 1b). KNAT7-GD (full length KNAT7 protein) binds to the GAL4 domain by itself and showed no significant repression of GUS activity in this assay. However, when an effector plasmid containing a hemagglutinin (HA)-tagged KNAT7 protein was co-transfected along with the GD-MYB75 and reporter plasmids, suppression of the GUS activity was observed (Fig. 1b), even though the KNAT7 protein is not able to bind to the promoter of the GUS reporter construct. This result is consistent with the proposed physical interaction between MYB75 and KNAT7, and also confirmed the previously reported transcriptional repressor activity of KNAT7 (Li et al. 2011).
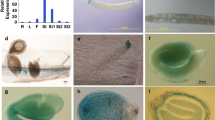
MYB75–KNAT7 interaction confirmed by protoplast transfection assay. a Effector and reporter constructs used in the transfection assays. b Protein–protein interaction. Effector genes included the GAL4 DNA-binding domain (GD) fused in-frame to MYB75, the GD alone and KNAT7 fused with hemagglutinin epitope (HA) tag. Effector gene plasmid DNA was co-transfected with reporter gene GAL4:GUS plasmid DNA into Arabidopsis leaf mesophyll protoplasts. GUS activity was assayed after the transfected protoplasts were incubated in darkness for 20–22 h. Shown are mean ± SE of three replicates. MYB75 and KNAT7 show interaction as seen by repression of the GUS activity
To identify the MYB75 domain(s) required for its interaction with KNAT7, separate regions of MYB75 were cloned, fused to the GAL4-AD, and tested for interaction with the full-length KNAT7 (FL-KNAT7) transcription factor in directed Y2H assays [Fig. 2a(i)]. A known and strong interaction between MYB75 and TT8 transcription factors (Zimmermann et al. 2004) was used as one positive control in this experiment, while FL-KNAT7 and full-length MYB75 (FL-MYB75) co-transformation provided another. The appropriate empty vector, along with one candidate protein, served as negative controls in each experiment. Positive interaction in the Y2H system was observed only between FL-KNAT7 and constructs expressing either the MYB75 R3 domain or FL-MYB75 [Fig. 2a(ii)]. We next used BiFC assays in Arabidopsis protoplasts to confirm in vivo the interactions observed in the Y2H system. After transformation with different fusion constructs, YFP fluorescence was detected in protoplasts co-transfected either with plasmids encoding the MYB75 R3 domain and FL-KNAT7 or with plasmids encoding the MYB75 R2–R3 domain and FL-KNAT7 [Fig. 2a(iii), Fig. S1a]. Together, these assay results suggest that the R3 domain, at least, of MYB75 is necessary and sufficient for the interaction of this transcription factor with KNAT7.
a Identification of MYB75 domain(s) involved in protein–protein interaction with KNAT7. i Different domains of MYB75 used to test the interaction with full length (FL) KNAT protein using yeast two-hybrid and BiFC assays. ii Yeast two-hybrid assay to test the interaction. Different domains of the MYB75 protein were fused to the GAL4 activation domain (AD) in yeast two-hybrid assays and tested for interaction with KNAT7-full length protein fused to the DNA-binding domain (BD). iii BiFC assays using split-YFP to test the same interactions in vivo using an Arabidopsis protoplast system. b Identification of KNAT7 domain(s) involved in protein–protein interaction with MYB75. i Different domains of KNAT7 were used to test the interaction with the MYB75 R2–R3 domain using yeast two-hybrid and BiFC assay. ii Yeast two-hybrid assay to test the interaction. Different domains of MYB75 protein were fused to the GAL4 activation domain (AD) and were tested for interaction with different domains of KNAT7 protein fused to the DNA-binding domain (BD) in yeast two-hybrid assays. iii BiFC was used to test the same interactions in vivo in the Arabidopsis protoplast system
In a reciprocal experiment designed to identify the regions of KNAT7 involved in its interaction with MYB75, separate KNAT7 domains were used to generate GAL4BD fusions [Fig. 2b(i)] and tested for their ability to interact with the MYB75R3 and R2–R3 domains in directed Y2H assays. The interaction between FL-KNAT7 and FL-MYB75 was used as a positive control. Neither the KNAT7 KNOX1 (K1) nor the tandem KNOX1–KNOX2 (K1–K2) domain supported interaction with MYB75 or its separate domains [Fig. 2b(ii)], aside from a weak interaction (growth of co-transformants on SC-Leu-Trp-His + 3AT but not on the more stringent selection medium, SC-Leu-Trp-Ura) between the MYB75 R2–R3 domain and the K2 domain of KNAT7 [Fig. 2b(ii)]. When these interactions were tested in vivo using BiFC, on the other hand, interactions were indeed observed between the R2–R3 domain of MYB75 and either the KNAT7–K2 or the KNAT7–K1–K2 domains (Fig. 2b(iii), Fig. S1b), but not with the KNAT7–K1 domain. No interactions were detected when the different KNAT7 domains were tested against only the R3 domain of MYB75 (Fig. S2). Overall, however, these domain interaction results imply that the physical interaction between KNAT7 and MYB75 is dependent primarily on the R3 domain of MYB75 and the KNOX2 domain of KNAT7.
