Abstract
Main conclusion
In alfalfa, the B form of Sucrose phosphate synthase synthesizes sucrose in the leaves while the A form participates in regulatory cycles of synthesis/breakdown of sucrose/starch in the root nodules.
Abstract
Sucrose (Suc) is the major stable product of photosynthesis that is transported to all heterotrophic organs as a source of energy and carbon. The enzyme sucrose phosphate synthase (SPS) catalyzes the synthesis of Suc. Besides the leaves, SPS is also found in heterotrophic organs. There are two isoforms of SPS in alfalfa (Medicago sativa): SPSA and SPSB. While SPSA is expressed in the vasculature of all the organs and in the N2-fixing zone in the nodules, SPSB is exclusively expressed in the photosynthetic cells. Two classes of alfalfa transformants were produced, one with a gene construct consisting of the alfalfa SPSA promoter and the other with the SPSB promoter—both driving the maize SPS coding region—referred to as SPSA-ZmSPS and SPSB-ZmSPS, respectively. Both classes of transformants showed increased growth compared to control plants. The SPSB-ZmSPS transformants showed increased SPS protein levels and activity along with a significant increase in the Suc levels in the leaves. The SPSA-ZmSPS transformants showed an increase in the SPS protein level and enzyme activity both in the leaves and the nodules with no increase in Suc content in the leaves but a substantial increase in the nodules. Both SPSA and SPSB have unique roles in the nodules (sink) and leaves (source). SPSB is responsible for the synthesis of Suc in the photosynthetic cells and SPSA participates in a regulatory cycle in which Suc is simultaneously degraded and re-synthesized; both these functions contribute to plant growth in rhizobia nodulated alfalfa plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sucrose (Suc) is the major stable product of photosynthesis that is transported from the photosynthetic tissues via the phloem into all heterotrophic tissues and is a source of energy and carbon (C) metabolites (Lunn and MacRae 2003; Stitt et al. 2010). The C that is fixed during the day is either exported from the plastid as triose phosphate to the cytoplasm, which is used for the synthesis of Suc or is retained in the chloroplast and used for the synthesis of starch. The starch is mobilized in the dark to provide the substrates for Suc synthesis. Sucrose phosphate synthase (SPS; EC 2.3.1.14) catalyzes the synthesis of Suc-6-phosphate (Suc-6P) from fructose-6-phosphate (Fru-6P) and UDP-glucose (UDP-Glc). The enzyme sucrose phosphate phosphatase (SPP; EC 3.1.3.24), in a subsequent reaction, hydrolyzes Suc-6P to produce Suc. In this two-step reaction, SPS plays a key role in the synthesis of Suc (Stitt et al. 1988).
Based on its function, it is not surprising that SPS is found mostly in the photosynthetic tissues. Several groups, however, have reported that SPS is also expressed in heterotrophic sink tissues, such as potato tubers and cotton fibers (Geigenberger et al. 1997; Babb and Haigler 2001; Im 2004). More recently, we have shown SPS synthesis and accumulation in the root nodules of alfalfa and pea plants (Aleman et al. 2010; Mohmed 2014; Kaur et al. 2019). The root nodules formed in leguminous plants are the result of the symbiotic interaction between the host and the N2-fixing bacteria, rhizobia (Graham and Vance 2003). It is the site for N2-fixation where the symbiont can convert free nitrogen into NH3, which can then be used in the synthesis of glutamine (Gln). The Gln is transported from the nodules to the aerial parts of the plant where it acts as the starting point for the synthesis of all other amino acids, nucleic acids, and other N containing compounds. The nodule, thus, acts as a large sink for photosynthetic products required to fuel the N2 fixation process and the assimilation of nitrogen.
Two possible roles for SPS in tissues that import and degrade Suc have been proposed: SPS could allow resynthesis of Suc after import via apoplastic cleavage or SPS could be involved in a regulatory cycle in which Suc is simultaneously degraded and resynthesized. This cycle has been shown to operate in potato tubers (Geigenberger and Stitt 1991) and tomato fruits (Nguyen-Quoc and Foyer 2001), and could facilitate sensitive regulation of Suc mobilization in response to changes in the supply of, and the demand for, Suc. Studies suggest that the SPS activity in heterotrophic organs may have a role in Suc unloading in the sink tissues. Tomato transformants expressing SPS transgene in the fruits showed elevated SPS activity in the fruits accompanied by an increase in the Suc level (Nguyen-Quoc et al. 1999).
Sucrose phosphate synthase is encoded by a small multigene family and the members, besides showing differences in tissue-specific expression at the transcription level (Privat et al. 2008; Roy Choudhury et al. 2008; Choudhury et al. 2010; Verma et al. 2011; Yonekura et al. 2013), are also regulated at the posttranslational level through covalent modification in response to metabolic and environmental stimuli and allosteric regulation via metabolites, Glc-6P, and inorganic phosphate (Huber 2007; Stitt et al. 2010). SPS proteins cluster into at least three families, A, B, and C (Langenkamper et al. 2002) and at least one representative member of each family is expressed for each gene family present in the genome of a given dicot species (Reimholz et al. 1997; Lunn and MacRae 2003; Chen et al. 2005). Monocotyledonous plants contain an additional SPS gene family referred to as the D family (Castleden et al. 2004; Lutfiyya et al. 2007). Analysis of alfalfa (Medicago sativa), M. truncatula, and pea (Pisum sativum) (Aleman et al. 2010; Mohmed 2014), however, has shown that only the A and B families are present in their genome with the SPSB family having two gene members, SPSB3 and SPSB5.
