Abstract
Sucrose phosphate synthase (SPS) catalyzes the first step in the synthesis of sucrose in photosynthetic tissues. We characterized the expression of three different isoforms of SPS belonging to two different SPS gene families in alfalfa (Medicago sativa L.), a previously identified SPS (MsSPSA) and two novel isoforms belonging to class B (MsSPSB and MsSPSB3). While MsSPSA showed nodule-enhanced expression, both MsSPSB genes exhibited leaf-enhanced expression. Alfalfa leaf and nodule SPS enzymes showed differences in chromatographic and electrophoretic migration and differences in V max and allosteric regulation. The root nodules in legume plants are a strong sink for photosynthates with its need for ATP, reducing power and carbon skeletons for dinitrogen fixation and ammonia assimilation. The expression of genes encoding SPS and other key enzymes in sucrose metabolism, sucrose phosphate phosphatase and sucrose synthase, was analyzed in the leaves and nodules of plants inoculated with Sinorhizobium meliloti. Based on the expression pattern of these genes, the properties of the SPS isoforms and the concentration of starch and soluble sugars in nodules induced by a wild type and a nitrogen fixation deficient strain, we propose that SPS has an important role in the control of carbon flux into different metabolic pathways in the symbiotic nodules.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Legumes have the capability to establish symbiotic associations with compatible rhizobia (Graham and Vance 2003). The legume–rhizobia symbiosis involves very complex interactions, which are initiated by the exchange of signal molecules between the partners followed by the infection of the plant by the microbe (Trevaskis et al. 2002). Biological reduction of atmospheric nitrogen (N2) takes place in the root nodules, the new organ formed by this interaction. Symbiotic N2 fixation requires a complex metabolic interdependence of each symbiotic partner (Lodwig et al. 2003). The plant provides carbon (C) in the form of dicarboxylic acids to the differentiated bacteroids for oxidation to provide ATP and reducing power (Reich et al. 2006). The bacteroids, in turn, provide the plant with ammonia derived from the reduction of N2 by the bacterial enzyme nitrogenase.
Sucrose (Suc) is the main form in which C is translocated from the leaves to the sink tissues like the nodules. Suc unloaded into the nodules is hydrolyzed by sucrose synthase (SucS; EC 2.4.1.13) and alkaline/neutral invertase (EC 3.2.1.26). Whereas SucS plays a critical role in nodule metabolism, invertase does not seem to be crucial for nodule function (Baier et al. 2007; Welham et al. 2009). Carbon derived from the metabolism of Suc in the nodules is used for several physiological processes, including plant and bacterial respiration, assimilation of fixed N2, and also starch and cellulose biosynthesis. Besides its role as a substrate in metabolic processes, Suc is a storage compound and also acts as a signal molecule (Loreti et al. 2001). The allocation and metabolism of Suc in the nodules is dictated by the specific metabolic requirements of each cell type (Hohnjec et al. 2003; Flemetakis et al. 2003, 2006).
The formation of Suc involves a two-step reaction: the synthesis of sucrose-6-phosphate (Suc6P) from UDP-glucose (UDPGlc) and fructose-6-phosphate (Fru6P), catalyzed by the enzyme sucrose phosphate synthase (SPS; EC 2.3.1.14), followed by the irreversible hydrolysis of Suc6P, catalyzed by the enzyme sucrose phosphate phosphatase (SPP; EC 3.1.3.24), pulling the reaction catalyzed by SPS in the direction of net Suc synthesis (Lunn et al. 2000). SPS can be regulated by a hierarchy of several interacting mechanisms, including regulation of gene expression, covalent modification and allosteric regulation via metabolites, glucose-6-phosphate (Glc6P) and inorganic phosphate (Pi). Reversible phosphorylation on distinct serine residues is involved in the regulation of SPS in response to different environmental conditions. The dephosphorylated protein has a higher affinity for its activator Glc6P and a lower affinity for the inhibitor Pi, pointing to a connection between covalent modification and allosteric regulation (Huber and Huber 1996; Toroser and Huber 1997; Lunn and MacRae 2003; Huber 2007).
SPS is encoded by multigene families which are expressed in photosynthetic and also in heterotrophic tissues, including potato tubers (Geigenberger et al. 1997), cotyledons (Geigenberger and Stitt 1991), fruits (Hubbard et al. 1991; Komatsu et al. 1996), roots and flowers (Fung et al. 2003), developing and germinating embryos (Im 2004), and cotton fibers (Xu et al. 2007). The metabolism of Suc in heterotrophic tissues is characterized by a continuous process of utilization and synthesis (Roby et al. 2002). Nodule metabolism has been well characterized, but so far there have been no reports on the expression of SPS in symbiotic nodules. In this paper, we show that SPS is encoded by a small gene family in alfalfa (Medicago sativa L.) and one of the gene member shows nodule-enhanced expression. Based on the expression pattern of SPS along with those of SPP and SucS genes, SPS activity and the metabolite profile in N sufficient and N deficient nodules, we propose what functional role SPS might play in the root nodules.
Materials and methods
Plant material
Alfalfa (Medicago sativa L. cv. Mesa) seeds were surface sterilized and planted on sterile vermiculite in double Magenta box setup simulating a Leonard jar (Wacek and Alm 1978; Supplementary material Fig. S1). Four days after sowing, seedlings were inoculated with either a wild type S. meliloti strain 2011 or a nif H::Tn5 derived mutant strain 1491 (Hirsch et al. 1983). Plants were maintained under sterile conditions in the greenhouse (16 h light and 8 h dark) with 0.25X Hoagland’s nutrient solution supplemented with 0.125 mM NH4NO3. The plants inoculated with Fix-strain showed some yellowing of the leaves compared to the plants inoculated with the WT strain. For a similar proportion of harvested material, nine boxes of plants inoculated with the Fix-strain, and only five boxes of plants inoculated with the WT strain were used (Supplementary material Fig. S1). All experiments described in this paper were repeated five different times. For Western and Northern data, only a representative experiment in each case is presented. Alfalfa plant tissues were harvested at midday, always at the same time of the day, 28 day after inoculation with S. melioti, frozen in liquid nitrogen and kept at −80°C.