MYB75 and KNAT7 regulate secondary cell wall formation in both the Arabidopsis stem and seed coat
Both MYB75 and KNAT7 have been shown to individually regulate secondary cell wall deposition in Arabidopsis inflorescence stems, where they each appear to work as negative regulators of wall deposition in interfascicular fiber cells (Bhargava et al. 2010; Li et al. 2012). In brief, light and transmission electron microscopic analysis of cross-sections from the basal part of inflorescence stems of the mutants showed collapsed vessels and increased thickness of interfascicular fiber cell walls. Comparison of the cell wall chemical composition of WT lower inflorescence stems with those of the single mutants showed higher acid-insoluble lignin content in the mutants, and expression of genes encoding enzymes known to be involved in secondary cell wall biosynthesis (lignin-, cellulose- and hemicellulose-associated genes) was also affected in the mutants (Bhargava et al. 2010; Li et al. 2012).
In light of the impact on secondary cell wall development in the inflorescence stem of manipulating either MYB75 or KNAT7 expression, we asked whether these transcription factors also helped regulate the same process in other tissues, such as the seed coat. The mature Arabidopsis seed is surrounded by a rigid coat that derives much of its strength from secondary cell wall polymers that are intensively deposited on the thick radial walls and the central columella of the epidermal cells as the seed matures. A recent study found that loss-of-function at the KNAT7 locus affected both seed coat structure and mucilage production (Romano et al. 2012), but the possible influence of MYB75 on this phenotype was not examined.
We first examined the seed coat surface and cell shape phenotypes of myb75 and knat7 mutants by scanning electron microscopy (SEM). This revealed that the surface structures of the mature myb75 seed coat were irregular. The angle between the radial wall junctions was distorted in this mutant, and in general, only four radial walls were visible, in contrast to the symmetrical six-walled structures that dominate the surface of WT seed coats (Fig. 3a). No change in the hexagonal epidermal cell wall pattern was evident in the knat7-1 seed, but the radial walls were thicker in both myb75-1 and knat7-1 seed coats, as compared to their respective WT seeds (Fig. 3a). Although the dimensions of the radial walls of epidermal cells in the pap1-d (gain-of-function MYB75 mutant) appeared different (smaller and thinner) than in WT (Columbia), those differences were not significant at p < 0.05 (Fig. S3). To quantify the phenotypes observed by SEM, radial wall heights and widths were measured from a total of ten cells for each genotype in Toluidine blue-stained sections (Fig. 3b, c), and those results confirmed the visual observations.
