Abstract
Genes encoding two new isoforms of sucrose synthase from barley, HvSs3 and HvSs4, have been characterised and their expression patterns compared with those previously described for HvSs1 and HvSs2, in different organs and during seed maturation and germination. Their response to several abiotic stimuli has also been investigated in leaves: HvSs1 is up-regulated by anoxia and HvSs3 by water deprivation while no response is observed to 150 mM NaCl treatment; HvSs1 and HvSs3 are also induced by cold temperatures. Using translational fusions and transient expression analyses, the four isozymes have been localised not only to the cytoplasm but also along several cytoplasmic tracks and at the inner side of the cell membrane; besides, HvSS1 is also associated with mitochondria, a localisation that has been predicted in silico with the TargetP and Predotar programmes. These data suggest distinct although partially overlapping roles, for the four barley sucrose synthase isoforms, in the channelling of carbon towards different metabolic pathways within the cell.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In higher plants, sucrose produced in photosynthetic sources is transported to sink tissues where it is cleaved as a first step for its utilisation in various metabolic pathways. This process can be catalysed by invertases (EC 3.2.1.26) that irreversibly hydrolyse this disaccharide into glucose and fructose and by the sucrose synthases (SUS; EC 2.4.1.13) that catalyse the reversible reaction: sucrose + UDP ↔ UDP-glucose + fructose. The SUS isozymes play an important role due to their dual capacity of directing carbon towards polysaccharide biosynthesis (cellulose, starch, fructans or lipids) or towards the adenylate-conserving pathway of respiration (Koch 2004).
A six member multigene family has been found to encode different SUS isoforms in several plant species: Arabidopsis thaliana (Barratt et al. 2001), Oryza sativa (Harada et al. 2005) and Lotus japonicus (Horst et al. 2007). In Arabidopsis and rice, these six genes have distinct although partially overlapping expression patterns and they respond differentially to various abiotic stresses such are oxygen deprivation or dehydration (Baud et al. 2004; Hirose et al. 2008).
In maize, three paralogous genes Sh1, Sus1 and Sus3 encoding different SUS isozymes SH1, SUS1 and SUS3, respectively, are highly expressed in seeds (Carlson et al. 2002). Genetic evidence shows that the two first SUS isozymes influence the starch content of the endosperm, since the mutant sh1 Sus1 and the double mutant sh1 sus1-1 accumulate 78 and 53%, respectively, of the total starch content found in the wild type seed (Chourey et al. 1998). These authors have inferred that the SH1 isozyme plays the dominant role in providing the substrate for cellulose biosynthesis whereas the SUS1 protein is needed for generating precursors for starch biosynthesis. The third maize Sus3 gene is predominantly expressed in embryos (Carlson et al. 2002). Barratt et al. (2001) studying a rug4 mutant in peas, which carry mutations in PsSus1, have assigned a role to this gene in starch accumulation in seeds. A major role of SUS for sink strength, has been inferred from the antisense inhibition of the SUS genes in transgenic potatoes that lead not only to a decrease in starch accumulation but also to a reduction of soluble proteins and total dry weight in tubers; noteworthy, an enhancement of SUS activity in transgenic potatoes increased the starch content and total yield in tubers (Zrenner et al. 1995; Baroja-Fernández et al. 2009).
Despite the accepted importance of SUS isoforms, a systematic analysis in Arabidopsis of single mutants, lacking individual SUS isoforms, failed to discover an obvious phenotype or a specific role for any of these six isoforms. Although none of the sucrose synthases individually appear to be essential for normal growth of Arabidopsis under well-aerated conditions, AtSUS1 and AtSUS4 are together necessary for tolerance to hypoxic conditions (Bieniawska et al. 2007). A more detailed biochemical analysis of single knock-out mutants of the two isoforms expressed strongly and preferentially in Arabidopsis seeds (AtSus2 and AtSus3), has indicated that SUS is important for metabolite homeostasis and for the timing of seed development (Angeles-Nuñez and Tiessen 2010). In addition, the double mutant Atsus5 Atsus6 has a phenotype with reduced amounts of callose in the sieve plates (Barratt et al. 2009). The over-expression of the Gossypium hirsutum sucrose synthase in hybrid poplar results in an increase of secondary cell wall cellulose content, thus suggesting an association of sucrose synthase with cellulose synthesis (Coleman et al. 2009). Furthermore, a recent study in Azuki beans has demonstrated that SUS is part of the catalytic complex for cellulose synthesis in the plasma membrane-associated rosette structure (Fujii et al. 2010), thus corroborating previous data of SUS involvement in cotton fibre formation (Ruan et al. 2003). Additional SUS functions have been proposed in other important metabolic processes such are nitrogen fixation in legume nodules and maintenance of the respiratory requirements for sucrose loading and unloading in vascular tissues (Nolte and Koch 1993; Gordon et al. 1999).
Two genes HvSs1 and HvSs2, located respectively in chromosomes 7H and 2H, encoding two distinct isoforms of SUS, HvSS1 and HvSS2 respectively, have been characterised in barley (Hordeum vulgare, Sánchez de la Hoz et al. 1992; Martínez de Ilarduya et al. 1993). Both mRNAs are strongly expressed in developing barley endosperm, but while HvSs2 is seed specific, HvSs1 is also detected by Northern blot analysis in vegetative tissues. Western blot analysis (Guerin and Carbonero 1997) demonstrated that the prevalent protein in roots is the homotetramer (SS1)4, and immuno-cytological studies indicated that SS1 is present in the vascular region and in the root cap cells. In barley seeds, all five possible tetramer combinations made from HvSS1 and HvSS2 proteins, [(SS1)4; (SS1)3SS2; (SS1)2(SS2)2; SS1(SS2)3; (SS2)4] are detected in non-denaturing Western blots. HvSS1 is immuno-localised to the assimilate-unloading tissues of seeds, like the nucellar projection, the chalazal region, and the vascular bundle at the base of the endosperm, suggesting a significant role of HvSS1, but not HvSS2, in the sucrose unloading process into the endosperm (Guerin and Carbonero 1997).