Isolation of SPS and SPP genes
Two classes of alfalfa SPS clones were amplified by PCR reactions. One SPS clone was amplified from nodule cDNA with primers designed from conserved regions based on multiple sequence alignment of SPS sequences. It had strong identity to the SPSA family and corresponded to a previously reported alfalfa SPS cDNA (NCBI accession number AF322116), it was designated MsSPSA. The primers used to isolate MsSPSA cDNA were 5′-ATG GCA GGA AAT GAT TGG TT-3′ (forward) and 5′-ATA CTT AAC CTG ACC ACC CG-3′ (reverse). Two different PCR fragments with close similarity to the SPSB family were amplified from leaf cDNA using primers derived from a M. truncatula genomic EST clone, 5′-ACA TGG AGC TTG GTA GAG ATT CTG A-3′ (forward) and 5′-GTC CCC ATT GTT CAT CAA TTT CTT G-3′ (reverse). The two members were designated MsSPSB and MsSPSB3 (NCBI accession numbers EU234514 and FJ790495). The MsSPSB3 cDNA was further amplified using three different set of primers based on the M. truncatula genomic sequencing database (http://medicago.org). The primer pairs used were: 5′-GGT TGT GGC AAG TGT TGA TG-3′ (forward), 5′-TGT TCA CCC AAT GCT TTT GA-3′ (reverse); 5′-AGA ACA TGT GTT GGC GTA TTT G-3′ (forward), 5′-ACA TTT AAA GCA CCC GAA AGA A-3′ (reverse), and 5′- AAG AGT TTG TGG ATG GAG CAT T-3′ (forward), 5′-TGC TGC AGC CTC AAT TAA AGT A-3′ (reverse). A partial alfalfa SPP clone (MsSPP, NCBI accession number AY651774) was obtained by RT-PCR amplification from leaf and nodules cDNA using primers derived from a reported M. truncatula SPP cDNA clone. The primers used were 5′-AAT CGA TGG TTC CCG ATG ATG G-3′ (SPP forward) and 5′-GCA CAA CGC TCT GAG GCA TGC-3′ (SPP reverse). The MsSPP and MsSPSB clones were extended by 5′- and 3′-RACE from 1 μg of RNA from the leaves using 5′- and 3′-cDNA synthesis primers and SMART II oligonucleotides (Clontech, Mountain View, CA, USA). The reverse transcription reaction was performed using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). A total of four libraries were generated: 5′leaf RACE, 3′leaf RACE, 5′nodule RACE and 3′nodule RACE. The amplification of the 5′- and 3′ ends was performed with Advantage 2 genomic DNA polymerase (Clontech) as recommended by the users manual. PCR products were subcloned and subjected to DNA sequencing.
RNA analysis
Total RNA was isolated from leaves and nodules by LiCl precipitation (De Vries et al. 1982). RNA samples were fractionated in 1.33% agarose/formaldehyde gels, transferred to nylon membranes (BioRad, Hercules, CA, USA) and hybridized to 32P labeled probes for the coding regions of alfalfa MsSPP, MsSucS, and SPS genes, a 588 bp MsSPSA fragment, a 641 bp MsSPSB and a 765 bp MsSPS3 fragment. The nodule-enhanced alfalfa SucS clone (NCBI accession no. AF049487) was kindly provided by Dr Carroll Vance (University of Minnesota, St. Paul, MN). Membranes were hybridized as previously reported (Ortega et al. 2006). Following hybridization, the filters were washed at either low stringency (3 times with 2X SSC, 0.5% [w/v] SDS and once with 0.5X SSC, 0.5% [w/v] SDS at 54°C) or high stringency (3 times with 2X SSC, 0.5% [w/v] SDS, once with 0.5X SSC, 0.5% [w/v] SDS and once with 0.1X SSC, 0.5% [w/v] SDS at 65°C), and exposed to X-ray film. Band intensities were quantified and plotted.
Protein extraction
Plant tissues were ground in liquid N and proteins extracted in a buffer containing 50 mM Hepes, pH 7.5, 20% (v/v) glycerol, 5% (v/v) ethylene glycol, 5 mM EDTA, 5 mM Magnesium acetate, 0.5% Triton X-100, 10 mM DTT, and a protease inhibitors cocktail (Roche, Indianapolis IN). Protein extracts were desalted through Sephadex G25 columns equilibrated in desalting buffer (25 mM Hepes pH, 20% [v/v] glycerol, 5% [v/v] ethylene glycol, 2.5 mM Magnesium acetate; 5 mM DTT and protease inhibitors). For anion exchange chromatography, protein extracted from alfalfa leaves (3.5 g) and nodules (5 g) was precipitated with polyethylene glycol (PEG) to a final concentration of 35% (w/v).
SPS activity determination and fractionation
Data on SPS protein analysis are the results from five independent experiments. Activity measurements are the average ± SD from at least five different data points. Activity assays were performed as reported by Baxter et al. (2003). Reaction mixtures contained 10 mM Fru6P, 12 mM UDPGlc, 2.5 mM Magnesium acetate, 25 mM Hepes (pH 7.2), and increasing concentrations of Glc6P. The SPS maximum catalytic activity (V max) and the activation constant (K a) for Glc6P were calculated by linear regression analysis of double reciprocal plots of the Glc6P concentration versus enzyme activity. The effect of Pi on SPS activity was assayed in a reaction mixture containing 5 mM Fru6P, 6 mM UDPGlc, 20 mM Glc6P, 2.5 mM Magnesium acetate, 25 mM Hepes (pH 7.2), and increasing concentrations of potassium phosphate. Enzyme activity was calculated by measuring the absorbance at 625 nm, subtracting the absorbance of control reactions containing the same amounts of substrates and effectors. Glc6P activation and Pi inhibition were compared to controls containing the same amount of substrates but no effectors. The amount of Suc6P produced was calculated against a standard curve of Suc.
For anion exchange chromatography, 10 ml DEAE Sephacel columns were equilibrated in running buffer (10 mM Hepes pH 7.2, 10% [v/v] glycerol, 5% [v/v] ethylene glycol, 1 mM EDTA). The columns were loaded with the 35% (w/v) PEG protein fractions, washed with running buffer and eluted with a 0–0.5 M NaCl linear gradient in running buffer at flow rate of 35 ml h−1, at 4°C. Fractions were assayed for SPS activity using 10 mM Fru6P, 12 mM UDPGlc, and 40 mM Glc6P in the reaction mixture.
Protein immunodetection
Proteins were analyzed by SDS PAGE in 10% acrylamide gels, using a Mini-Protean 3 electrophoresis apparatus (BioRad) followed by western blotting as described by Ortega et al. (2006). Spinach SPS and SPP antibodies were kindly provided by Dr. Uwe Sonnewald (Institut für Biologie, Universität Erlangen-Nürnberg, Erlangen, Germany). SucS antibodies were kindly provided by Dr. Raymond Chollet (University of Nebraska, Lincoln, NE). The SPS antibodies were selected through immunoaffinity purification against spinach SPS following procedures described (Harlow and Lane 1998). Cross reacting polypeptides were visualized with an alkaline phosphatase linked secondary antibody. Experiments for protein analysis were performed five different times with the same results. Only representative experiments are presented. Immunoreactive bands were quantified and plotted. Molecular mass was estimated against prestained protein standards (Biorad).