Mutations in MYB75 result in distorted cell shape and thickening of radial walls in seed coat epidermal cells. a Scanning electron microscopy of dry seeds of wild type (Columbia and Nossen), myb75-1 and knat7-1 (as indicated). Note the prominent hexagonal cell walls and central volcano-shaped columella in the center of WT cells. Radial wall and columella are the regions of secondary cell wall deposition. Blue arrows indicate changes in radial wall and red arrows indicate columella. b Epidermal cell morphology of wild type (Columbia and Nossen), myb75-1 and knat7-1 Toluidine blue-stained sections of aqueous (3 % v/v) glutaraldehyde-fixed mature seeds (as indicated). (c) Quantification of the radial wall height, width, and columella length of wild-type and mutant seed coat cells (μm). Error bars are SE from the mean. Square indicates significant difference from WT-Col. Asterisk indicates significant difference from WT-Nos. myb75 is in Nossen background and knat7 is in Columbia background
Seed coat maturation involves not only deposition of massive secondary cell walls in the epidermal cells but also synthesis of large quantities of pectinaceous mucilage as well as proanthocyanidins (Dixon et al. 2005). Since mucilage deposition is closely linked developmentally with secondary cell wall formation in these cells, we examined the possibility that either MYB75 or KNAT7 might also play a role in mucilage biosynthesis. Defects in mucilage accumulation, structure, or release from hydrated epidermal cells can be easily detected following imbibition of mature seeds (Fig. S4). When WT Arabidopsis seeds are submerged in water, the epidermis of the seed coat cells ruptures, and the mucilage released from the mucilage pockets expands to form a gelatinous coat over the seeds (Western et al. 2004). This mucilage coat can be visualized by staining with the dye Ruthenium Red, which stains negatively charged polymers such as pectin (Koornneef 1981) (Fig. S4).
When knat7 seeds were placed in water without agitation and stained with Ruthenium Red, a dense layer of mucilage was apparent. By contrast, when the seeds were stained after shaking in water (a treatment that disrupts the pectin network of the mucilage), only a barely visible layer of stained mucilage could be observed around knat7 seeds, whereas WT seeds retained an adherent layer of mucilage after shaking (Fig. S4). This indicates that loss of KNAT7 interferes with some aspect of mucilage adherence to the seed coat following mucilage release. In myb75 seeds, on the other hand, a mucilage layer similar to WT was observed for imbibed seeds, either with or without shaking (Fig. S4). The role of MYB75 in developing seeds thus appears to be more focused on secondary cell wall formation.
MYB75 and KNAT7 are part of the transcriptional network regulating secondary cell wall formation and mucilage formation in the Arabidopsis seed coat
In light of the ability of MYB75 and KNAT7 to interact physically, and the similarities in seed coat phenotype associated with their loss-of-function mutants, we anticipated that MYB75 and/or KNAT7 might form a part of the genetic network regulating seed coat maturation. If this were true, we predicted that the expression of the corresponding genes in developing seeds would be most prominent during the period 7–11 days post-anthesis (DPA) (Haughn and Chaudhury 2005). Quantitative real-time PCR (qRT-PCR) assays of MYB75 and KNAT7 expression in WT seed coats confirmed that this was indeed the case (Fig. 4a, b), as did the publicly available expression data for MYB75 and KNAT7 (Winter et al. 2007; Le et al. 2010; Dean et al. 2011) (Fig. S5 and Fig. S6).
Expression of MYB75 and KNAT7 in mutants of putative regulatory transcription factors during seed coat development. Total RNA was isolated from 3-DPA, 7-DPA, and 11-DPA stages of seed coat development of wild-type, ap2-1, gl2-1, ttg1-1, ttg2-1, and myb75-1 and knat7-1 siliques. cDNA was used for quantitative real-time PCR (q-RT-PCR) using gene-specific primers for MYB75 (a) and KNAT7 (b). Gene expression values are arbitrary units and were calculated as described previously (Gutierrez et al. 2008). DPA days post-anthesis
Previous studies have identified a number of transcription factors whose activity influences either secondary cell wall formation, mucilage formation, or both. In developing Arabidopsis seeds (Haughn and Chaudhury 2005; Gonzalez et al. 2009; Romano et al. 2012), We asked whether loss-of-function at any of these loci would impact the timing or intensity of KNAT7 or MYB75 expression in developing seed coats. For this purpose, we quantified the expression of both genes in plants harboring knock-out mutations of APETALA2 (AP2), TRANSPARENT TESTA GLABRA 1 (TTG1), GLABRA2 (GL2), and TRANSPARENT TESTA GLABRA 2 (TTG2). Among the mutants tested, only loss of TTG2 strongly affected MYB75 transcript abundance (Fig. 4a), indicating that TTG2 activity is controlling MYB75 expression. Since no changes in MYB75 expression were observed in the ap2, gl2, or ttg1 mutant backgrounds, this potential MYB75–TTG2 regulatory node may be independent from pathways previously shown to be regulated by TTG2 (Johnson et al. 2002; Dilkes et al. 2008).