Here, we report the molecular characterization of two new SUS encoding genes in barley (HvSs3, and HvSs4) and a phylogenetic comparison of the four barley genes with the Sus genes previously described in rice and maize, as well as, with the six members of this family annotated in the model plant Brachypodium distachyon. A comprehensive expression profile by RT-qPCR of the four barley genes (HvSs1, HvSs2, HvSs3 and HvSs4) has been performed, including time-scale studies in developing and germinating seeds, as well as their responses to several abiotic stresses. The subcellular localisation has been also established by confocal microscopy, using translational fusion constructs of HvSS1, HvSS2, HvSS3 and HvSS4 to the green fluorescent protein (GFP). Taken together, these data suggest diverse, although partially overlapping roles for the four distinct SUS isozyme genes in barley.
Materials and methods
Bioinformatic search and phylogenetic dendrogram
To identify further putative members of the SUS gene family in barley, a search was done at the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov). The cDNAs encoding two new SUS isozymes (HvSs3 and HvSs4) were annotated from the collection of CREST, The Barley Germplasm Centre from Japan (Sato et al. 2009). To determine the exon–intron structure, genomic DNA purified from barley leaves was amplified by a PCR strategy, using primers flanking the putative exon sites.
The nucleotide sequences HvSs1, HvSs2, HvSs3 and HvSs4, were used as a query to identify potential SUS genes in the genome of Brachypodium distachyon through TBLASTN at the Brachypodium Genome Database (http://www.brachybase.org). A further search for conserved domains in the corresponding SUS proteins was done with the InterPro program using the Pfam database (Bateman et al. 2002).
The complete deduced amino acid sequences of the genes from barley, wheat, maize, rice, Brachypodium and Arabidopsis were used to construct a phylogenetic dendrogram. The alignment of these sequences was carried out by means of the CLUSTAL W program (Thompson et al. 1994) prior to the phylogenetic analysis by the Neighbour-Joining method with the MEGA software, version 4.0 (Tamura et al. 2007), using a bootstrap analysis with 1000 replicates, pairwise deletion and the Jones–Taylor–Thornton matrix as settings.
The bioinformatic search for prediction of the subcellular localisation of the SUS proteins was done by using the TargetP (Emanuelsson et al. 2007) and the Predotar (Small et al. 2004) programmes.
Plant material and growth conditions
Seeds of Hordeum vulgare cv. Bomi (supplied by Risö Experimental Station, Denmark) were surface-sterilised and germinated on wet filter paper for 2 days. Seedlings were then transferred to sterilised compost in individual pots and grown at 22°C under a photoperiod of 16/8 h (light/dark) for 8 days, vernalised at 4°C for 7 days in the dark, and then transferred again to the greenhouse at 22°C and a 16/8 h (light/dark) photoperiod. For studies in time course expression, developing endosperms at 10, 14, 18, 22 and 26 days after pollination (dap), immature embryos at 14 dap and mature embryos were isolated. Germinating seeds of barley cv. Himalaya were used for the preparation of aleurone layers at 8, 16, 24 and 48 h after imbibition (hai); germinating embryos at 16 hai were also collected.
RNA extraction and cDNA preparation
Total RNA was purified from frozen leaves and roots of 10-day-old plants and from developing seeds by the phenol/chloroform extraction method and precipitated with 3 M LiCl (Lagrimini et al. 1987). RNA from aleurones of germinating seeds was purified according to Chang et al. (1993) a procedure that includes polyvinylpyrrolidone (PVP-K30) and β-mercaptoethanol (β-ME) in the extraction buffer. Aliquots (2 μg) of RNA, previously incubated with DNAse (DNase I, RNAase-free; Roche Diagnostics, Mannheim, Germany), were used as templates to synthesise the first-strand of cDNA, according to the manufacturer’s instructions (High-Capacity cDNA Reverse Transcription Kit, Applied Biosystems, Foster City, CA, USA).
Real-time quantitative PCR analyses
The RT-qPCR analyses were performed (in triplicate biological samples) using a SYBR-green detection procedure and a 7300 Real Time PCR System from Applied Biosystems. Quantification was standardised to the barley α-Actin2 mRNA level (Moreno-Risueno et al. 2007, 2008) with the exception of the abiotic stress response experiments where the UbiquitinE2 (UbiE2) gene was used as a reference, since the α-Actin2 gene was shown to be expressed in a variable manner in response to these stresses, while the UbiE2 gene remained constant, under the conditions assayed (Fig. S1).
The specific primer sets designed for the RT-qPCR analyses for each gene appear in Supplementary Table S1.
Subcellular localisation
Translational fusions of HvSS1, HvSS2, HvSS3 and HvSS4 to the GFP reporter were generated by cloning the corresponding ORFs into the BamHI restriction site of a psmRS-GFP plasmid, that was obtained from a pUC118 vector (U07649) in which the 35S-psmRS-GFP-Nos had previously been cloned into the HindIII–EcoRI sites (psmRS-GFP is a soluble modified red-shifted variant of GFP; Davis and Vierstra 1998). A similar construct strategy was used to get fusions to the mCherry fluorescent protein reporter (mCherryFP). The generated constructs are: P 35S ::HvSs1-GFP, P 35S ::HvSs2-GFP, P 35S ::HvSs3-GFP, P 35S ::HvSs4-GFP, P 35S ::HvSs1-mCherryFP, P 35S ::HvSs2-mCherryFP, P 35S ::HvSs3-mCherryFP and P 35S ::HvSs4-mCherryFP; P 35S ::GFP and P 35S ::mCherryFP were used as controls. Plasmids containing several organelle markers tagged to the mCherryFP for co-localisation experiments (Nelson et al. 2007) were provided by the Arabidopsis Biological Resource Centre. The organelle markers employed were: endoplasmic reticulum (ER-rk CD3-959), plasma membrane (PM-rk CD3-1007), Golgi apparatus (G-rk CD3-967), plastids (PT-rk CD3-999) and mitochondria (MT-rk CD3-991).