Analysis of carbohydrates
Soluble carbohydrates were extracted from frozen tissues by grinding in 10 volumes (v/w) of 80% (v/v) ethanol and incubating at 70°C for 90 min. Carbohydrate determination was performed in a Waters Acquity UPLC system with a Q-TOF mass detector. Carbohydrates were separated on a 2.1 × 150 mm YMC amino column with a 80–50% (v/v) acetonitrile gradient in 1 mM LiCl, and detected by negative electrospray ionization. Carbohydrate concentrations were calculated from standards run under the same conditions. The pellets from the soluble carbohydrate extraction were used for enzymatic starch determination following the method described in Barsch et al. (2006). The amount of glucose (C6 units) released by enzymatic hydrolysis of starch was determined by HPLC using a Shodex SC1011 column and quantified by comparison to a glucose standard. The Glc6P concentration in alfalfa tissues was determined by an enzymatic assay as described in Baxter et al. (2003).
Results
Three functional SPS genes representing the A and B families were identified in alfalfa
Phylogenetic analysis of SPS genes has defined four different SPS gene families designated A, B, and C, found in both monocot and dicot species, and D family, found only in monocots (Reimholtz et al. 1997; Langenkamper et al., 2002; Lunn and MacRae 2003; Castleden et al. 2004; Chen et al. 2005). Functional SPS gene members belonging to different families were obtained by RT-PCR performed on leaf and nodule cDNA using gene family-specific primer sets. Several clones obtained using nodule RNA contained a gene sequence with stronger identity to the SPSA family which was designated MsSPSA, while the other two gene sequences obtained using leaf RNA, showed closer similarity to the SPSB family and were designated MsSPSB and MsSPSB3. Clones were 5′ and 3′ extended and further characterized. The alfalfa SPS gene sequences were compared to the corresponding sequences in the M. truncatula genome sequence database (http://medicago.org). The analysis showed 96% sequence identity of the MsSPSA gene to the corresponding M. truncatula SPS gene on chromosome 8, while the MsSPSB and the MsSPSB3 genes showed 95% identity to the corresponding genes of M. truncatula (located in the chromosomes 5 and 3 of the M. truncatula genome, respectively). The MsSPSB and MsSPSB3 share 83% nucleotide sequence identity between them. In our attempts to amplify other SPS genes from alfalfa, we did not succeed in isolating any SPSC gene members using either genomic DNA or RNA isolated from leaves and nodules.
MsSPSB genes exhibit leaf-enhanced expression while MsSPSA shows nodule-enhanced expression
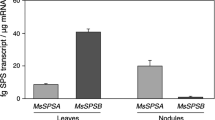
To determine the expression pattern of the SPS genes in the different organs of alfalfa, total RNA isolated from leaves, stem, roots and nodules was subjected to northern blot analysis using gene fragments of the MsSPSA and MsSPSB clones and washed at high stringency. The MsSPSA probe showed hybridization to a band in all the lanes (Fig. 1), however, the hybridization signal with the nodule RNA was threefold higher than with the leaf RNA. The MsSPSB probe showed strong hybridization to leaf RNA and very weak hybridization to stem RNA. MsSPSB3, used as a probe showed the same pattern of hybridization as MsSPSB except that the hybridization signal was slightly higher in the stems (data not shown). Nevertheless, no hybridization signal with either of the MsSPSB probes to RNA from roots or nodules was detected. These results indicate that both MsSPSB forms are expressed in a leaf-enhanced or photosynthetic cell-specific manner, while the MsSPSA gene is transcribed constitutively with higher accumulation of the transcript in the nodules. We also analyzed the expression pattern of other genes involved in Suc metabolism, SPP and SucS. Since SucS in legume plants is encoded by a small gene family (Barratt et al. 2001; Horst et al. 2007; Wienkoop et al. 2008), as may also be the case for SPP, we used the coding regions of these genes as probes at low stringency. This should allow for the detection of the transcripts for all gene family members. The signal with the MsSPP probe was higher in the leaves but it was not substantially different from the signals in the other tissues (Fig. 1). The MsSucS hybridization signal was predominant in the roots and nodules with the highest signal in the nodules. While the stems showed some level of hybridization to the MsSucS probe, the leaves exhibited very low expression of MsSucS. The data indicate that while SPP is expressed at similar levels in all the organs tested, the SucS is expressed mostly in the heterotrophic tissues.
Transcript analysis of genes related to Suc metabolism in different organs of alfalfa. a Total RNA (18 μg) from leaves (L), stems (S), roots (R) and nodules (N) of alfalfa was subjected to northern blot hybridization using probes for genes encoding enzymes involved in Suc metabolism: sucrose phosphate synthase (MsSPSA, MsSPSB), sucrose phosphate phosphatase (MsSPP) and sucrose synthase (MsSucS). The stained gel is shown as a control for loadings. b mRNA band intensities from at least five different blots were quantified and the average ± standard error of the mean (SEM) were plotted
Nodule SPS protein and activity show differences in electrophoretic and chromatographic patterns, and in allosteric regulation compared to leaf SPS
Extracts from leaves and nodules of alfalfa were subjected to Western blot analysis, in addition to ion exchange chromatography. Since SPS enzyme activity is known to be regulated allosterically (Trevanion et al. 2004), we also compared the allosteric properties of SPS extracted from the leaves and the nodules.
Protein extracts from leaves and nodules of alfalfa plants inoculated with a wild type (WT) Sinorhizobium meliloti strain, along with a spinach leaf extract, were analyzed by Western blotting using antibodies against spinach SPS. A relatively broad immunoreactive band was seen in the alfalfa leaf extract that comigrated with the immunoreactive band seen in the spinach leaf extract (Fig. 2a). The broad immunoreactive band in the leaves may be reflective of multiple isoforms. The nodule extract showed a much sharper immunoreactive band migrating slightly faster than the immunoreactive band in the spinach leaf sample. The nodules also showed additional immunoreactive bands. Since the SPS antibodies were selected through immunoaffinity purification against spinach SPS, the smaller molecular weight immunoreactive bands probably correspond to SPS degradation products (Fig. 2a), suggesting that nodule SPS is more unstable than the leaf isoform.
Analysis of alfalfa leaf and nodule SPS protein and activity. a Protein extracts (50 μg) from leaves (L) and WT nodules (N) of alfalfa plants, along with a Spinach leaf extract (15 μg) were subjected to Western blot analysis using anti-SPS antibodies; the migration of the molecular weight standards is as shown. b Protein extracted from alfalfa leaves and nodules was separated by ion exchange chromatography. SPS activity (solid lines) was determined in 1-ml fractions eluted with a linear gradient of 0–0.5 M NaCl (dashed lines)
Anion exchange chromatography showed that SPS from both leaves and nodules eluted as symmetrical peaks in fractions corresponding to different salt concentrations. The activity peak of the leaf SPS eluted at a lower salt concentration than the nodule SPS. The overlap in the elution profiles of the leaf and the nodule SPS (Fig. 2b) may reflect the heterogeneous nature of homomeric and heteromeric SPS dimers resulting from the expression of both SPSA and SPSB forms in the leaves, and only the SPSA in the nodules.