While SPSB is the major leaf-specific isoform in alfalfa, SPSA is expressed in all organs but with the highest expression level in the nodules followed by the stem (Aleman et al. 2010). Similarly, SPSB is only expressed in pea leaves, while the expression of SPSA is the highest in the pod wall, seeds, and nodules in a pea (Mohmed 2014). The exclusive expression of SPSB genes in the leaves would suggest a role in the biosynthesis of sucrose during photosynthesis (Aleman et al. 2010; Stitt et al. 2010), but the role of SPSA is still speculative. The focus of this paper is to determine the role of SPSA in alfalfa plants grown under N2-fixing conditions. Towards this objective, our experimental approach has been to check the outcome of modulating the expression of SPS in cells where SPSA is expressed. Also, for the purpose of delineating the function of SPSA from SPSB, we have produced plants with an increased expression of SPS in cells where SPSB is expressed. Alfalfa plants were transformed with three different gene constructs: the maize SPS gene (ZmSPS) driven either by the alfalfa SPSB gene promoter or the SPSA gene promoter and the SPSA promoter driving the SPSA coding region in an antisense orientation. Analysis of these three classes of transformants suggests that while SPSB has a role in the synthesis of Suc in the photosynthetic cells, SPSA is involved in the synthesis of Suc in nonphotosynthetic cells including the nodules. Both SPSA and SPSB are critical for the functioning of nodules and for the development of alfalfa plants grown under symbiotic conditions.
Materials and methods
Isolation of the alfalfa SPSB and SPSA promoters and construction of SPSA-ZmSPS, SPSB-ZmSPS, SPSA-antiSPSA, and SPSA-GUS
The promoter regions corresponding to the SPSA and SPSB5 genes were isolated by the PCR amplification of alfalfa genomic DNA using primer sets that were designed based on the MsSPSA (GenBank Accession no. AF322116.2) and MsSPSB (Aleman et al. 2010; GenBank Accession no. EU234514.1) gene sequences. By using the appropriate restriction enzymes, the alfalfa SPS promoters were ligated to the maize SPS1 cDNA (Worrell et al. 1991) followed by the NOS terminator. All the cloning was done in the pUC19 vector and the whole ligated fragments (promoter-ZmSPS-terminator) were then inserted into Cambia 2300, which has the NPTII gene cassette for the selection of transformed plants on kanamycin.
The SPSA-antisense SPSA (SPSA-Anti) was made by inserting the alfalfa SPSA cDNA in reverse orientation behind the SPSA promoter. The SPSA-GUS gene construct was made using pBI121, which contains the CaMV 35S promoter driving the GUS gene (CLONETECH). Using the pertinent restriction enzymes, the CaMV 35S promoter was replaced by the SPSA promoter. The NPTII gene cassette was maintained in the vector. The plasmids with the different gene constructs were mobilized into the Agrobacterium tumefaciens strain GV3101 by a freeze–thaw method.
Plant transformation and growth conditions
Agrobacterium-mediated alfalfa plant transformations were carried out as described by Gebril et al. (2015). Three randomly selected PCR positive transformants from tissue culture representing each class, SPSA-GUS, 35S-GUS, SPSA-ZmSPS, SPSB-ZmSPS, SPSA-anti SPSA and alfalfa plants obtained by regeneration (Control) were used for all the analysis. The tissue culture plants were acclimated to the greenhouse conditions and once established, the plants were clonally propagated to make biological replicates (Kaur et al. 2019). Plants were inoculated with Sinorhizobium meliloti to initiate nodule development and were then fed with N-free Hoagland’s nutrient solution, weekly. For each control plant and individual transformant, three to five clones were analyzed as the biological replicates and averaged or pooled for experiments. The plants were grown in the greenhouse with full sunlight during the day along with supplemental LED grow lights (LIFTED, Rio Rancho, NM), for an extended light period during the winter.
DNA isolation and genomic PCR
DNA was isolated from the leaves of alfalfa plants using the DNeasy plant mini kit (Qiagen, Germantown, MD).
RNA extraction and qRT PCR
For quantitative real-time reverse transcription-PCR (qRT-PCR) analysis, total RNA was isolated from the leaves and nodules of alfalfa plants using an RNAeasy kit (Qiagen, Germantown, MD) and the cDNA was synthesized using Superscript III reverse transcriptase (Life Technologies, Carlsbad, CA). The cDNA was subjected to PCR using an IsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions with primers based on the MsSPSA (forward: 5′-GAG TGA TGT TTC TGC TCA TGG TGG TG-3′; reverse: 5′-CGA CAT ACT TAA CCT GAC CAC CCG TAT C-3′) and MsSPSB (forward: 5′-GGA GCT TGG TAG AGA TTC TGA TAC TGG TG-3′ reverse: 5′-CTC TCC GGT GCT ATC ATC CTC ATC ATT-3′) sequences. The amount of MsSPS mRNA per total mRNA was calculated using qPCR standard curves with the MsSPSA and MsSPSB cDNAs as targets.
In situ GUS localization
Expressions of the gene constructs with the GUS coding region were monitored using histochemical staining according to Jefferson et al. (1987).
Protein isolation and analysis
Leaf and nodule tissues from biological replicates were harvested for each of the three independent transformants for each class, and were immediately placed in liquid N and stored at − 80 °C until the experiments were done. The extractions were done as described by Gebril et al. (2015). Protein concentration was measured using the Bradford protein assay (BioRad, Hercules, CA) with bovine serum albumin as standard.