Gold particles (1 μm), coated with DNA from the recombinant constructs described above, were bombarded into freshly prepared onion epidermal peels with a biolistic Helium gun device (DuPont PDS-1000; BioRad Laboratories, Inc., Hercules, CA, USA). Plasmids containing the organelle markers were similarly processed. The expression of the fluorescent proteins in onion epidermal cells was observed after 24 h of incubation at 22°C in the dark. Images were captured with a confocal spectral microscope LEICA TCS-SP2-AOBS-UV (Leica, http://www.leica.com) under the Ar laser excitation line of 488 nm and the He–Ne laser excitation line of 543 nm, to detect the fluorescence signals of GFP and CherryFP, respectively. Confocal images were analysed using the Image J program (http://rsbweb.nih.gov/ij/). The co-expression of each sucrose synthase tagged to GFP and to CherryFP was analysed on a Zeiss Axiophot fluorescence microscope with the filters 450-490, FT 510, LP 520 and BP 546, FT 580, LP 590, respectively, the images captured with a Leica DFC 300FX CCD colour camera under the Leica Application Suite 2.8.1 build 1554 acquisition software, and the final figures composited on Adobe Photoshop.
Accession numbers
The accession numbers for the cDNAs containing the coding regions of the barley genes described in this article are: HvSs1 (X65871), HvSs2 (X69931), HvSs3 (AK249450) and HvSs4 (AK251329); the corresponding genomic sequences are: HvSs1 (FN400939), HvSs2 (Y15802), HvSs3 (FN421447) and HvSs4 (HQ650888).
Predicted SUS amino acid sequences from Brachypodium distachyon were obtained from the Brachypodium Genome Database and annotated by us: BdSUS1 (Bradi1g46670.1), BdSUS2 (Bradi1g60320.1), BdSUS3 (Bradi1g20890), BdSUS4 (Bradi1g62960.1), BdSUS5 (Bradi1g29570), BdSUS6 (Bradi3g60690).
Results
Characterization of two new Sucrose Synthase encoding genes from barley
In order to find new paralogous sucrose synthase genes in barley, several public databases were searched for cDNA sequences using the deduced protein sequences derived from the cDNAs of HvSs1 and HvSs2 (BLAST program). Only two new cDNA sequences putatively encoding two different sucrose synthase isozymes (hereafter HvSs3, AK249450; HvSs4, AK251329) were found (CREST collection; The Barley Germplasm Centre from Japan, Sato et al. 2009).
An alignment of the deduced amino acid sequences of sucrose synthase proteins were performed using the Needle program (Needleman and Wunsch 1970). The percentage of sequence identity (Table 1) was higher between HvSS1 and HvSS2, 78.4% identical residues, while HvSS3 and HvSS1 had only 68.8% identity and HvSS3 and HvSS2 were 67.6% identical. These percentages decreased when HvSS4 was used in the comparison; <55% of identical residues. The molecular weights for HvSS1, HvSS2 and HvSS3 were in the range of 92 kDa, whereas HvSS4 had a higher molecular weight of approximately 98 kDa due to its longer C-terminal. The four barley SUS proteins shared the two characteristic sucrose synthase and glucosyl-transferase motifs, identified in the Pfam database (Bateman et al. 2002). Bioinformatic prediction using the TargetP (Emanuelsson et al. 2007) and the Predotar (Small et al. 2004) programmes detected a putative mitochondrial targeting peptide of 33 amino acid residues at the N terminus of HvSS1 (mTP; >90% specificity). This mTP signal is not predicted in the other members of the family.
The comparison between the cDNA sequences and the PCR fragments amplified from genomic DNA established that all the genes had between 14 and 16 introns. A schematic representation of the exon–intron structure of the four genes is presented in Fig. 1; introns, although different in size, appeared at equivalent positions and were flanked by typical GT/AG boundaries. However, HvSs3 and HvSs4 lacked the first intron and HvSs2 lacked the last intron and, consequently, exons XV and XVI were fused into a larger exon; HvSs4 had a 3′-extension, interrupted by two introns, giving rise to exons XVI, XVII and XVIII. The sucrose synthase domain spanned exons II to XII; the glucosyl-transferase domain was codified from exons XIII and XIV in the four barley SUS isozymes.
Exon–intron structure of the H. vulgare sucrose synthase genes. Exons are represented with roman numbered boxes and introns by connecting lines; number of exon nucleotides are indicated above the boxes and the numbers in parenthesis indicate transcribed but not translated sequences. Black boxes indicate the open reading frames
Phylogenetic analysis of sucrose synthase proteins
In order to establish the phylogenetic relationships between the four barley SUS proteins and those described from rice and maize (Carlson et al. 2002; Hirose et al. 2008), a multi-alignment analysis using the MEGA 4.0 software has been performed. The six genes encoding different putative isoforms of sucrose synthase in Brachypodium distachyon, annotated after a detailed analysis of the published genome data available at the Brachypodium Genome Database (US Department of Energy Joint Genome Institute http://jgi.doe.gov/), were also included in the dendrogram and designated as BdSUS1-6, according to their degree of similarity with their rice putative orthologous. The six Arabidopsis SUS proteins were also included for comparison (Barratt et al. 2001).