It is noteworthy that while KNAT7 expression was only modestly reduced in myb75 seed coats, compared to WT (Fig. 4b), it was strongly elevated in 3 DPA and 11 DPA seed coats in the ttg2 background. This response would be indicative of negative regulation (repression) of KNAT7 expression by TTG2 at these particular stages of seed coat development (Fig. 4b). On the other hand, expression of KNAT7 was reduced in the ap2 and gl2 backgrounds, which suggests that KNAT7 expression may be positively regulated by AP2 and GL2, and potentially places KNAT7 downstream of these transcription factors in the seed coat regulatory network. Other transcriptional regulators may also be involved in modulating KNAT7 function in the seed coat, since the KNAT7 promoter has recently been shown to be a direct target of the MYB61 transcription factor, and myb61 loss-of-function mutants possess both aberrant seed coat morphology and mucilage release properties (Romano et al. 2012).
MYB75 and KNAT7 interact physically with other seed coat-associated transcription factors
To identify potential protein–protein interactions between MYB75/KNAT7 and other TFs known to regulate seed coat differentiation (MYB5 and TT8), we used directed Y2H assays (Fig. 5). Each of the tested candidates was fused to either the GAL4 DNA-binding domain (BD) or the GAL4 activation domain (AD), and they were then assayed for their ability to bind to themselves, or to each other. However, because of the inherent transcription activating function of MYB proteins, it was not possible to use the MYB constructs as BD fusions in this assay.
In vitro protein–protein interactions among potential secondary cell wall-associated and seed coat regulating proteins as determined by yeast two-hybrid assay (a) and confirmation of interactions in vivo by BiFC assay in Arabidopsis protoplasts (b). Known interaction as indicated in text (Zimmermann et al. 2004) was used as a positive control and co-transformed empty vectors were used as a negative control
Positive Y2H interactions were observed between KNAT7 and MYB5 (Fig. 5a), a transcription factor previously shown to regulate seed coat mucilage biosynthesis (Li et al. 2009), and between MYB75 and TT8. The latter interaction had been observed in other studies (Zimmermann et al. 2004), as had the MYB75–KNAT7 interaction (Bhargava et al. 2010). On the other hand, no interaction was observed between KNAT7 and TT8, despite the ability of both to interact with MYB75 (Fig. S7). BiFC assays confirmed that each of the observed Y2H interactions could also be detected in vivo, as could MYB5–TT8 interaction (Fig. 5b).
Discussion
No functional information is available about physical interaction between members of the plant homeodomain and MYB transcription factor families, with the exception of one report (Timmermans et al. 1999) of MYB and KNOX interaction in maize. The observation that KNAT7 (Li et al. 2011; Brown et al. 2005; Zhong et al. 2008; Romano et al. 2012) and MYB75 (Bhargava et al. 2010) both play roles in secondary wall formation, and physically interact (Bhargava et al. 2010), suggested that these two proteins might function as a part of a KNOX-MYB complex to help regulate secondary wall formation in Arabidopsis. We confirmed the physical interaction between MYB75 and KNAT7 using in vivo assays (Figs. 1, 2), and identified the MYB-R3 domain of MYB75 as an important region for binding to the KNOX domain of KNAT7. This same MYB domain has been shown to mediate interaction with bHLH proteins (Zimmermann et al. 2004), but it has also been proposed that the MYB domain might contribute to DNA binding, as well (Pooma et al. 2002). The KNOX domain, on the other hand, may support higher-order complex formation for regulation of target genes (Dubos et al. 2010).
In addition to the physical interaction between these two TF proteins, the genes encoding them in Arabidopsis are expressed with similar spatial and temporal patterning, both in inflorescence stems (Bhargava et al. 2010; Li et al. 2011) and in developing seeds (present work). In seeds, both MYB75 and KNAT7 exhibit their highest transcript abundance at 11 DPA, the stage in seed coat maturation during which secondary cell wall deposition takes place. Loss-of-function at either locus results in inflorescence stem phenotypes that include characteristic enhanced thickening of the interfascicular fiber cell walls (Li 2009; Brown et al. 2005; Romano et al. 2012), and also small but measurable differences in the structure and/or dimensions of the seed coat epidermal cell walls.