Sucrose phosphate synthase enzyme activity was assayed by quantifying the fructosyl moiety of Suc using the Anthrone test (Seger et al. 2015) and the activity is expressed as nmol Suc-P mg protein−1 min−1. For western blot analysis, the protein extracts used for enzyme activities were subjected to SDS PAGE followed by western blotting. The fractionated protein from these gels was electroblotted on Immobilon-P PVDF membrane (Millipore, Bedford, MA). Detection of polypeptides was performed using polyclonal antibodies raised against maize SPS (#AS06 185; Agrisera, Sweden). The immunoreactive bands were visualized with alkaline phosphatase linked secondary antibodies using nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indoyl phosphate (BCIP) as substrates. The immunoreactive bands were quantified using an image analysis system (CARESTREAM). Experiments were performed at least 3–4 times and only representative results are presented.
Carbohydrate analysis
Soluble carbohydrates were extracted from the leaf and nodule tissues by grinding in 10 volumes (v/w) of 80% (v/v) ethanol and incubating at 70 °C for 90 min. Sucrose determination was done by using the Anthrone reagent (Seger et al. 2015). Sucrose content was calculated from a Suc standard curve.
Statistical analysis
All the statistical significance analysis was done using Student’s t test andcomputed using Microsoft Excel. Each bar on the graphs is the average of three biological replicates and the bars represent the value of standard deviation. Significant differences were evaluated using a t test and are shown by asterisks. Single asterisk (*) indicates 0.01 < P < 0.05 and double asterisks (**) indicate 0.001 < P < 0.01.
Results
While SPSA is expressed in both the leaves and nodules, SPSB is expressed specifically in the leaves of alfalfa
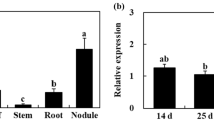
To check the expression pattern of SPSA and SPSB in alfalfa, RNA was isolated from both the leaves and nodules of nontransformed alfalfa plants and subjected to qRT PCR using primer sets specific for the alfalfa SPSA and SPSB genes. As seen in Fig. 1, SPSA showed expression in both the leaves and the nodules, with the transcript level being > twofold higher in the nodules. Transcripts for MsSPSB, however, were seen only in the leaves and the level was > fourfold higher than the SPSA transcript level in the leaves.
Confirmation of the presence of the transgene in the SPSA-ZmSPS and SPSB-ZmSPS transformants
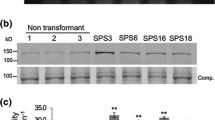
Alfalfa was transformed with the two gene constructs: the ZmSPS coding region driven by the MsSPSA gene promoter (SPSA-ZmSPS) or the MsSPSB gene promoter (SPSB-ZmSPS). The putative transformants were confirmed for the presence of the transgene by genomic PCR using the primer set for NPTII, the gene conferring resistance to kanamycin, and ZmSPS—the coding region of the two constructs (Table 1). All the independent transformants tested showed amplification products with both sets of primers (Fig. 2). Three independent transformants for both SPSA-ZmSPS and SPSB-ZmSPS classes along with three control plants were selected for further analysis.
Analysis of SPSA-ZmSPS and SPSB-ZmSPS transformants to check for the integration of the gene constructs. DNA isolated from three independent transformants for each class: SPSA-ZmSPS and SPSB-ZmSPS and control plants were isolated and subjected to genomic PCR using NPTII and ZmSPS specific primer sets, and the products were then fractionated on agarose gels. The amplicon size with the NPTII gene primer set was determined to be 678 bp and with the ZmSPS primer set was 886 bp
SPSA-ZmSPS and SPSB-ZmSPS transformants showed both an increase in the steady-state level of the protein and SPS enzyme activity in their leaves
To check if the SPSA-ZmSPS and SPSB-ZmSPS gene constructs are expressed in the leaves, proteins were extracted from the leaves of the same maturity, harvested from three clonally propagated plants for each independent transformant, and subjected to western blot analysis using SPS antibodies (Fig. 3a). The intensity of the individual immunostained bands were quantified and plotted graphically as band intensity in pixels (Fig. 3b, c). As seen in Fig. 3a, all the lanes including the ones with samples from control plants showed an immunoreactive band (138 kD), but the level of accumulation was significantly higher in the two classes of transformants. The SPS protein level in the two classes of transformants in excess of the level seen in the control plants can be attributed to ZmSPS, a product of the transgene construct. It is important to note that the level of SPS accumulation was the same in the leaves of both classes of transformants.
Analysis of SPS protein levels and SPS enzyme activity in the leaves of the SPSA-ZmSPS and SPSB-ZmSPS transformants. a 75 μg of the total protein extracted from the leaves of three independent transformants representing the two classes and control plants was subjected to SDS PAGE (7.5% acrylamide) followed by western blot analysis using SPS antibodies. A representative blot is shown here. The size of the immunoreactive band was determined to be 138 kD based on the migration of proteins of known molecular weight. b The immunoreactive bands from the western blot were quantified using the Kodak image analysis software and plotted as band intensity in pixels. c The average relative band intensity for each class of plants was calculated from b. d The same nodule extracts used for western blot analysis were used for enzyme activity measurement by quantifying the synthesis of Suc-6P from UDP-Glc and Fru-6P. SPS enzyme activity values are plotted as nmol Sucrose-P mg−1 protein min−1. Values are the mean ± SD of samples from three independent transformants for each class and the control plants. Significant differences from the average value obtained for the control plants were evaluated by t test and are shown by asterisks (*P < 0.05 or ** < 0.01)
To check if the increased accumulation of SPS protein in the leaves of the transformants is translated to a higher level of SPS enzyme activity, the same protein extract used for western blot analysis was used for the measurement of SPS enzyme activity. The activities obtained for each of the three independent transformants representing each class and the three control plants were averaged and subjected to statistical analysis and are presented graphically in Fig. 3d. The two classes of transformants exhibited a significantly higher level of activity compared to control plants. Taken together, the results suggest that there is an increased SPS protein level and enzyme activity in the leaves of both classes of transformants.