The dendrogram shown in Fig. 2 highlights the presence of four clades: (1) monocot-specific Group-1, containing barley HvSS1 and its putative orthologous from maize ZmSH1, rice OsSUS1 and Brachypodium BdSUS1, as well as HvSS2 and its putative orthologous OsSUS3 and BdSUS3; (2) dicot-specific Group-1, comprising only the two Arabidopsis proteins AtSUS1 and AtSUS4; (3) Group-2, including both monocot and dicot species, where HvSS3 was placed together with its putative orthologous ZmSUS3, OsSUS4 and BdSUS4, as well as, Arabidopsis AtSUS2 and AtSUS3; and (4) Group-3, that included HvSS4, OsSUS5 and OsSUS6, BdSUS5 and BdSUS6, and AtSUS5 and AtSUS6. Thus, at least one SUS member of barley was found in the three monocot-phylogenetic dendrogram clades.
Phylogenetic dendrogram of the deduced amino acid sequences of cereal sucrose synthases. The Arabidopsis SUS data are included for comparison. CDS accession numbers are: (1) Hordeum vulgare, HvSS1, X65871; HvSS2, X69931; HvSS3, AK249450; HvSS4, AK251329; (2) Zea mays, ZmSh1, X02400; ZmSUS1, L22296; ZmSUS3, AY124703; (3) Oryza sativa, OsSUS1, AK100334; OsSUS2, AK072074; OsSUS3, AK100306; OsSUS4, AK102158; OsSUS5, AK063304; OsSUS6, AK065549; (4) Brachypodium distachyon, Bradi1g46670.1, BdSUS1; Bradi1g60320.1, BdSUS2; Bradi1g20890, BdSUS3; Bradi1g62960.1, BdSUS4; Bradi1g29570, BdSUS5; Bradi3g60690, BdSUS6; (5) Arabidopsis thaliana, AtSUS1, X70990; AtSUS2, Q00917; AtSUS3, AL161494; AtSUS4, AL353871, AtSUS5, BAB11375; AtSUS6, AAG30975. Bootstrapping values are indicated as percentages
Expression analyses of HvSs1, HvSs2, HvSs3 and HvSs4 genes
HvSs1 and HvSs2 gene expression had been previously analysed by Northern blot and the corresponding proteins were immuno-localised using appropriate antibodies (Martínez de Ilarduya et al. 1993; Guerin and Carbonero 1997). In this study, a more detailed expression analysis of the four HvSs genes has been performed by RT-qPCR. For this purpose, four sets of specific primers from the 3′-non-coding regions were designed (Suppl. Table S1), and the relative expression levels referred to that of the α-Actin2 gene (Fig. 3). While transcripts of HvSs1 were detected in all the samples analysed, HvSs2 was mainly expressed in the developing endosperm. HvSs3 was the prevalent transcript in mature embryos, and HvSs4 was faintly expressed in all the tissues analysed. In the developing endosperm (14 dap), the most abundant transcripts were those of HvSs1 and HvSs2 (≥3,000% that of α-Actin2). In mature embryos, HvSs2 and HvSs4 were barely detected while HvSs1 and HvSs3 were expressed at 600 and 1,300% that of α-Actin2, respectively. In leaves of 10-day-old plants, HvSs1 and HvSs3 transcripts were expressed approximately at the same level than that of α-Actin2, and in roots of these plants the most prevalent transcript was that of HvSs1 (Fig. 3).
RT-qPCR global expression analyses of the four HvSs genes. Plant material was collected from flowers (Fl); developing endosperm (dEn) and embryo (dEm) at 14 days after pollination (dap); mature embryos (mEm), germinating embryo (gEm) and aleurone (gAl) at 16 h after imbibition (hai); leaves (Le) and roots (Ro) from 10-day-old plants. Relative transcript levels were standardised to the constitutive α-Actin2 gene expression. Values are the average ± SE of at least three independent experiments
The expression profile of the SUS genes was also investigated in developing endosperms collected at 10, 14, 18, 22 and 26 days after pollination (dap), and in aleurones of germinating seeds at different times after water imbibition (8, 16, 24 and 48 hai) as shown in Fig. 4a and b. HvSs1 and HvSs2 were abundantly expressed in seeds from early developmental stages. While the transcripts of HvSs3 and HvSs4 were barely detected, the maximum level of expression for HvSs1, accounting to 5,000% that of α-Actin2, is reached at 18 dap, while HvSs2 reaches maximum levels at the first two stages analysed, 10 and 14 dap (~3,000% that of α-Actin2); these transcripts decrease progressively in the last steps of the seed maturation phase (Fig. 4a).
Expression kinetic profile (RT-qPCR) of the four HvSs genes in developing seeds. a Endosperms were harvested at 10, 14, 18, 22 and 26 dap. b Aleurone layers were isolated from germinating seeds at 8, 16, 24 and 48 hai. Relative transcript levels are standardised to the constitutive α-Actin2 gene expression. Values are the average ± SE of at least three independent experiments
The aleurone layers from germinating kernels were separated from the rest of the seed at 8, 16, 24 and 48 h hai. As shown in Fig. 4b, HvSs1 was the prevalent sucrose synthase transcript upon germination, peaking at 24 hai (4,000% that of α-Actin2) and declining thereafter, while the HvSs3 transcript peaked at 16 h (600% that of α-Actin2), and HvSs2 and HvSs4 were barely detected (Fig. 4b).
Differential response of HvSs1, HvSs3 and HvSs4 to abiotic stimuli in leaves
The response of the four barley SUS encoding genes to several abiotic stresses, such are anaerobiosis, drought and salt conditions, and to low temperatures, were further investigated. The leaves of 10-day-old plants, subjected to hypoxia by completely submerging them into degassed water, were collected at 24 and 48 h for RT-qPCR analyses. Under these conditions, only the HvSs1 gene was up-regulated (more than three times) after 24 h (Fig. 5a).