These correlations are collectively consistent with a model in which these two transcription factors act together to modulate secondary cell wall formation both during seed coat maturation and stem vasculature development. At the same time, however, it is clear that each TF also makes unique contributions to these developmental processes. For instance, the knat7-1 mutant shows an irx phenotype indicative of secondary cell wall changes in vessel elements (Li et al. 2012), whereas myb75-1 does not (Bhargava et al. 2010). In addition to this evidence of tissue specificity, loss of KNAT7, but not of MYB75, results in altered seed coat mucilage composition or secreted mucilage amount. Since, this knat7-1 phenotype is similar to the seed phenotype of a cesa5 mutant (Harpaz-Saad et al. 2011; Mendu et al. 2011; Sullivan et al. 2011), where reduced mucilage adherence was associated with a decrease in the cellulose content of the mucilage, it is possible that KNAT7 acts directly or indirectly as a positive regulator of the activity of CESA5, or of another gene required for synthesis of mucilage cellulose (Sullivan et al. 2011; Stork et al. 2010).
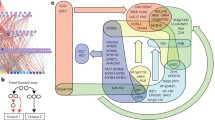
Based on the available evidence, KNAT7 function seems to be primarily restricted to developing secondary walls (Li 2009; Zhong et al. 2008; Li et al. 2011), in contrast to the broader regulatory functions (anthocyanin biosynthesis, senescence, nitrogen metabolism, secondary cell wall deposition) associated with MYB75 (Tohge et al. 2005; Teng et al. 2005; Lea et al. 2007; Bhargava et al. 2010). The ability of MYB75 to participate in regulation of secondary wall formation could be envisioned as being mediated through its interaction with KNAT7 within larger complexes. Although MYB75 itself shows weak transcriptional activation, and acts as a positive regulator of anthocyanin accumulation when over-expressed (Gonzalez et al. 2008), its physical interaction with KNAT7 results in enhancement of KNAT7’s repression of secondary cell wall deposition (Fig. 6). In this model, KNAT7, a transcriptional repressor (Li et al. 2011), may directly bind to the promoter of secondary cell wall-specific genes and form larger regulatory complexes by recruiting MYB75 and other components such as ovate family proteins (OFPs; Li et al. 2011) to the promoter region. These complexes could have different constituents in different cell and tissue types, such as stem interfascicular fiber cells (Fig. 6a) and seed coat epidermal cells (Fig. 6b), and might also be re-configure at different stages of tissue development. For instance, the bimodal pattern of KNAT7 expression in developing seeds might reflect two distinct roles for KNAT7 during differentiation of the seed coat epidermis; initially, during mucilage synthesis/secretion and later, during the formation of the secondary cell wall in the epidermal cells. Such an alternation of regulatory function during development has been reported for both plant (Ishida et al. 2007; Zheng et al. 2007) and animal systems (Greenbaum et al. 2004).
Model depicting the regulatory role of a putative MYB75–KNAT7 complex in secondary cell wall formation. In this model, KNAT7 (which is typically a transcriptional repressor) directly binds to the promoter of secondary cell wall-specific genes, while MYB75 and other components interact with KNAT7 to form development-specific complexes that modulate secondary cell wall synthesis in different tissues
Both our genetic interaction and protein–protein interaction data point to the possibility that MYB75 and KNAT7 form distinct complexes specifically with transcription factors of the TTG1 complex. The TTG1 protein has been shown to have roles in multiple pathways in seed coat development and other processes (Gonzalez et al. 2009; Debeaujon et al. 2003; Ishida et al. 2007) by acting as a WD40 scaffold protein to recruit different proteins into a functional complex. For example, in the regulation of proanthocyanidin biosynthesis, TTG1 interacts with TT2 (MYB family) and TT8 (bHLH family) (Debeaujon et al. 2003; Marles et al. 2003), while during mucilage biosynthesis it recruits MYB5 and EGL3/TT8 (Gonzalez et al. 2009), and in trichome development it interacts with GL1 and GL3/EGL3 (Zhao et al. 2008). Recently, a protein complex was described that involves multiple Arabidopsis TFs (including MYB75, TT8, GL3, EGL3, and GL1) interacting with another transcriptional regulator from the jasmonate ZIM-domain family (COI1). Proteolytic degradation of COI1 induced by jasmonate is proposed to release the other TFs from the COI1 scaffold, enabling them to activate downstream genes that regulate anthocyanin accumulation and trichome initiation (Qi et al. 2011). In an analogous fashion, TTG1 could act as a scaffold protein to help bring a MYB75–KNAT7 complex together and regulate secondary cell wall formation, but may recruit only KNAT7, together with different mucilage-specific regulators like MYB5 (Gonzalez et al. 2009; Li et al. 2009), to form a putative mucilage regulatory complex (Fig. S8).