Only the SPSA-ZmSPS transformants showed higher accumulation of SPS protein in the nodules, but both sets of transformants showed an increase in the SPS enzyme activity
To check if the SPSA-ZmSPS and SPSB-ZmSPS gene constructs function in a manner similar to the corresponding endogenous genes with regard to the expression pattern in the nodules (Fig. 1), we checked for the SPS protein levels in the nodules of the two classes of transformants along with control plants. Nodules were harvested from the same plants as those used for the leaf samples and the total protein extract from these nodules was subjected to western blot analysis using SPS antibodies. As seen in Fig. 4a, all the lanes showed an immunoreactive band with the SPSA-ZmSPS transformants showing a significantly higher level of SPS accumulation (~ 60%) compared to control and the SPSB-ZmSPS transformants (Fig. 4b, c).
Analysis of SPS protein levels and SPS enzyme activity in the nodules of the SPSA-ZmSPS and SPSB-ZmSPS transformants. a 50 μg of the total protein extracted from the nodules of three independent transformants representing the two classes and control plants was subjected to SDS PAGE (7.5% acrylamide) followed by western blot analysis using SPS antibodies. A representative blot is shown here. The size of the immunoreactive band was determined to be 138 kD based on the migration of proteins of known molecular weight. b The immunoreactive bands from the western blot were quantified using Kodak image analysis software and plotted as band intensity in pixels. c The average relative band intensity for each class of plants calculated from b. d The same nodule extracts used for western blot analysis were used for enzyme activity measurement by quantifying the synthesis of Suc-6P from UDP-Glc and Fru-6P. SPS enzyme activity values are plotted as nmol Sucrose-P mg−1 protein min−1. Values are the mean ± SD of samples from three independent transformants for each class and the control plants. Significant differences from the average value obtained for the control plants were evaluated by t test and are shown by asterisks (*P < 0.05 or ** < 0.01)
The same extracts used for the western blot analysis were used for the SPS enzyme activity measurements. The activities obtained for each of the three independent transformants representing each class and the three control plants were averaged and subjected to statistical analysis. The activities are presented graphically in Fig. 4d. The SPSA-ZmSPS transformants showed a significant increase (~ 2.5-fold) in enzyme activity in the nodules while the nodules of the SPSB-ZmSPS transformants showed a smaller but significant (~ 0.5-fold) increase over control plants.
Only the SPSB-ZmSPS transformants showed a higher level of Suc accumulation in the leaves while the Suc level in the nodules was higher in both the SPSA-ZmSPS and SPSB-ZmSPS transformants
With the rationale that an increase in the SPS enzyme activity could translate into higher rates of Suc synthesis, the Suc content was measured in the leaves and nodules of the SPSA-ZmSPS, SPSB-ZmSPS, and control plants. Sucrose was extracted from the tissues harvested from the same plants as used for protein analysis (Figs. 3, 4). The Suc concentration obtained for each of the three independent transformants representing each class and the three control plant were averaged and subjected to statistical analysis. The concentration (mmoles Suc/g tissue) is presented graphically in Fig. 5. While the SPSB-ZmSPS transformants showed ~ twofold increase in the Suc concentration in the leaves compared to the control plants, the SPSA-ZmSPS transformants showed no significant change. When comparing the Suc content in the nodules, the SPSA-ZmSPS transformants showed a > twofold increase compared to the control plants and the SPSB-ZmSPS transformants showed a ~ 0.5-fold increase.
Sucrose content in the leaves and nodules of the two classes of transformants. Sucrose content was measured in the leaves (a) and nodules (b) as described in “Materials and methods”. Sucrose content was plotted as nmoles Suc mg−1 fresh weight. Values for three independent transformants representing each one of the two classes and three control plants were measured and the mean value ± SD was calculated for each transformant. Significant differences between each class of transformants and the control plants were evaluated by t test and are shown by asterisks (*P < 0.05 or ** < 0.01)
SPSA is expressed in the vasculature of leaves, stem, roots, and nodules and in the N2-fixing zone of nodules
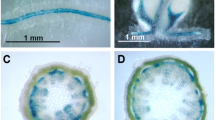
Based on the data obtained from the analysis of the two classes of transformants, it would appear that SPSB is the only isoform that has a role in the synthesis of Suc from the photosynthate. However, the role of SPSA is still conjectural. In planta localization of the site of synthesis of the SPSA protein could shed some light on its function. Towards this goal, we engineered the β-glucuronidase (GUS) gene behind the SPSA promoter (SPSA-GUS) and introduced it into alfalfa. The different parts of the plant were subjected to GUS staining. Alfalfa plants transformed with the 35S-GUS gene construct and control plants were also subjected to GUS staining. As seen in Fig. 6, the control plants showed no staining in any of the tissues tested. The 35S-GUS transformants showed uniform staining all over the leaves with the most intense staining in the borders of the cells, uniform staining of the stem, and staining restricted to the vasculature in the roots. The nodules of the 35S-GUS transformants displayed staining throughout with the maximum staining in the vasculature. In the SPSA-GUS transformants, staining in the leaves, stem, and roots was localized in the vasculature, while in the nodules, the highest staining was in the N2-fixation zone and the expansion zone. The meristematic region and the senescence zone in these nodules showed no staining.