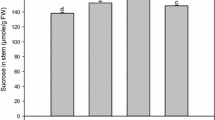
HvSs expression response to abiotic stresses in leaves. a Hypoxia treatment; RNA was extracted from leaves of plants submerged into degassed water after 0, 24 and 48 h. b Drought response and c salt treatment (150 mM NaCl; RNA was extracted from leaves of 10-day-old plants deprived of water or irrigated with NaCl solution, at 0, 3, 7 and 10 days of treatment. Treated plants (open circles) and control plants (closed circles). The transcript level of each gene was analysed by RT-qPCR and standardised to the constitutively expressed UbiE2 gene. Values are the average ± SE of at least three independent experiments
The drought response was investigated in 10-day-old plants not watered for up to 10 days, and leaf samples taken at 0, 3, 7 and 10 days without irrigation. As shown in Fig. 5b, only the HvSs3 transcripts were induced by drought (~four times after 10 days), as compared with watered controls.
The transcriptional response of barley sucrose synthases to salt stress was investigated by irrigating 10-day-old plants with a 150 mM NaCl solution every 3 days for up to 10 days; the controls were irrigated with the same amount of water. In leaf samples, collected after 0, 3, 7 and 10 days of treatment, no significant induction was observed for any of the transcripts analysed although the plants started to fade (Fig. 5c).
The expression of the HvSs genes was then studied in leaf samples of 10-day-old plants in response to cold temperatures (4°C for 24 h; Fig. 6). While HvSs1 and HvSs3 were significantly induced (1.5- and 2-fold, respectively), the HvSs4 transcripts remain practically constant.
HvSs expression response to cold temperatures. RNA was extracted from leaves of 10-day-old plants transferred to 4°C for 24 h (white bars) and from leaves of plants maintained at 22°C as controls (black bars). The transcript level of each gene has been analysed by RT-qPCR and standardised to the constitutively expressed UbiE2 gene. Values are the average ± SE of at least three independent experiments
The expression of HvSs2 transcripts were neither detected in the leaves of control plants nor induced under any of the abiotic stresses considered here.
Subcellular distribution of the HvSs genes
Although SUS isozymes were traditionally considered to be soluble cytosolic proteins, recent studies have demonstrated that some of them can exist as membrane-bound in a complex with cellulose synthase (Fujii et al. 2010). Since the subcellular localisation of genes could help to elucidate their function, all four barley sucrose synthase ORFs have been translationally fused to the GFP, and these constructs cloned under the control of the constitutive 35S promoter (P 35S ::HvSs1-GFP, P 35S ::HvSs2-GFP, P 35S ::HvSs3-GFP, P 35S ::HvSs4-GFP) to analyse their subcellular localisation in transient expression experiments, in bombarded onion epidermal cells.
As shown in Fig. 7, a similar subcellular pattern was observed for the four constructs. The green fluorescence signal appeared as a rim around the cell nucleus (arrowheads), along several cytoplasmic tracks (double arrows) and at the inner side of the cell membrane forming aggregates (single arrow; Fig. 7a–d). All the possible binary combinations of the four sucrose synthases fused to GFP and to the mCherryFP were used for transient co-expression experiments; the co-localisation was observed by a yellowish colour; an example with P 35S ::HvSs2-GFP and P 35S ::HvSs3-mCherryFP is presented in Supplementary Fig. S2.
Subcellular localisation of the four barley SUS proteins. Translational fusion constructs to the reporter GFP were transiently expressed in onion epidermal layers: HvSS1 (a), HvSS2 (b), HvSS3 (c) and HvSS4 (d). The fluorescence signal found around the cell nucleus (arrowhead), at the periphery of the cell (single arrow) and on the cytoplasmic tracks (double arrow) are indicated
In order to accurately determine the subcellular distribution of these sucrose synthases, known organelle markers for endoplasmic reticulum (ER), plasma membrane (PM), Golgi apparatus (G), plastids (PT) and mitochondria (MT) were tagged to mCherryFP (pattern of localisation in onion epidermal cells shown in Suppl. Fig. S3) and used for further co-localisation studies with each barley P 35S ::HvSs-GFP. The overlay projections of confocal stacks spanning co-transformed epidermal cells of each sucrose synthase combined with each subcellular marker is shown in Fig. 8. A major co-localisation (yellowish colour) was observed when the HvSS::GFP constructs were co-expressed with the ER marker (Fig. 8a–d). Regions of co-localisation were also observed in the plasma membrane, specifically with the sucrose synthases HvSS1, HvSS2 and HvSS3 that localise forming patches in the inner side of the membrane (Fig. 8e–g). No co-localisation was observed, at the optical level of resolution, with the Golgi apparatus (Fig. 8i–l) or with the plastid markers (Fig. 8m–t), although a partial co-localisation with the mitochondria marker and HvSS1 was found (Fig. 8q–t). A mitochondrial signal peptide had been predicted for HvSS1 in silico with the TargetP and Predotar programmes (see Table 1). Plasmolysis experiments after 1 M mannitol treatment confirmed the localisation pattern in the wall-detached cytoplasm, with no fluorescence observed either in the cell wall or in the apoplast (see as an example P 35S ::HvSs2-GFP in Fig. 9).
Overlay projections of confocal stacks spanning epidermal cells co-transformed with each sucrose synthase tagged to the GFP reporter (HvSS1: a, e, i, m, q; HvSS2: b, f, j, n, r; HvSS3: c, g, k, o, s; HvSS4: d, h, l, p, t) and each of the five different organelle markers tagged to mCherryFP, which are specific for endoplasmic reticulum (a–d), plasma membrane (e–h), Golgi apparatus (i–l), plastids (m–p) and mitochondria (q–t)
Discussion
In this study, the barley genes HvSs3 and HvSs4, encoding two new isoforms of SUS (HvSS3 and HvSS4) were annotated from the CREST collection (Sato et al. 2009) and further characterised. The overall intron/exon structure of these genes, together with those corresponding to HvSs1 and HvSs2, was established. Considering the sequence similarity and the intron–exon organisation, it can be postulated that the HvSs1 and HvSs2 genes may have evolved after the divergence of the HvSs3 and HvSs4 from their common ancestor and that they have most probably originated after a gene duplication event followed by a new divergent evolution (Fig. 1). The SUS family in Brachypodium distachyon was also annotated and, used, together with the SUS previously described in rice and maize, to derive a phylogenetic dendrogram. As shown in Fig. 2, the four barley isoforms are distributed into three separate clades of this dendrogram: HvSS1 and HvSS2 are found in the monocot Group-1, HvSS3 is located in the Group-2 while HvSS4 is included in the Group-3. These clades correspond to the Groups Sus1, SusA and NG, respectively, as described by Komatsu et al. (2002). Although the barley genome most probably contains six SUS paralogous, since rice and Brachypodium do have six members in the SUS family, only four barley isoforms were found searching the EST public databases (December 2010; http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html).