Conserved regulatory roles but different interacting partners within transcription factor complexes have been found in invertebrate and mammalian systems (Dallman et al. 2004). Although in plants it is now well established that some protein interactions are conserved in different tissue types during transcriptional regulation, it is likely that participation of many other interacting protein partners contributes to the final pattern of tissue specificity. Therefore, such differentiated complexes could explain how MYB75 and KNAT7 might regulate secondary cell wall formation in both the seed coat and flowering stem. The expression of KNAT7 in the ttg1 and ttg2 backgrounds shows a bimodal distribution of expression across seed coat development stages (Fig. 4b), which would be consistent with the existence of distinct transcript populations in 3 DPA and 11 DPA seed coats, and with their differential regulation by TTG1 and TTG2. TTG2 and TTG1 are believed to operate sequentially in the same regulatory cascade (Johnson et al. 2002) and therefore would be anticipated to share some biological functions. TTG1 is thought to form a multi-protein transcriptional complex with GLABROUS1 (GL1) and GL3/EGL3, and to be involved in controlling epidermal trichome development (Ishida et al. 2007). In view of the increased expression of KNAT7 in the ttg1 and ttg2 3DPA seed coat, it seems possible that an analogous TTG1/2-KNAT7 complex might be contributing to early epidermal development in the seed. TTG1 has also been shown to regulate seed coat mucilage production by interacting with MYB61, EGL3 and TT8 (Penfield et al. 2001), and heightened expression of KNAT7 in 11 DPA seed coats might indicate that both TTG1/2 and KNAT7 are also involved in this function. We could speculate that KNAT7-associated transcripts (and their derived proteins) may play different roles in each stage, with one set (3 DPA) involved in early growth and development of the epidermal layers, while the other (11 DPA) would be more specific for functions (e.g. mucilage and secondary wall deposition) associated with later stages of differentiation.
In summary, our data are not only consistent with conservation of an overall role of MYB75 and KNAT7 as negative regulators of secondary cell wall formation in Arabidopsis but also suggest modulation of this role, perhaps through their participation in different functional complexes whose composition would be governed, in part, by the spatial and temporal expression of other TFs. In planta identification of the relevant protein complexes will be necessary to provide a fuller understanding of the dynamics of this regulatory network.