Protein extracts from alfalfa leaves and nodules were also subjected to SPS activity determination. SPS activity measurements were performed in crude extracts, immediately after extraction and desalting (Fig. 2a). We standardized the reaction conditions for the alfalfa leaf SPS by assaying the spinach leaf and alfalfa leaf extracts in parallel, and then used the same assay conditions to compare the activity of the alfalfa leaf and nodule enzymes. We found that the response of the alfalfa leaf SPS activity to the different substrate concentrations was identical to that of the spinach leaf SPS (data not shown). Optimal substrate concentrations determined were 10 mM for Fru6P and 12 mM for UDPGlc, which are the same or closely similar to those reported for tobacco, maize and wheat leaf SPS activity assays (Baxter et al. 2003; Im 2004; Trevanion et al. 2004).
The allosteric effect of Glc6P on the alfalfa leaf and nodule enzyme activities, under optimal substrate concentrations is shown in Fig. 3. The alfalfa leaf enzyme showed higher activity than the nodule enzyme in the absence of the activator (13.6 ± 1.0 and 9.2 ± 0.7 nmol Suc6P min−1 mg protein−1, respectively). Alfalfa leaf SPS activity showed an increase of 1.6 and 1.7-fold at 20 and 40 mM Glc6P, respectively. The specific activities for the alfalfa and spinach leaf SPS enzymes at 20 mM Glc6P were 21.5 ± 1.6 and 19.1 ± 0.6, respectively. Nodule SPS showed a striking increase in activity in response to Glc6P. The SPS activity in alfalfa nodule extracts was six times higher at 40 mM Glc6P, compared to the activity without the activator. It is noteworthy that the activity curve for nodule extract does not saturate at this high concentration of Glc6P (Fig. 3). The total catalytic SPS activity (V max) in leaf and nodule extracts, calculated by linear regression analysis of double reciprocal plots of Glc6P concentration against enzyme activity, showed that the SPS V max in the WT nodules could be as much as 3.7-fold higher than in the leaves under high concentrations of Glc6P (Table 1).
SPS activity in leaf and nodule extracts of alfalfa plants inoculated with a wild type Sinorhizobium melioti. a Suc6P synthesis by SPS activity was measured in reactions containing increasing concentrations of the activator Glc6P as indicated. b The inhibitory effect of inorganic phosphate on SPS activity was assayed in reactions containing suboptimal concentrations of the substrates and increasing concentrations of potassium phosphate. Values are the average ± SD of five independent experiments
Figure 3 shows the effect of the Pi on the leaf and nodule SPS activity under suboptimal substrates and activator concentrations. The nodule enzyme showed a sharp decline in activity at increasing concentrations of Pi, losing up to 86.3% of its activity at 5 mM Pi, whereas, the leaf enzyme was inhibited only by 15.7% at 2 mM Pi and 25.9% at 5 mM Pi (Fig. 3). We found that the inhibitory effect of Pi on the leaf enzyme was similar at 10 and 20 mM Glc6P, in consequence, the assay conditions reported in this paper are the same for leaf and nodule samples. The allosteric response of SPS to Glc6P and Pi inhibition showed that the leaf and the nodule enzymes are different isoforms and also show that the SPS in the nodules is more active at high concentrations of Glc6P.
MsSPSA transcript levels in the nodules are not affected by the N2 fixing ability of the symbiont
To determine how N2-fixation or the C/N status affect the expression of MsSPS genes in alfalfa, plants were inoculated with either a WT S. meliloti (strain 2011) or a Fix− mutant (strain 1491, Hirsch et al. 1983). Leaf and nodule RNA from the two sets of plants were subjected to northern blot hybridization using the MsSPSA, MsSPSB, MsSPP and MsSucS probes. As seen in Fig. 4, MsSPSA showed higher level of hybridization signal to nodule RNA and the signal intensity was only 10% higher in the Fix− nodules compared to the WT nodules. MsSPSB showed hybridization only to leaf RNA and the mRNA levels were 50% higher in the leaves from the plants inoculated with the Fix− strain. The hybridization signals with the MsSPP probe were similar in the leaves and 33% lower in the WT nodules compared to the Fix− nodules. The hybridization signals with the MsSucS probe were lower in the leaves and nodules of the plants inoculated with the Fix− strain compared to the hybridization signals in the corresponding tissues from the WT inoculated plants. Differences in the hybridization signals obtained by northern blot analysis were consistently seen in each of the five experiments that were done with different biological samples. The results suggest that the absence of N2-fixation in the nodules is accompanied by an increase in the steady state level of MsSPP transcript, a drop in the MsSucS transcript level, and only a slight change in the MsSPSA mRNA levels.
Transcript analysis of genes related to Suc metabolism in alfalfa nodulated with a WT and a Fix-mutant. a Total RNA (18 μg) from leaves and nodules of plants inoculated with a wild type Sinorhizobium meliloti (WT) or a N2 fixation deficient strain (Fix−) was subjected to Northern blot hybridization with probes for genes encoding sucrose phosphate synthase (MsSPSA, MsSPSB), sucrose phosphate phosphatase (MsSPP), and sucrose synthase (MsSucS). The stained gel is shown as a control for loadings. b mRNA band intensities from at least five different blots were quantified and the average ± standard error of the mean (SEM) were plotted
Steady state accumulation of SPS, SPP and SucS proteins in plants inoculated with WT and Fix− S. meliloti strains correlates with the corresponding transcript profile
Protein extracts from the leaves and nodules of plants inoculated with either the WT or Fix− strains of S. meliloti were subjected to SDS PAGE followed by western blotting. (Fig. 5). The leaves showed one major SPS immunoreactive band, and no difference was seen in the level of SPS protein in the leaves from the two sets of plants. The nodules showed multiple immunoreactive bands and while the ~120 kDa band showed slightly higher level in the WT nodules, the Fix-nodules showed more of the lower molecular weight immunoreactive bands.
SPS, SPP, and SucS protein accumulation in leaves and nodules of N deficient and N sufficient plants. a Protein extracts from plants inoculated with either a WT or a Fix− Sinorhizobium meliloti were subjected to Western blot analysis using antibodies against SPS, SPP, and SucS. The amount of protein loaded was 50 μg for SPS, and 25 μg for SPP and 15 μg for SucS immunodetection. The migration of molecular weight standards (mwm) is as indicated. b Protein band intensities were quantified and plotted. SPS band intensities represent only the ~120 kDa band. SPP band intensities correspond to the 42 kDa leaf SPP band and to the more abundant 45 kDa band in the nodules
The anti-SPP reacting protein profile showed major differences between the leaves and the nodules. While one major 42 kDa SPP band was immunodetected in the leaves, the nodules showed two bands, one comigrating with the band in the leaves and a major 45 kDa isoform. The immunoreactive 50 kDa band seen in the leaf samples most likely represents a nonspecific reaction against the Rubisco large subunit. No significant difference in the steady level of SPP protein in the leaves or nodules was seen between plants inoculated with the WT or Fix− bacteria. A minor immunoreactive band corresponding to SucS could be detected in the leaves while the nodules showed significant accumulation of the SucS protein, the level being twofold higher in the WT nodules compared to the Fix− nodules.