HvSs1, together with HvSs2, presents the highest level of expression in developing seeds endosperms (peaking at 14 and 18 dap, respectively). HvSs1 is also the predominant transcript in germinating aleurone, with a maximum expression at 24 hai. This gene is inducible by oxygen deprivation and less so by cold temperatures in leaves. Interesting parallels can be drawn between anaerobiosis and cold induction of HvSs1 and analogous data concerning its corresponding cereal orthologous, like wheat TaSs1 (Maraña et al. 1990; Crespi et al. 1991), maize ZmSh1 (Zeng et al. 1998) and rice OsSUS1 (Ricard et al. 1991). Arabidopsis AtSus1 and AtSus4, two genes belonging to this group, appear to have also a role in the anaerobiosis response since the double mutant sus1/sus4 has a marked growth retardation and accumulation of sugars when roots are subjected to hypoxia (Bieniawska et al. 2007).
As previously described by Martínez de Ilarduya et al. (1993) and Guerin and Carbonero (1997), the HvSs2 transcripts, and corresponding HvSS2 proteins, are highly expressed in the early stages of seed development, and they predominate in the starchy endosperm cells, when the starch synthase activity is at its highest level; there is a strong correlation between sucrose synthase transcript levels and sucrose synthase activity, as demonstrated in rice by Hirose et al. (2008). The rice isoform OsSUS3, orthologous of HvSS2, presents a similar spatial and temporal expression pattern (Wang et al. 1999). Genetic evidence for the involvement of SUS in starch biosynthesis was found in the double maize mutant sh1sus1 where the starch content in endosperm is approximately half of that found in wild type kernels (Chourey et al. 1998); and in the pea mutant rug4, lacking isoform PsSus1 that has a reduced seed weight and a lower starch content than the wild type (Barratt et al. 2001).
HvSs3 that belongs to Group-2 of the dendrogram is mainly expressed in dry embryos and is induced in leaves in response to dehydration. Its orthologs in maize ZmSUS3 (Carlson et al. 2002) and rice OsSUS4 (Hirose et al. 2008) are also expressed in drying kernels. Transcripts of AtSus3 from Arabidopsis (Baud et al. 2004) and PsSus2 from pea (Barratt et al. 2001) are found at the end of the maturation phase in the cotyledons, and AtSus3 is induced in leaves under water depletion. Similarly, the two SUS isoforms described in Craterostigma plantagineum (Kleines et al. 1999) are induced in leaves in response to desiccation. All these genes may have a role in the plant desiccation response, both in stressed leaves and in the final stages of seed maturation. Interestingly, the HvSs3 transcripts respond to cold temperatures that is in part a desiccation process.
The HvSs4 transcripts are faintly detected in mature embryos, germinating aleurones and leaves, and their expression is on the limit of detection in the other tissues analysed. This gene does not respond to any of the environmental stresses studied in this work, in contrast with data reported for its two rice orthologous genes (OsSus5 and OsSus6) that were described as down-regulated in response to anaerobiosis (Hirose et al. 2008). It has been previously reported that the Arabidopsis genes included in this same Group 3 (AtSus5 and AtSus6) are expressed in the phloem sieve elements where they may be involved in the sieve-plate callose synthesis (Bieniawska et al. 2007; Barratt et al. 2009).
Although barley is considered a salt-tolerant crop species, Walia et al. (2006) described more than 600 unigenes differentially expressed at 100 mM NaCl concentration in cv Morex; at 150 mM NaCl we have not found any change of expression in any of the four sucrose synthase genes in our Bomi cultivar.
Although SUS isozymes are water-soluble and supposed to be present in the cytoplasm, an important fraction of them is localised to the endoplasmic reticulum and the inner side of the plasma membrane. The association of these proteins with the membranes is corroborated by the presence of two trans-membrane domains in their sequences, as predicted by the program TMPred (our data not shown). Membrane-associated forms of sucrose synthase have been detected in cotton and maize, and have been implicated in both cellulose and callose biosynthesis (Amor et al. 1995; Carlson and Chourey 1996). Recently, it has been demonstrated that SUS is a component of the cellulose-synthesising machinery in the plasma membrane that is associated with the rosette structures in Azuki beans (Vigna angularis; Fujii et al. 2010), and SUS is expressed concomitantly with the activation of the synthesis of cellulose in the secondary cell walls during wood formation in poplar (Coleman et al. 2009). By contrast, Arabidopsis mutants lacking four of the six SUS isoforms are not deficient in cellulose biosynthesis, suggesting that an alternative pathway must supply carbon for this process in this species (Barratt et al. 2009).
Some authors associated SUS to the tonoplast in Beta vulgaris (Etxeberria and Gońzález 2003), and to the cell walls of tobacco pollen tubes (Persia et al. 2008). However, in our plasmolysis experiments after 1 M mannitol treatment, when the plasma membrane retracts from the cell wall, the fluorescent GFP signal remains associated with the plasma membrane and fluorescence is not observed in the cell wall or in the periplasmic space (Fig. 9).