Expression pattern of the SPSA gene in alfalfa as revealed by the analysis of SPSA promoter-GUS plants. GUS histochemical staining of the stem, leaf, roots, and nodules of nodulated alfalfa plants (SPSA-GUS, 35S-GUS and control) in vitro. Tissues were harvested from the transformants and control plant and stained with X-gluc overnight, then cleared of chlorophyll by incubating tissues with ethanol. Tissues were then fixed in 0.1% glutaraldehyde and visualized through digital images using a stereofluorescence microscope
SPSA-ZmSPS and SPSB-ZmSPS transformants showed increased growth and nodule numbers
Replicate clones for each independent transformant representing both classes and control plants were inoculated with S. meliloti, and 10 weeks later the plants were photographed (Fig. 7). The two classes of transformants showed increased growth when compared to control plants. A closer look at the two classes of plants, however, showed a difference in growth habit—the SPSA-ZmSPS transformants showed more erect and longer internodes and a thicker stem when compared to the SPSB-ZmSPS transformants. Both sets of transformants flowered about 3 weeks earlier than the control plants.
Comparing the growth pattern of the two classes of SPSA-ZmSPS and SPSB-ZmSPS transformants with the control plants. Established transformants were used to obtain shoots for propagation. The cut shoots were planted on vermiculite, and once established (~ 10 days), the cuttings were inoculated with S. meliloti and allowed to grow for a period of 60 days. Plants representing each of the three independent transformants for each class along with control plants were photographed
To check if the expression of the transgene had an effect on nodulation, the plants were uprooted and all the nodules were harvested, counted, and weighed. The number and weight of the nodules per plant were calculated and presented as a table. As seen in Table 2, both classes of transformants showed a significant increase in the number and weight of the nodules. However, when comparing the two classes of transformants, the nodules in the SPSA-ZmSPS showed a bigger increase in weight per plant while the SPSB-ZmSPS transformants showed a bigger increase in the number per plant.
SPSA-antiSPSA transformants showed a decrease in growth
SPSA-ZmSPS transformants showed an increase in growth when compared to control plants, suggesting that SPSA plays an important role in plant growth and development, in spite of the fact that it does not synthesize Suc in the photosynthetic tissues. For further validation of its importance in plant growth, we used a loss-of-function approach. Alfalfa plant was transformed with a gene construct consisting of the SPSA gene promoter driving the alfalfa SPSA cDNA in an antisense orientation (SPSA-AntiSPSA), with the rationale that SPS will be down-regulated specifically in the cells where SPSA is normally expressed. Three independent SPSA-AntiSPSA transformants and control plants were grown in replicate and inoculated with S. meliloti, and 10 weeks following inoculation they were tested for the SPS protein level and for their phenotype. Proteins were extracted from the leaves and nodules and subjected to western blot analysis using SPS antibodies and a measurement of the SPS enzyme activity. As seen in Fig. 8, SPSA-AntiSPSA transformants showed a small but significant drop in the level of SPS protein in both the leaves and nodules when compared to control plants. However, at the level of enzyme activity, a significant drop was seen in the leaves of the SPSA-AntiSPSA but not in the nodules. With regard to the growth pattern, the SPSA-antiSPSA transformants exhibited a thinner stem and narrower leaves when compared to control plants. Moreover, flowering was delayed in the SPSA-AntiSPSA transformants (Fig. 9).
Analysis of SPS protein levels and SPS enzyme activity in the leaves and nodules of the SPSA-antisense SPSA transformants and control plants. a 70 μg of the total protein extracted from the leaves and 50 μg of the nodule protein of three independent transformants and control plants were subjected to SDS PAGE (7.5% acrylamide) followed by western blot analysis using SPS antibodies. A representative blot is shown here. The size of the immunoreactive band was determined to be 138 kD based on the migration of proteins of known molecular weight. b The immunoreactive bands from the western blot were quantified using the Kodak image analysis software and plotted as band intensity in pixels. c The average relative band intensity for each class of plants calculated from b. d The same extracts used for western blot analysis were used for enzyme activity measurement by quantifying the synthesis of Suc-6P from UDP-Glc and Fru-6P. SPS enzyme activity values are plotted as nmol Sucrose-P mg−1 protein min−1. Values are the mean ± SD of samples from three independent transformants for each class and the control plants. Significant differences from the average value obtained for the control plants were evaluated by t test and are shown by asterisks (*P < 0.05 or ** < 0.01)
Comparing the growth pattern of the SPSA-antisense SPSA transformants with control plants. Established transformants were used to obtain shoots for propagation. The cut shoots were planted on vermiculite, and once established (~ 10 days), the cuttings were inoculated with S. meliloti and allowed to grow for a period of 60 days. Three plants representing each of the three independent transformants and control plants were photographed
Discussion
There is enough evidence in the literature indicating that SPS in plants is encoded by members of small gene families but the functional significance of the individual members in most cases is not well understood. The expression pattern of SPS genes has been studied in a few cases and based on their site of expression and/or the external cues that regulate their expression, particular functions have been assigned to the individual members in particular plant systems (Reimholz et al. 1997; Chavez-Barcenas et al. 2000; Fung et al. 2002; Komatsu et al. 2002; Im 2004; Okamura et al. 2011). Some studies have utilized loss-of-function mutants to assign functions to particular SPS isoforms (Chen et al. 2005; Sun et al. 2011; Hirose et al. 2014; Volkert et al. 2014; Bahaji et al. 2015). However, specific functions cannot be assigned to an individual class broadly because there is no consistent pattern observed across species.