SPS from WT and Fix− nodules show differences in activity and in allosteric regulation
We examined the activity and the allosteric effects of Glc6P and Pi on the SPS in protein extracts from the leaves and nodules of plants inoculated with the WT and Fix− strains of S. meliloti. The total catalytic activity (V max) of the SPS extracted from the N deficient leaves was slightly higher (37%) than the V max of the SPS extracted from the N sufficient leaves, while the V max of the SPS from the N deficient nodules was slightly lower (13%) than the V max of the enzyme from WT nodules (Table 1). SPS from Fix− nodules showed a 3.3-fold activation at 40 mM Glc6P, while SPS from WT nodules was activated 6.2-fold, two times higher when compared to that with the Fix− nodule enzyme. Moreover, the activation curve of SPS from the N deficient nodules showed saturation at 40 mM Glc6P (data not shown), while the SPS from WT nodules was not completely activated at this concentration (Fig. 3). The analysis showed a significant difference in the Glc6P activation constant (K a) for the SPS enzyme from the WT and the Fix− nodules, 22.2 and 7.0 mM, respectively (Table 1), however, the difference in the Glc6P activation constant for the leaf enzyme was similar between the two N conditions. The inhibitory effect of Pi on the leaf SPS was similar in both sets of inoculated plants (WT and Fix− S. meliloti). However, Pi, inhibited Fix− nodule SPS to a smaller extent compared to its inhibitory effect on the SPS from WT nodules. The differences in the allosteric activation of the nodule SPS by Glc6P and inhibition by Pi indicate that the N status may have an effect on the activation state or the covalent modification of the nodule enzyme.
The leaves and nodules of alfalfa plants inoculated with either WT or Fix− S. meliloti show differences in starch and soluble carbohydrate content
Suc is the main photosynthate used for nodule respiration. The pathways of starch and Suc synthesis compete for a common pool of substrates (Flemetakis et al. 2006). We compared the accumulation of starch and Suc in the nodules and leaves of N sufficient and N deficient alfalfa plants and the results were compared to the SPS activity (Table 1). All the tissues were harvested at midday to avoid differences due to diurnal changes in Suc and starch synthesis and degradation and the experiment was repeated three times. Analysis showed that Suc account for most of the soluble carbohydrates in leaves and nodules (data not shown). The results also showed that nodules accumulate more Suc than the leaves, whereas the leaves accumulate three times more starch than the nodules (Table 2). The leaves and the nodules of N deficient plants showed a threefold and fivefold increase in the accumulation of starch, respectively, compared to the leaves and nodules from plants inoculated with the WT bacteria. On the other hand, the leaves and nodules of plants inoculated with the Fix− mutant showed a ~50% drop in the accumulation of Suc than the tissues from the plants inoculated with the WT bacteria. The amount of hexoses in the Fix− nodules was higher compared to the WT nodules, while the leaves of the N deficient plants showed a drop in the hexose level compared to the leaves of N sufficient plants (Table 2). Glc6P was 33% higher in the Fix-nodules than in the WT nodules, it was lower in the WT leaves and below the detection limits in the Fix− leaves. Our results showed a correlation between the accumulation of Suc, SPS activity calculated on a tissue weight basis, and the N status in the different organs (Tables 1, 2), suggesting that SPS may have a significant role in nodule C and N metabolism.
Discussion
The data presented here demonstrates the nodule-enhanced expression of a SPS gene member, MsSPSA, in alfalfa. The nodules showed higher accumulation of SPS protein than the leaves and also exhibited higher SPS activity indicating that Suc synthesis takes place in the nodules. This is a surprising observation since the nodules obtain their C from the leaves in the form of Suc. However, there have been reports of expression of SPS in different kinds of heterotrophic tissues (Hubbard et al. 1991; Geigenberger et al. 1997; Winter and Huber 2000; Nguyen-Quoc and Foyer 2001; Roby et al. 2002; Fung et al. 2003; Im 2004; Xu et al. 2007). Moreover, a recent report of the presence of SPS in N2 fixing, heterotrophic heterocysts of an Anabaena sp. (Cumino et al. 2007) suggests that Suc synthesis in the nodules may be crucial with regards to N2 fixation and N assimilation as it is in the heterocysts. The presence of SPS in nodules of other legume species (Supplementary material, Fig. S2) further emphasizes the ubiquity of SPS in the root nodules and the importance of SPS in nodule physiology.
We have identified two different SPS gene families that are differentially expressed in alfalfa, one functional MsSPSA gene member and two functional MsSPSB genes. BLAST searches against the M. truncatula genome sequence database (http://medicago.org) using SPSA, SPSB and SPSC sequences from several plant species resulted in the identification of only three SPS gene sequences in M. truncatula that correspond to the same genes we have identified in alfalfa, with a 95% similarity between the orthologous genes. The high level of synteny between M. truncatula and M. sativa genomes (Choi et al. 2004) would suggest that alfalfa, as in the case of M. truncatula, probably does not have other SPS gene members. A phylogenetic analysis of the predicted SPS protein sequences from alfalfa and M. truncatula showed the clustering of the SPSA forms with the Arabidopsis and tobacco sequences belonging to the SPSA family (Langenkamper et al. 2002; Castleden et al. 2004; Chen et al. 2005), the analysis also showed the clustering of the alfalfa and the M. truncatula SPSB forms with the class B sequences (Supplementary material Fig. S3).
The MsSPSA gene form was found to be constitutively expressed in alfalfa, as is the case for the SPSA forms in sugarcane, citrus, Oncidium goldiana, Craterostigma plantagineum, tobacco and wheat (Komatsu et al. 1996; Ingram et al. 1997; Li et al. 2003; Castleden et al. 2004; Chen et al. 2005; Grof et al. 2006), but its expression in alfalfa is many fold higher in the nodules compared to the other organs. The expression of the MsSPSA gene in the nodules, however, does not seem to be regulated by N2 fixation or other factors caused by N defficiency. Since we did not analyze any other sink tissues like the seeds, we do not know whether SPSA is a sink-enhanced member in alfalfa. The alfalfa MsSPSB and MsSPSB3 forms showed leaf-enhanced expression, unlike tobacco, where the predominant form in the leaves is SPSC (Chen et al. 2005; Lutfiyya et al. 2007). The expression of the leaf-enhanced MsSPSB genes seems to be regulated by N availability (Fig. 4). However, the changes in expression of MsSPS genes may also result from indirect effects caused by the reduction in growth under N starvation conditions.