Abbreviations
- Y2H:
-
Yeast two-hybrid
- SEM:
-
Scanning electron micrograph
- PCR:
-
Polymerase chain reaction
References
An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10:107–121
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380
Beeckman T, De Rycke R, Viane R, Inzé D (2000) Histological study of seed coat development in Arabidopsis thaliana. J Plant Res 113:139–148
Bellaoui M, Pidkowich MS, Samach A, Kushalappa K, Kohalmi SE, Modrusan Z, Crosby WL, Haughn GW (2001) The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13:2455–2470
Bhargava A, Mansfield SD, Hall HC, Douglas CJ, Ellis BE (2010) MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol 154:1428–1438
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2394
Broun P (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol 8:272–279
Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17:2281–2295
Burglin TR (1997) Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids 25(21):4173–4180
Dallman JE, Allopenna J, Bassett A, Travers A, Mandel G (2004) A conserved role but different partners for the transcriptional corepressor CoREST in fly and mammalian nervous system formation. J Neurosci 24:7186–7193
Dean G, Cao Y, Xiang D, Provart NJ, Ramsay L, Ahad A, White R, Selvaraj G, Datla R, Haughn G (2011) Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol. Plant 4(6):1074–1091
Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15:2514–2531
Demura T, Ye ZH (2010) Regulation of plant biomass production. Curr Opin Plant Biol 13:299–304
Dilkes BP, Spielman M, Weizbauer R, Burkart-Waco D, Watson B, Scott RJ, Comai L (2008) The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PLoS Biol 6:2707–2720
Dixon R, Xie DY, Sharma S (2005) Proanthocyanidins—a final frontier in flavonoid research? New Phytol 165:9–28
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 10:573–581
Frampton J, Gibson TJ, Ness SA, Doderlein G, Graf T (1991) Proposed structure for the DNA-binding domain of the Myb oncoprotein based on model building and mutational analysis. Protein Eng 4:891–901
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Gonzalez A, Mendenhall J, Huo Y, Lloyd A (2009) TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev Biol 325:412–421
Greenbaum S, Lazorchak AS, Zhuang Y (2004) Differential functions for the transcription factor E2A in positive and negative gene regulation in pre-B lymphocytes. J Biol Chem 279:4502–4503
Guimil S, Dunand C (2006) Patterning of Arabidopsis epidermal cells: epigenetic factors regulate the complex epidermal cell fate pathway. Trends Plant Sci 11:601–609
Gutierrez L, Mauriat M, Pelloux J, Bellini C, Van Wuytswinkel O (2008) Towards a systematic validation of references in real-time RT-PCR. Plant Cell 20:1734–1735
Hackbusch J, Richter K, Muller J, Salamini F, Uhrig JF (2005) A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci U S A 102:4908–4912
Harpaz-Saad S, McFarlane HE, Xu S, Divi UK, Forward B, Western TL, Kieber JJ (2011) Cellulose synthesis via the FEI2 RLK/SOS5 pathway and CELLULOSE SYNTHASE 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J 68:941–953
Haughn G, Chaudhury A (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10:472–477
Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kato T, Tabata S, Okada K, Wada T (2007) Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19:2531–2543
Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14(6):1359–1375
Kanei-Ishii C, Sarai A, Sawazaki T, Nakagoshi H, He DN, Ogata K, Nishimura Y, Ishii S (1990) The tryptophan cluster: a hypothetical structure of the DNA-binding domain of the myb protooncogene product. J Biol Chem 265:19990–19995
Kanei-Ishii C, Tanikawa J, Nakai A, Morimoto RI, Ishii S (1997) Activation of heat shock transcription factor 3 by c-Myb in the absence of cellular stress. Science 277:246–248
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:236–242
Koornneef M (1981) The complex syndrome of ttg mutants. Arabidopsis Inf Serv 18:45–51
Kumar R (2006) The roles of BEL1-like proteins in organ morphogenesis in Arabidopsis thaliana. PhD thesis, University of British Columbia (UBC), Vancouver
Le BH et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. PNAS 107:8063–8070
Lea US, Slimestad R, Smedvig P, Lillo C (2007) Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 225:1245–1253
Li E (2009) Identification and characterization of regulatory genes associated with secondary wall formation in Populus and Arabidopsis thaliana. PhD thesis, University of British Columbia
Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, Koltunow AM, Parish RW (2009) The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21:72–89
Li E, Wang S, Liu Y, Chen JG, Douglas CJ (2011) OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J 67:328–341
Li E, Bhargava A, Qiang W, Friedmann MC, Forneris N, Savidge RA, Johnson LA, Mansfield SD, Ellis BE, Douglas CJ (2012) The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol 194:102–115
Marles MA, Ray H, Gruber MY (2003) New perspectives on proanthocyanidin biochemistry and molecular regulation. Phytochemistry 64:367–383