In both tobacco and Arabidopsis, SPSA and SPSC are the two isoforms found in the leaves, both having a role in Suc synthesis (Chen et al. 2005; Volkert et al. 2014; Bahaji et al. 2015). However, while in tobacco, NtSPSC is specifically involved in the synthesis of Suc during starch mobilization in the dark and NtSPSA in the synthesis of Suc from the photosynthate (Chen et al. 2005), both the AtSPSC and AtSPSA in Arabidopsis participate in both the synthesis of Suc from the photosynthate and the mobilization of starch (Sun et al. 2011; Volkert et al. 2014; Bahaji et al. 2015). SPSB in both tobacco and Arabidopsis shows expression in the reproductive organs and SPSA is expressed constitutively (Chen et al. 2005; Volkert et al. 2014). Alfalfa, on the other hand, has only two gene families (A and B), unlike all the other dicot plants that have the additional C family (Langenkamper et al. 2002). While SPSC is expressed exclusively in the source leaves in tobacco and Arabidopsis, SPSB is the isoform whose expression is confined to the leaves in alfalfa. SPSA, as in Arabidopsis and tobacco, exhibits constitutive expression in alfalfa with the highest expression of SPSA in the nodules, followed by the stem (Aleman et al. 2010). Pea plants also exhibit a similar kind of expression pattern as alfalfa. In pea plants where we checked for expression of the two isoforms in other organs, the pod wall and the seeds showed exclusive expression of SPSA with no visible expression of SPSB, notwithstanding that the pod walls are green and photosynthetic. Analysis of the site of expression of the gene for sucrose synthase (SucS) showed that its expression coincided with the site of SPSA expression in both alfalfa and pea—SucS expression was the highest in the nodules, pod walls, and seeds, with little to no expression in the leaves where SPSB was exclusively expressed (Aleman et al. 2010; Mohmed 2014). Since SucS activity is associated with sink strength (Stein and Granot 2019), we would conclude that SPSA has a role in the sink tissues and SPSB in the source tissues.
Sucrose in the leaves is transported through the veins into various sink tissues and the CO2 fixed in the excess Suc that is exported is either stored in the vacuoles or is used to make starch in the chloroplasts. The starch is then remobilized during the night to continuously supply Suc to the sink tissues (Smith and Stitt 2007), which would necessitate the involvement of SPS. In planta expression analysis of alfalfa transformed with an SPSA-GUS gene construct showed that the expression of SPSA was confined to the vasculature in the leaves, suggesting that it does not have a primary role either in the synthesis of Suc from the photosynthate or starch breakdown. It would appear that, in alfalfa, SPSB in the leaves has a role both is the synthesis of Suc from the photosynthate during the daytime and starch mobilization in the dark.
The flow of Suc through the vasculature is driven by an osmotically generated difference in the pressure between the source and the sink tissues (Bihmidine et al. 2013; Braun et al. 2014). As the Suc is being transported in the vasculature in the stem, it also acts as the source of hexoses for the synthesis of cellulose needed for the synthesis of primary and secondary cell walls. Thus, the cycle of Suc breakdown and synthesis occurs in the stem as in the case of other heterotrophic organs (Geigenberger and Stitt 1991; Nguyen-Quoc and Foyer 2001), and the SPSA in alfalfa plays the role in the synthesis of Suc in the stem. Higher activity of SPS in the stem would favor Suc synthesis in the SPSA-ZmSPS transformants, creating a steeper difference in pressure between the stem and the nodule, and would thus increase the transport of Suc into the nodules.
Alfalfa plants transformed with the 35S-ZmSPS gene construct showed increased growth and we attributed it to the increased transport of Suc to the nodules from the leaves, which in turn resulted in an increase in N2-fixation and ammonia assimilation (Gebril et al. 2015; Kaur et al. 2019). The present study, however, shows that SPS in heterotrophic organs, including the nodules, plays just as important a role in plant growth as SPS in photosynthetic organs. One explanation for increased growth of the SPSA-ZmSPS transformants could be that increased SPS activity in the nodules of these transformants is accompanied by an increase in sink strength and a corresponding increase in the import of Suc to the nodules. This is not without precedence—overexpression of SPS in a fruit-specific manner in tomato showed an increase in Suc unloading and content in the transformed fruits, suggesting that increased SPS activity in this instance had increased the sink strength (Nguyen-Quoc et al. 1999).
At a qualitative level, the pattern of SPS protein accumulation in the leaves and nodules of the two sets of transformants, SPSA-ZmSPS and SPSB-ZmSPS, was in keeping with the expression pattern of the endogenous MsSPSA and MsSPSB genes. Both the SPS protein accumulation and enzyme activity showed an increase in the leaves of the SPSA-ZmSPS and SPSB-ZmSPS transformants when compared to control plants. With respect to the nodules, only the SPSA-ZmSPS transformants showed an increase in the SPS protein level over control plants. However, while the ~ twofold increase in SPS enzyme activity in the nodules of the SPSA-ZmSPS transformants was in keeping with the increase in the protein level, the enzyme activity in the nodules of the SPSB-ZmSPS transformants was not. The increase in SPS enzyme activity with no increase in protein level in the nodules of the SPSB-ZmSPS transformants could be attributed to enzyme activation. There is ample evidence in the literature showing that SPS is subject to regulation by phosphorylation/dephosphorylation, the dephosphorylated form being the active version (Huber and Huber 1996; Winter and Huber 2000), implying that SPS phosphatases are induced in the nodules. One could envision the higher Suc level in the nodules acts to induce the expression of genes that have a role in the dephosphorylation of SPS in the nodules. Sucrose is known to function as a signaling molecule (Wind et al. 2010; Ruan 2012).