Like SPS, there appears to be more than one isoform of SPP. Western blot analysis showed two immunoreactive bands for SPP and one of the isoforms present in the nodules is absent in the leaves. This would suggest that SPP is encoded by a small gene family in alfalfa, as in other plants like Arabidopsis and rice (Lunn and MacRae 2003). However, we have identified only one SPP gene in alfalfa, and only one gene has been identified in M. truncatula (Lunn et al. 2000). BLAST searches of the M. truncatula sequencing database using SPP nucleotide sequences from different plant species have resulted in the identification of only one SPP gene.
The expression pattern of SucS in alfalfa supports the notion that SucS activity is required for supplying the carbohydrates needed for N2-fixation and N assimilation, with much higher accumulation of MsSucS mRNA and protein in the nodules (sink) compared to the leaves (source). SucS is also important for regulating C metabolism and N2 fixation (Gordon et al. 1999; Barratt et al. 2001; Colebatch et al. 2002; Hohnjec et al. 2003). Downregulation of SucS by mutations in pea or antisense strategy in L. japonicus and M. truncatula has dramatic negative effects on the establishment and maintenance of an efficient N2 fixing symbiosis (Barratt et al. 2001; Baier et al. 2007; Horst et al. 2007). Furthermore, the SucS transcript and protein level are higher in the WT nodules compared to Fix− nodules (Figs. 4, 5).
The pathways for Suc utilization in alfalfa nodules are shown in Fig. 6. While the major part of the pathway is known to occur in the nodules, we introduced the reactions catalyzed by SPS, SPP and SucS based on the information obtained from this study. Suc is metabolized by either SucS to produce UDPGlc and Fru, or by invertase to produce Glc and Fru, which after phosphorylation by hexokinases, either enter the glycolytic or the oxidative pentose phosphate pathways and are metabolized to phosphoenol pyruvate (PEP), which is converted to oxaloacetic acid and then to l-malate, to fuel nitrogen fixation in the bacteroids, and to α-ketoglutarate for the assimilation of ammonia by the GS-GOGAT pathway (Barsch et al. 2006; Tesfaye et al. 2006). The relative contribution of SucS and alkaline/neutral invertase in the partitioning of C between the different pathways in the nodules is not clear, but the evidence indicates that invertase contributes to the provision of hexoses for starch biosynthesis (Flemetakis et al. 2006). However, a more recent report on the analysis of mutants for the nodule isoform of alkaline/neutral invertase (LjNV1), showed no change in the number of starch granules compared to the nodules on WT plants (Welham et al. 2009). Moreover, since the LjNV1 mutants were capable of forming functional root nodules, it would follow that LjNV1 does not play a crucial role in nodule formation or function. In contrast, however, the nodule-enhanced isoforms of SucS are required for nodule function in different legumes (Gordon et al. 1999; Baier et al. 2007; Horst et al. 2007). The spatial expression patterns of SucS in M. truncatula nodules and SucS and invertase in L. japonicus nodules, suggest that Suc is primarily hydrolyzed in the vascular tissues and that hexoses are transported to the central part of the nodules (Flemetakis et al. 2003; Hohnjec et al. 2003).
Starch content in the Fix− nodules is almost fourfold higher than in WT nodules and there is higher accumulation of hexoses in the Fix− nodules indicating a change in the allocation of carbon. The hexoses derived from breakdown of Suc are channelled more into starch synthesis in the Fix− nodules. Higher SPS activity along with low starch levels in the WT nodules may suggest that one of the roles for SPS in nodules is to synthesize Suc from the breakdown of starch, as in the case of Anabaena heterocysts, where it has been proposed that SPS functions to synthesize Suc from the degradation of glycogen (Cumino et al. 2007).
An inverse correlation in the accumulation pattern of Suc and starch along with the hexoses in the WT and Fix− nodules, suggests that SPS might have a regulatory role in C cycling in the nodules, as in other sink tissues. Suc cycling occurs in photosynthetic tissues when the C uptake exceeds the capacity for carbohydrate utilization (Reich et al. 2006), but Suc cycling in heterotrophic tissues has additional physiological roles (Geigenberger et al. 1997; Rae et al. 2005). Unloaded Suc can undergo this cycle that could facilitate sensitive regulation of Suc mobilization in response to changes in the supply and the demand of Suc (Xu et al. 2007). Thus, SPS in the root nodules might function to maintain the concentration of the different carbohydrates for optimum functioning of the nodules.
Differences in SPS activity and accumulation of SucS between the WT and Fix− nodules, suggest that the process of Suc breakdown and re-synthesis is more active in the WT nodules engaged in the processes of N2 fixation and ammonia assimilation. This is an indication of an important role for SPS in nodule physiology. Our results on the characterization of SPS activity in alfalfa show that nodule SPS activity is very low at low concentrations of the activator Glc6P, but its activity increases to high levels at higher concentration of Glc6P. This type of allosteric response to Glc6P activation would make the enzyme very active when hexose phosphate pools become high, allocating more C to Suc synthesis, and modulating the amount of C available for respiration and other physiological processes. The allosteric differences between the Fix− and WT nodule SPS indicate that the N status has a role in the activation state of the enzyme, resulting in differences in modulation by the Glc6P concentration in the nodules. However, it is also possible that the differences in the allosteric properties of SPS between the WT and Fix-nodules may not be due to differences in the N status but could be attributed to differences in the sucrose levels. The Glc6P concentration is higher in the Fix− than in the WT nodules, which is expected if less Glc6P is allocated for the production of energy and C skeletons for N2 fixation and N assimilation. The excess of C in the Fix− nodules is allocated to the synthesis of starch instead of Suc synthesis as in the WT nodules. The differences in the allosteric response of SPS to Glc6P in the nodules may be a determinant of allocating C to either Suc or starch synthesis. The SPS protein in the Fix− nodules appears more labile, as shown by the presence of protein degradation products reacting with the anti-SPS antibody (Fig. 6). The control of nodule SPS activity by protein turnover may be of physiological significance in the nodules as has been suggested in general for SPS (Lunn and MacRae 2003). The differences in the allosteric properties of nodule SPS suggest differences in protein modification, like phosphorylation, which may also render the enzyme a target for degradation (Zuk et al. 2005). It has been shown that the phosphorylation state of SPS is the largest determinant of its catalytic activity (Winter and Huber 2000; Huber 2007).