Our data corroborate that the four barley SUS isozymes described so far, exist both in soluble and in membrane-associated forms, a prominent fraction of them being associated with the endoplasmic reticulum (ER). Since there is direct evidence for a structural association of actin filaments with ER tubules (Staehelin 1997), an association with the cytoskeleton is plausible. In this context, Winter et al. (1998) reported that approximately 15% of the total maize SUS protein is associated with the actin cytoskeleton.
Although SUS association with other subcellular organelles was previously reported, such as the Golgi apparatus (Buckeridge et al. 1999), plastids (Nuñez et al. 2008) and mitochondria (Subbaiah et al. 2006), no co-localisation with these organelles for three of the barley isoforms analysed is observed; however, HvSS1 has a predicted mitochondrial targeting peptide at its N terminus (Table 1), as occurs with its orthologous in maize ZmSh1, and it co-localises with the mitochondrial marker (Fig. 7q). This mitSUS localisation was proposed by Subbaiah et al. (2006) to have a non-sucrolytic role, being important in the regulation of solute fluxes into and out of the mitochondria.
The subcellular distribution of the four HvSS isoforms is consistent with the idea that some of them would be present in several subcellular compartments where they may fulfil different physiological functions. However, this does not exclude than when present in the same subcellular localisation in the same tissue and at the same developmental stage, they may perform similar roles, as shown in the developing barley endosperm where all the possible homotetramers and heterotetramers from HvSS1 and HvSS2 are present (Guerin and Carbonero 1997).
Abbreviations
- dap:
-
Days after pollination
- ER:
-
Endoplasmic reticulum
- GFP:
-
Green fluorescent protein
- G:
-
Golgi apparatus
- hai:
-
Hours after imbibition
- mCherryFP:
-
mCherry fluorescent protein
- MT:
-
Mitochondria
- mTP:
-
Mitochondrial targeting peptide
- PM:
-
Plasma membrane
- PT:
-
Plastids
- SUS:
-
Sucrose synthase
- UbiE2:
-
UbiquitinE2
References
Amor Y, Haigler C, Johnson S, Winscott M, Delmer D (1995) A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc Natl Acad Sci USA 92:9353–9357
Angeles-Nuñez JG, Tiessen A (2010) Arabidopsis sucrose synthase 2 and 3 modulate metabolic homeostasis and direct carbon towards starch synthesis in developing seeds. Planta 232:701–718
Baroja-Fernández E, Muñoz F, Montero M, Etxeberria E, Sesma M, Ovecka M, Bahaji A, Ezquer I, Li J, Prat S, Pozueta-Romero J (2009) Enhancing sucrose synthase activity in transgenic potato (Solanum tuberosum L.) tubers results in increased levels of starch, ADPglucose and UDPglucose and total yield. Plant Cell Physiol 50:1651–1662
Barratt DH, Barber L, Kruger N, Smith A, Wang T, Martin C (2001) Multiple, distinct isoforms of sucrose synthase in pea. Plant Physiol 127:655–664
Barratt DH, Derbishire P, Findlay K, Pike M, Wellner N, Lunn J, Feil R, Simpson C, Maule A, Smith A (2009) Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc Natl Acad Sci USA 106:13124–13129
Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy S, Griffiths-Jones S, Howe K, Marshall M, Sonnhammer E (2002) The Pfam protein families database. Nucleic Acid Res 30:276–280
Baud S, Vaultier MN, Rochat C (2004) Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J Exp Bot 55:397–409
Bieniawska Z, Barratt DH, Garlick A, Thole V, Kruger N, Martin C, Zrenner R, Smith A (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49:810–828
Buckeridge M, Vergara C, Carpita N (1999) The mechanism of synthesis of a mixed-linkage (1 → 3),(1 → 4)β-d-glucan in maize. Evidence for multiple sites of glucosyl transfer in the synthase complex. Plant Physiol 120:1105–1116
Carlson S, Chourey P (1996) Evidence for plasma membrane-associated forms of sucrose synthase in maize. Mol Gen Genet 252:1432–1874
Carlson SJ, Chourey PS, Helentjaris T, Datta R (2002) Gene expression studies on developing kernels of maize sucrose synthase (SuSy) mutants show evidence for a third SuSy gene. Plant Mol Biol 49:15–29
Chang S, Puryear J, Carney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116
Chourey P, Taliercio E, Carlson S, Ruan YL (1998) Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol Gen Genet 259:88–96
Coleman HD, Yan J, Mansfield S (2009) Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc Natl Acad Sci USA 106:13118–13123
Crespi M, Zabaleta E, Pontis H, Salerno G (1991) Sucrose synthase expression during cold acclimation in wheat. Plant Physiol 96:887–891
Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36:521–528
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protoc 2:953–971
Etxeberria E, Gońzález P (2003) Evidence for a tonoplast-associated form of sucrose synthase and its potential involvement in sucrose mobilization from the vacuole. J Exp Bot 54:1407–1414
Fujii S, Hayashi T, Mizuno K (2010) Sucrose synthase is an integral component of the cellulose synthesis machinery, 2010. Plant Cell Physiol 51:294–301
Gordon A, Minchin F, James C, Komina O (1999) Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol 120:867–878
Guerin J, Carbonero P (1997) The spatial distribution of sucrose synthase isozymes in barley. Plant Physiol 114:55–62
Harada T, Satoh S, Yoshioka T, Ishizawa K (2005) Expression of sucrose synthase genes involved in enhanced elongation of pondweed (Potamogeton distinctus) turions under anoxia. Ann Bot 96:683–692
Hirose T, Scofield G, Terao T (2008) An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci 174:534–543
Horst I, Welham T, Kelly S, Kaneko T, Sato S, Tabata S, Parniske M, Wang T (2007) TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiol 144:806–820