While a twofold increase in Suc content was seen in the leaves of the SPSB-ZmSPS transformants, the SPSA-ZmSPS transformants showed no significant increase when compared to control plants. Since both sets of transformants exhibited a similar increase in SPS levels and enzyme activity in the leaves, the difference in the Suc concentration between the two sets of plants can only be accounted for by the site of accumulation of the enzyme. Since MsSPSA in the leaves is exclusively expressed in the vasculature (Fig. 6), it does not likely have a role in synthesizing Suc from the photosynthate, thus accounting for no measurable increase in the Suc level in the leaves of the SPSA-ZmSPS transformants. An increase in Suc level in the leaves of the SPSB-ZmSPS transformants would reaffirm that SPSB functions in synthesizing Suc from the photosynthate. Besides the leaves, SPSB-ZmSPS transformants also showed an increase in Suc levels in their nodules. This increase in Suc level could be an attribute of increased export of Suc from the leaves to the nodules. However, the more than twofold increase in Suc level in the nodules of the SPSA-ZmSPS transformants has to be ascribed solely to the increased expression of SPS in the nodules.
The Suc unloaded in the nodules is acted upon by SucS to produce hexoses that are utilized for the synthesis of starch and cellulose, and to provide a source of C-skeletons and energy for the process of N2-fixation and ammonia assimilation (Oldroyd et al. 2011). The assimilated N in the form of amino acids is then transported to the aerial parts of the plant. However, as in most heterotrophic organs, a cycle of Suc synthesis and breakdown into hexoses probably operates in the nodules (Nguyen-Quoc and Foyer 2001). An increase in the SPS activity in the nodules of the SPSA-ZmSPS transformants would favor the synthetic pathway resulting in an increase in the Suc level and nodule function.
Both classes of transformants exhibited increased growth. We suggest that an increased Suc concentration in the nodules is key to the increased growth of plants as has been proposed for the 35S-ZmSPS transformants (Gebril et al. 2015; Kaur et al. 2019). In spite of the fact that both classes of plants showed increased growth, some distinct but subtle differences could be discerned between the two (Fig. 7). Compared to the SPSB-ZmSPS transformants, the SPSA-ZmSPS transformants showed a thicker and more erect stem. Cellulose is the major structural polymer in the plant stems, and unlike starch, is an irreversible carbon sink. As Suc is being transported in the vasculature, some of it is cleaved by SucS to produce fructose and UDP-Glc—the latter acting as a substrate for cellulose synthesis (Haigler et al. 2001). Fructose, however, is inhibitory for SucS activity and using Fruc as a substrate, SPS alleviates the inhibitory effect of Fruc on SucS activity. SPS activity, thus, has a dual role—contributing to the pathway leading to the synthesis of UDP-Glc for cellulose deposition and for maintaining a constant supply of substrate by recycling the products from the initial photosynthate. Increased SPS activity in the stem, as seen in the SPSA-ZmSPS transformants, produces higher cellulose levels while maintaining the Suc levels needed for transport into the nodules. Thus, the greater stem thickness in the SPSA-ZmSPS transformants can be an attribute of higher SPS activity in the stem. The SPSA-antisense transformants showed a thinner stem and narrower leaves (Fig. 9), further supporting our premise that SPS has a role in the synthesis of cellulose in the stem. Using the same antisense RNA technology, Tian et al. (2010) showed that downregulating SPS in muskmelon resulted in the plants having smaller leaves, a thinner stem, and an overall smaller plant size. Tobacco plants overexpressing SPS showed an increase in the internode length, stem diameter, length of fibers, and total dry weight relative to control plants (Park et al. 2008). Transgenic poplar trees transformed with an Arabidopsis SPS gene showed an increased xylem fiber length but did not show an increase in growth (Park et al. 2009). Transgenic cotton over-producing SPS showed improved fiber quality (Haigler et al. 2007).
The SPSB-ZmSPS transformants flowered earlier than the SPSA-ZmSPS transformants and this can be attributed to the higher level of Suc in the aerial parts of the SPSB-ZmSPS transformants compared to the SPSA-ZmSPS transformants. Moreover, flowering time can also be related to plant maturity and the two classes of transformants showed much higher rates of growth when compared to control plants. In keeping with this trend, the SPSA-antisense transformants showed delayed flowering compared to control plants. Sucrose and/or its derivative trehalose 6-phosphate (T6P) have been implicated to be the trigger for flowering (Wahl et al. 2013). The nodule number and weight per plant in each of the two classes, SPSA-ZmSPS and SPSB-ZmSPS, exceeded that in controls while the SPSA-AntiSPSA transformants showed no change. The nodule number was highest in the SPSB-ZmSPS transformants, probably an attribute of higher Suc transported from the leaves into the roots during nodule initiation. Suc has been shown to have a role in nodule initiation (Liu et al. 2015).
To summarize, through the functional analysis of the SPS genes by using the promoter-ZmSPS gene constructs, we have demonstrated that the two SPS gene families SPSA and SPSB are transcriptionally regulated and exhibit an organ-specific expression pattern. This study has also established that there is regulation at the level of enzyme activity. Furthermore, we have shown that while SPSB in alfalfa plays a role in the synthesis of Suc from the photosynthate and mobilization of starch in the leaves, SPSA has direct involvement in the carbohydrate regulatory cycles in which Suc and/or starch are degraded and Suc is resynthesized from one or more products. We have also shown that both SPSB-ZmSPS and SPSA-ZmSPS transformants exhibit increased growth and we ascribe it to the increased Suc levels in the nodules. Further studies involving detailed carbohydrate analysis of the nodules and stem need to be performed to support our proposed assignment of function to the two isoforms of SPS.
Author contribution statement
SP, MMH and FMV carried out the experiments. JLO supervised the lab research. CSG conceived the idea, interpreted the data and wrote the MS.