If as we propose that SPS and SPP along with SucS are involved in cytosolic cycles of sucrose breakdown and synthesis to allow for maintaining sucrose concentration at regulatory levels, then the expression of the relevant genes should be co-localized. Experiments are in progress to localize the site of SPS expression in the nodules. Eventually taking a genetic manipulation approach will provide an answer as to the role of SPS in the nodules.
References
Baier MC, Barsch A, Kuster H, Hohnjec N (2007) Antisense repression of the Medicago truncatula nodule-enhanced sucrose synthase leads to a handicapped nitrogen fixation mirrored by specific alterations in the symbiotic transcriptome and metabolome. Plant Physiol 145:1600–1618
Barratt DHP, Barber L, Kruger NJ, Smith AM, Wang TL, Martin C (2001) Multiple, distinct isoforms of sucrose synthase in pea. Plant Physiol 127:655–664
Barsch A, Tellstrom V, Patschkowski T, Kuster H, Niehaus K (2006) Metabolite profiles of nodulated alfalfa plants indicate that distinct stages of nodule organogenesis are accompanied by global physiological adaptations. Mol Plant–Microbe Interact 19:998–1013
Baxter CJ, Foyer CH, Turner J, Rolfe SA, Quik WP (2003) Elevated sucrose-phosphate synthase activity in transgenic tobacco sustains photosynthesis in older leaves and alters development. J Exp Bot 54:1813–1820
Castleden CK, Aoki N, Gillespie VJ, MacRae EA, Quik WP, Buchner P, Foyer CH, Furbank RT, Lunn JE (2004) Evolution and function of the sucrose phosphate synthase gene families in wheat and other grasses. Plant Physiol 135:1–12
Chen S, Hajirezaei M, Börnke F (2005) Differential expression of sucrose-phosphate synthase isoenzymes in tobacco reflects their functional specialization during dark governed starch mobilization in source leaves. Plant Physiol 139:1163–1174
Choi HK, Kim D, Uhm T, Limpens E, Lim H, Mun JH, Kalo P, Penmetsa RV, Seres A, Kulikova O, Roe BA, Bisseling T, Kiss GB, Cook DR (2004) A sequence-based genetic map of Medicago truncatula and comparison of marker colinearity with M. sativa. Genetics 166:1463–1502
Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK (2002) Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Mol Plant–Microbe Interact 15:411–420
Cumino AC, Marcozzi C, Barreiro R, Salerno GL (2007) Carbon cycling in Anabaena sp. PCC 7120. Sucrose synthesis in the heterocysts and possible role in nitrogen fixation. Plant Physiol 143:1385–1397
De Vries SC, Springer J, Wessels JGH (1982) Diversity of abundant mRNA sequences and patterns of protein synthesis in etiolated and greened pea seedlings. Planta 156:120–135
Flemetakis E, Dimou M, Cotzur D, Efrose RC, Alvalakis G, Colebatch G, Udvardi MK, Katinakis P (2003) A sucrose transporter, LjSUT4, is up-regulated during Lotus japonicus nodule development. J Exp Bot 54:1789–1791
Flemetakis E, Efrose RC, Ott T, Stedel C, Alvalakis G, Udvardi MK, Katinakis P (2006) Spatial and temporal organization of sucrose metabolism in Lotus japonicus nitrogen-fixing nodules suggests a role for the elusive alkaline/neutral invertase. Plant Mol Biol 62:53–69
Fung RWM, Langenkamper G, Gardner RC, MacRae E (2003) Differential expression within an SPS gene family. Plant Sci 164:459–470
Geigenberger P, Stitt M (1991) A futile cycle of sucrose synthesis and degradation is involved in regulating partitioning between sucrose, starch and respiration in cotyledons of germinating Ricinus communis L. seedlings when phloem transport is inhibited. Planta 185:81–90
Geigenberger P, Reimholz R, Geiger M, Merlo L, Canale V, Stitt M (1997) Regulation of sucrose and starch metabolism in potato tubers in response to short-term water deficit. Planta 201:502–518
Gordon AJ, Minchin FR, James CL, Komina O (1999) Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol 120:867–878
Graham PH, Vance CP (2003) Legumes: importance and constrains to greater use. Plant Physiol 131:872–877
Grof CPL, So CTE, Perroux JM, Bonnett GD, Forrester RI (2006) The five families of sucrose-phosphate synthase genes in Saccharum spp. are differentially expressed in leaves and stem. Funct Plant Biol 33:605–610
Harlow E, Lane D (1998) Antibodies, a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Hirsch AM, Bang M, Ausubel FM (1983) Ultrastructural analysis of ineffective alfalfa nodules formed by nif:Tn5 mutants of Rhizobium meliloti. J Bacteriol 155:367–380
Hohnjec N, Perlick AM, Pühler A, Küster H (2003) The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol Plant–Microbe Interact 16:903–915
Horst I, Welham T, Kelly S, Kaneko T, Sato S, Tabata S, Parniske M, Wang TL (2007) TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiol 144:806–820
Hubbard NL, Pharr DM, Huber SC (1991) Sucrose phosphate synthase and other sucrose metabolizing enzymes in fruits of various species. Physiol Plant 82:191–196
Huber SC (2007) Exploring the role of protein phosphorylation in plants: from signalling to metabolism. Biochem Soc Trans 35:28–32
Huber SC, Huber JL (1996) Role and regulation of sucrose-phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47:431–444
Im KH (2004) Expression of sucrose-phosphate synthase (SPS) in non-photosynthetic tissues of maize. Mol Cells 17:404–409
Ingram J, Chandler JW, Gallagher L, Salamini F, Bartels D (1997) Analysis of cDNA clones encoding sucrose-phosphate synthase in relation to sugar interconversions associated with dehydration in the resurrection plant Craterostigma plantagineum Hochst. Plant Physiol 115:113–121
Komatsu A, Takanokura Y, Omura M, Akihama T (1996) Cloning and molecular analysis of cDNAs encoding three sucrose phosphate synthase isoforms from a citrus fruit (Citrus unshiu Marc). Mol Gen Genet 252:346–351
Langenkamper G, Fung RWK, Newcomb RD, Atkinson RG, Gardner RC, MacRae EA (2002) Sucrose phosphate synthase genes in plants belong to three different families. J Mol Evol 54:322–332
Li CR, Zhang XB, Hew CS (2003) Cloning of a sucrose-phosphate synthase gene highly expressed in flowers from the tropical epiphytic orchid Oncidium goldiana. J Exp Bot 54:2189–2191
Lodwig EM, Hosie AHF, Bourdes A, Findlay K, Allaway D, Karunakaran R, Downie JA, Poole PS (2003) Amino acid recycling drives nitrogen fixation in the legume–rhizobium symbiosis. Nature 422:722–726