Kleines M, Elster R-C, Rodrigo MJ, Blervacq A-S, Salamini F, Bartels D (1999) Isolation and expression analysis of two stressed-responsive sucrose-synthase genes from the resurrection plant Craterostigma plantagineum (Hochst.). Planta 209:13–24
Koch K (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7:235–246
Komatsu A, Moriguchi T, Koyama K, Omura M, Akihama T (2002) Analysis of sucrose synthase genes in citrus suggests different roles and phylogenetic relationships. J Exp Bot 53:61–71
Lagrimini LM, Burkhart W, Moyer M, Rothstein S (1987) Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: Molecular analysis and tissue-specific expression. Proc Natl Acad Sci USA 84:7542–7546
Maraña C, Garcia-Olmedo F, Carbonero P (1990) Differential expression of two types of sucrose synthase-encoding genes in wheat in response to anaerobiosis, cold shock and light. Gene 88:167–172
Martínez de Ilarduya O, Vicente-Carbajosa J, Sánchez de la Hoz P, Carbonero P (1993) Sucrose synthase genes in barley. cDNA cloning of the Ss2 type and tissue-specific expression of Ss1 and Ss2. FEBS Lett 320:177–181
Moreno-Risueno MA, Díaz I, Carrillo L, Fuentes R, Carbonero P (2007) The HvDOF19 transcription factor mediates the abscisic acid dependent repression of hydrolase genes in germinating barley aleurone. Plant J 51:352–365
Moreno-Risueno MA, Gonzalez N, Diaz I, Parcy F, Carbonero P, Vicente-Carbajosa J (2008) FUSCA3 from barley unveils a common transcriptional regulation of seed-specific genes between cereals and Arabidopsis. Plant J 53:882–894
Needleman SB, Wunsch CD (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48:443–453
Nelson B, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localisation studies in Arabidopsis and other plants. Plant J 51:1126–1136
Nolte K, Koch K (1993) Companion-cell specific localisation of sucrose synthase in zones of phloem loading and unloading. Plant Physiol 101:899–905
Nuñez JGA, Kronenberger J, Wuilleme S, Lepiniec L, Rochat C (2008) Study of AtSUS2 localisation in seeds reveals a strong association with plastids. Plant Cell Physiol 49:1621–1626
Persia D, Cai G, Del Casino C, Faleri C, Willemse M, Cresti M (2008) Sucrose synthase is associated with the cell wall of tobacco pollen tubes. Plant Physiol 147:1603–1618
Ricard B, Rivoal J, Spiteri A, Pradet A (1991) Anaerobic stress induces the transcription and translation of sucrose synthase in rice. Plant Physiol 95:669–674
Ruan YL, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation and seed development. Plant Cell 15:952–964
Sánchez de la Hoz P, Vicente-Carbajosa J, Mena M, Carbonero P (1992) Homologous sucrose synthase genes in barley (Hordeum vulgare) are located in chromosomes 7H (syn. 1) and 2H. Evidence for a gene translocation? FEBS Lett 310:46–50
Sato K, Shin-I T, Seki M, Shinozaki K, Yoshida H, Takeda K, Yamazaki Y, Conte M, Kohara Y (2009) Development of 5006 full-length cDNAs in barley: a tool for accessing cereal genomics resources. DNA Res 16:81–89
Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4:1581–1590
Staehelin L (1997) The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J 11:1151–1165
Subbaiah C, Palaniappan A, Duncan K, Rhoads D, Huber S, Sachs M (2006) Mitochondrial localisation and putative signalling function of sucrose synthase in maize. J Biol Chem 281:15625–15635
Tamura K, Dudley J, Nei M, Kumar C (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson J, Higgins D, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res 22:4673–4680
Walia H, Wilson C, Wahid A, Condamine P, Cui X, Close TJ (2006) Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Funct Integr Genomics 6:143–156
Wang AI, Kao MH, Yang WH, Sayion Y, Liu LF, Lee PD, Su JC (1999) Differentially and developmentally regulated expression of three rice sucrose synthase genes. Plant Cell Physiol 40:800–807
Winter H, Huber J, Huber S (1998) Identification of sucrose synthase as an actin-binding protein. FEBS Lett 430:205–208
Zeng Y, Wu Y, Avigne WT, Koch KE (1998) Differential regulation of sugar-sensitive sucrose synthases by hypoxia and anoxia indicate complementary transcriptional and posttranscriptional responses. Plant Physiol 116:1573–1583
Zrenner R, Salanoubat M, Willmitzer L, Sonnewald U (1995) Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J 7:97–107
Acknowledgments
This work was financially supported by grants to PC from the Spanish Ministerio de Educación y Ciencia (BFU2006-07258) and from the Spanish Ministerio de Innovación y Ciencia (BFU2009-11809); C.B.-S. holds a postdoctoral Juan de la Cierva-UPM contract; S.H.-A. is supported by a FPU doctoral fellowship from MEC.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1
α-Actin2 and UbiE2 expression in abiotically stressed leaves: hypoxia, salt, drought and cold temperatures as specified in the text. The 18S gene was used to normalize the RT-qPCR data (TIFF 107 kb)
Supplementary Fig. S2
Subcellular localisation of HvSS2 fused to GFP (a), HvSS3 fused to mCherryFP (b) and overlay projection of the two previous ones (c) (TIFF 2418 kb)
Supplementary Fig. S3
Fluorescence pattern of five organelle markers (mCherryFP) in bombarded epidermal onion cells, specific for: the endoplasmic reticulum (a), plasma membrane (b), Golgi apparatus (c), plastids (d) and mitochondria (e) (TIFF 7975 kb)
Rights and permissions
About this article
Cite this article
Barrero-Sicilia, C., Hernando-Amado, S., González-Melendi, P. et al. Structure, expression profile and subcellular localisation of four different sucrose synthase genes from barley. Planta 234, 391–403 (2011). https://doi.org/10.1007/s00425-011-1408-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-011-1408-x