Martin C, Glover BJ (2007) Functional aspects of cell patterning in aerial epidermis. Curr Opin Plant Biol 10:70–82
Matsui K, Tanaka H, Ohme-Takagi M (2004) Suppression of the biosynthesis of proanthocyanidin in Arabidopsis by a chimeric PAP1 repressor. Plant Biotechnol J 2:487–493
Mendu V, Griffiths J, Persson S, Stork J, Downie B, Vioniciuc C, Haughn G, Debolt S (2011) Subfunctionalization of cellulose synthases in seed coat epidermal cells mediate secondary radial wall synthesis and mucilage attachment. Plant Physiol 157:441–453
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12:1863–1878
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13:2099–2114
Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13:2777–2791
Pooma W, Gersos C, Grotewold E (2002) Transposon insertions in the promoter of the Zea mays a1 gene differentially affect transcription by the Myb factors P and C1. Genetics 161(2):793–801
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The Jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23:1795–1814
Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10:63–70
Romano JM, Dubos C, Prouse MB, Wilkins O, Hong H, Poole M, Kang K-Y, Li E, Douglas CJ, Western TL, Mansfield SD, Campbell MM (2012) AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytol. doi:10.1111/j.1469-8137.2012.04201.x
Schellmann S, Hulskamp M (2005) Epidermal differentiation: trichomes in Arabidopsis as a model system. Int J Dev Biol 49:579–584
Serna L (2005) Epidermal cell patterning and differentiation throughout the apical-basal axis of the seedling. J Exp Bot 56:1983–1989
Serna L, Martin C (2006) Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci 11:274–280
Stork J, Harris D, Griffiths J, Williams B, Beisson F, Li-Beisson Y, Mendu V, Haughn G, Debolt S (2010) CELLULOSE SYNTHASE9 serves a nonredundant role in secondary cell wall synthesis in Arabidopsis epidermal testa cells. Plant Physiol 153:580–589
Sullivan S, Ralet MC, Berger A, Diatloff E, Bischoff V, Gonneau M, Marion-Poll A, North HM (2011) CESA5 is required for the synthesis of cellulose with a role in structuring the adherent mucilage of Arabidopsis seeds. Plant Physiol 156:1725–1739
Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139:1840–1852
Timmermans MC, Hudson A, Becraft PW, Nelson T (1999) ROUGH SHEATH2: a MYB protein that represses KNOX homeobox genes in maize lateral organ primordia. Science 284:151–153
Tohge T, Matsui K, Ohme-Takagi M, Yamazaki M, Saito K (2005) Enhanced radical scavenging activity of genetically modified Arabidopsis seeds. Biotechnol Lett 27:297–303
Viola IL, Gonzalez DH (2006) Interaction of the BELL-like protein ATH1 with DNA: role of homeodomain residue 54 in specifying the different binding properties of BELL and KNOX proteins. Biol Chem 387:31–40
Wang S, Chang Y, Guo J, Chen JG (2007) Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J 50:858–872
Western TJ (2006) Changing spaces: the Arabidopsis mucilage secretory cells as a novel system to dissect cell wall production in differentiating cells. Can J Bot 84:622–630
Western TL, Skinner DJ, Haughn GW (2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122:345–356
Western TL, Young DS, Dean GH, Tan WL, Samuels AL, Haughn GW (2004) MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol 134:296–306
Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2(8):e718. doi:10.1371/journal.pone.0000718
Wolberger C (1999) Multiprotein-DNA complexes in transcriptional regulation. Annu Rev Biophys Biomol Struct 28:29–56
Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H, Demura T (2010) VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22:1249–1263
Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130:4859–4869
Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A (2008) The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135:1991–1999
Zheng Z, Mosher S, Fan B, Klessig D, Chen Z (2007) Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol 7:2
Zhong R, Ye ZH (2007) Regulation of cell wall biosynthesis. Curr Opin Plant Biol 10:564–572
Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20:2763–2782
Zhong R, Lee C, Ye ZH (2010) Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci 15:625–631
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40:22–34
Acknowledgments
We would like to thank Brad Ross and the UBC Bioimaging facility for SEM support. We gratefully acknowledge Dr Joachim Ührig (Botanical Institute, Universität Köln) for the kind gift of the pBatTL vectors used for the BiFC assays. Funding for this work was provided by Discovery Grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) to SDM, GWH, CJD, and BEE, the NSERC Green Crops Network and by NSERC scholarship support to AB.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bhargava, A., Ahad, A., Wang, S. et al. The interacting MYB75 and KNAT7 transcription factors modulate secondary cell wall deposition both in stems and seed coat in Arabidopsis. Planta 237, 1199–1211 (2013). https://doi.org/10.1007/s00425-012-1821-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-012-1821-9