Loreti E, Bellis LD, Alpi A, Perata P (2001) Why and how do plants sense sugars? Ann Bot 88:803–812
Lunn JE, MacRae EA (2003) New complexities in the synthesis of sucrose. Curr Opinion Plant Biol 6:208–214
Lunn JE, Ashton AR, Hatch MD, Heldt HW (2000) Purification, molecular cloning, and sequence analysis of sucrose-6F-phosphate phosphohydrolase from plants. Proc Natl Acad Sci USA 97:12914–12919
Lutfiyya LL, Xu N, D’Ordine RL, Morrell JA, Miller PW, Duff SMG (2007) Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. J Plant Physiol 164:923–933
Nguyen-Quoc B, Foyer CH (2001) A role for ‘futile cycles’ involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. J Exp Bot 52:881–889
Ortega JL, Moguel-Esponda S, Potenza C, Conklin CF, Quintana A, Sengupta-Gopalan C (2006) The 3′ untranslated region of a soybean cytosolic glutamine synthetase (GS1) affects transcript stability and protein accumulation in transgenic alfalfa. Plant J 45:832–846
Rae AL, Grof CPL, Casu RE, Bonnett GD (2005) Sucrose accumulation in the sugarcane stem: pathway and control points for transport and compartmentation. Field Crop Res 92:159–168
Reich PB, Hungate BA, Luo Y (2006) Carbon–nitrogen interactions in terrestrial ecosystems in response to rising atmospheric carbon dioxide. Annu Rev Ecol Evol Syst 37:611–636
Reimholtz R, Geiger M, Haake V, Deiting U, Krause KP, Sonnewald U, Stitt M (1997) Potato plants contain multiple forms of sucrose phosphate synthase, which differ in their tissue distribution, their levels during development, and their response to low temperature. Plant Cell Environ 20:291–305
Roby C, Cortes S, Gromova M, Le Bail JL, Roberts JK (2002) Sucrose cycling in heterotrophic plant cell metabolism: first step towards an experimental model. Mol Biol Rep 29:145–149
Tesfaye M, Samac DA, Vance CP (2006) Insights into symbiotic nitrogen fixation in Medicago truncatula. Mol Plant–Microbe Interact 19:330–341
Toroser D, Huber SC (1997) Protein phosphorylation as a mechanism for osmotic stress activation of sucrose-phosphate synthase in spinach leaves. Plant Physiol 114:947–955
Trevanion SJ, Castleden CK, Foyer CH, Furbank RT, Quick WP, Lunn JE (2004) Regulation of sucrose-phosphate synthase in wheat (Triticum aestivum) leaves. Funct Plant Biol 31:685–695
Trevaskis B, Colebatch G, Desbrosses G, Wandrey M, Wienkoop S, Saalbach G, Udvardi M (2002) Differentiation of plant cells during symbiotic nitrogen fixation. Comp Funct Genom 3:151–157
Wacek TJ, Alm D (1978) Easy to make ‘Leonard jar’. Crop Sci 18:514–515
Welham T, Pike J, Horst I, Flemetakis E, Katinakis P, Kaneko T, Sato S, Tabata S, Perry J, Parniske M, Wang TL (2009) A cytosolic invertase is required for normal growth and cell development in the model legume, Lotus japonicus. J Exp Bot 60:3353–3365
Wienkoop S, Larrainzar E, Glinski M, González EM, Arrese-Igor C, Weckwerth W (2008) Absolute quantification of Medicago truncatula sucrose synthase isoforms and N-metabolism enzymes in symbiotic root nodules and the detection of novel nodule phosphoproteins by mass spectrometry. J Exp Bot 59:3307–3315
Winter H, Huber SC (2000) Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. CRC Crit Rev Plant Sci 19:31–67
Xu Y, Li HB, Zhu YX (2007) Molecular biological and biochemical studies reveal new pathways important for cotton fiber development. J Int Plant Biol 49:69–74
Zuk M, Weber R, Szopa J (2005) 14-3-3 Protein down-regulates key enzyme activities of nitrate and carbohydrate metabolism in potato plants. J Agric Food Chem 53:3454–3460
Acknowledgments
This work was supported by the National Institutes of Health (Grant numbers GMO-8136, GMO-61222, GMO-7667), National Science Foundation (Grant numbers NSF-DBI0619747, NSF-0331446), and by the Agricultural Experiment Station at New Mexico State University.
Author information
Authors and Affiliations
Corresponding author
Additional information
J. L. Ortega is the joint first author.
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2009_1043_MOESM1_ESM.eps
Fig. S1 Phenotype of alfalfa plants grown in the greenhouse in Magenta boxes 28d after inoculation with either Sinorhizobium meliloti 2011 (WT) or S. meliloti 1491 (Fix−) strains. Five boxes of plants inoculated with the WT strain and 9 boxes of plants inoculated with the Fix- strain are shown. Supplementary material 1 (EPS 7945 kb)
425_2009_1043_MOESM2_ESM.eps
Fig. S2 Analysis of SPS protein in the leaves and nodules of different legumes. Protein (50 μg) from leaf and nodule extracts from alfalfa (Ms), Phaseolus vulgaris (Pv), Pisum sativum (Ps) and Lotus japonicus (Lj) was separated by SDS PAGE followed by Western blot analysis using anti-SPS antibodies. The SPS antibody was immunoselected against SPS immobilized to a nylon membrane. The migration of the molecular weight standards is as shown. Supplementary material 2 (EPS 2300 kb)
425_2009_1043_MOESM3_ESM.eps
Fig. S3 Phylogenetic analysis of the SPS predicted protein sequences from alfalfa, Medicago truncatula, Arabidopsis and tobacco. Protein sequences from alfalfa MsSPSA, MsSPSB, MsSPSB3 (NCBI accession numbers AAR31210, ABW89596 and ACN89831), and M. truncatula SPSA and SPSB forms (M. truncatula sequencing resources gene numbers AC144657_7, AC157648_2_3, and CU424494_10), were clustered together with the A, B and C isoforms from Arabidopsis and tobacco (AtSPSA1, AtSPSA2, AtSPSB, AtSPSC, NtSPSA, NtSPSB and NtSPSC; NCBI accession numbers AAK09427, ABW89596, NP_197528, NP_196672, NP_171984, NP_192750, AAF06792, ABA64521 and ABA64520, respectively). The tree was generated using Geneious Pro 4.7 (Biomatters) under default parameters. The numbers on each node are the bootstrap proportion values for 1000 replicates. Bar represents the Jukes-Cantor genetic distance. Supplementary material 3 (EPS 397 kb)
Rights and permissions
About this article
Cite this article
Aleman, L., Ortega, J.L., Martinez-Grimes, M. et al. Nodule-enhanced expression of a sucrose phosphate synthase gene member (MsSPSA) has a role in carbon and nitrogen metabolism in the nodules of alfalfa (Medicago sativa L.). Planta 231, 233–244 (2010). https://doi.org/10.1007/s00425-009-1043-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-009-1043-y