Abbreviations
- MsSPSA :
-
Alfalfa sucrose phosphate synthase A gene
- MsSPSB :
-
Alfalfa sucrose phosphate synthase B gene
- SPSA-ZmSPS :
-
SPSA promoter driving Zea mays SPS cDNA
- SPSB-ZmSPS :
-
SPSB promoter driving Zea mays SPS cDNA
- SPSA-antiSPSA :
-
SPSA promoter driving alfalfa SPSA coding region in antisense orientation
- SPSA-GUS :
-
SPSA promoter driving β-glucuronidase
- Suc:
-
Sucrose
- Glc:
-
Glucose
- Fru-6P:
-
Fructose 6-phosphate
- UDP-Glc:
-
Uridine diphosphate glucose
- SucS:
-
Sucrose synthase
References
Aleman L, Ortega JL, Martinez-Grimes M, Seger M, Holguin FO, Uribe DJ, Sengupta-Gopalan C (2010) Nodule-enhanced expression of a sucrose phosphate synthase gene member (MsSPSA) has a role in carbon and nitrogen metabolism in the nodules of alfalfa (Medicago sativa L.). Planta 231:233–244
Babb VM, Haigler CH (2001) Sucrose phosphate synthase activity rises in correlation with high-rate cellulose synthesis in three heterotrophic systems. Plant Physiol 127:1234–1242
Bahaji A, Baroja-Fernandez E, Ricarte-Bermejo A, Sanchez-Lopez AM, Munoz FJ, Romero JM, Ruiz MT, Baslam M, Almagro G, Sesma MT, Pozueta-Romero J (2015) Characterization of multiple SPS knockout mutants reveals redundant functions of the four Arabidopsis sucrose phosphate synthase isoforms in plant viability, and strongly indicates that enhanced respiration and accelerated starch turnover can alleviate the blockage of sucrose biosynthesis. Plant Sci 238:135–147
Bihmidine S, Hunter CT III, Johns CE, Koch KE, Braun DM (2013) Regulation of assimilate import into sink organs: update on molecular drivers of sink strength. Front Plant Sci 4:177
Braun DM, Wang L, Ruan Y-L (2014) Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signaling to enhance crop yield and food security. J Exp Bot 65:1713–1735
Castleden CK, Aoki N, Gillespie VJ, MacRae EA, Quick WP, Buchner P, Foyer CH, Furbank RT, Lunn JE (2004) Evolution and function of the sucrose-phosphate synthase gene families in wheat and other grasses. Plant Physiol 135:1753–1764
Chavez-Barcenas AT, Valdez-Alarcon JJ, Martinez-Trujillo M, Chen L, Xoconostle-Cazares B, Lucas JW, Herrera-Estrella L (2000) Tissue-specific and developmental pattern of expression of the rice sps1 gene. Plant Physiol 124:641–653
Chen S, Hajirezaei M, Bornke F (2005) Differential expression of sucrose phosphate synthase isoenzymes in tobacco reflects their functional specialization during dark-governed starch mobilization in source leaves. Plant Physiol 139:1163–1174
Choudhury SR, Roy S, Singh SK, Sengupta DN (2010) Understanding the molecular mechanism of transcriptional regulation of banana sucrose phosphate synthase (SPS) gene during fruit ripening: an insight into the functions of various cis-acting regulatory elements. Plant Signal Behav 5:553–557
Fung RWM, Langenkamper G, Gardner RC, MacRae E (2002) Differential expression within an SPS gene family. Plant Sci 164:459–470
Gebril S, Seger M, Villanueva FM, Ortega JL, Bagga S, Sengupta-Gopalan C (2015) Transgenic alfalfa (Medicago sativa) with increased sucrose phosphate synthase activity shows enhanced growth when grown under N2-fixing conditions. Planta 242:1009–1024
Geigenberger P, Stitt M (1991) A “futile” cycle of sucrose synthesis and degradation is involved in regulating partitioning between sucrose, starch, and respiration in cotyledons of germinating Ricinus communis L. seedlings when phloem transport is inhibited. Planta 185:81–90
Geigenberger P, Reimholz R, Geiger M, Merlo L, Canale V, Stitt M (1997) Regulation of sucrose and starch metabolism in potato tubers in response to short-term water deficit. Planta 201:502–518
Graham PH, Vance CP (2003) Legumes: importance and constraints to greater use. Plant Physiol 131:872–877
Haigler CH, Ivanova-Datcheva M, Hogan PS, Salnikov VV, Hwang S, Martin K, Delmer DP (2001) Carbon partitioning to cellulose synthesis. Plant Mol Biol 47:29–51
Haigler CH, Singh B, Zhang D, Hwang S, Wu C, Cai W, Hequent EF (2007) Transgenic cotton over-producing spinach SPS showed enhanced leaf sucrose synthesis and improved fiber quality under controlled environmental conditions. Plant Mol Biol 63:815–832
Hirose T, Hashida Y, Aoki N, Okamura M, Yonekura M, Ohto C, Terao T, Ohsugi R (2014) Analysis of gene-disruption mutants of a sucrose phosphate synthase gene in rice, OsSPS1, shows the importance of sucrose synthesis in pollen germination. Plant Sci 225:102–106
Huber SC (2007) Exploring the role of protein phosphorylation in plants: from signaling to metabolism. Biochem Soc Transact 35:28–32
Huber SC, Huber JL (1996) Role and regulation of sucrose phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47:431–444
Im K-H (2004) Expression of sucrose phosphate synthase (SPS) in non-photosynthetic tissues of maize. Mol Cells 17:404–409
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Kaur H, Peel A, Acosta K, Gebril S, Ortega JL, Sengupta-Gopalan C (2019) Comparison of alfalfa plants overexpressing glutamine synthetase with those overexpressing sucrose phosphate synthase demonstrates a signaling mechanism integrating carbon and nitrogen metabolism between the leaves and nodules. Plant Direct 1:e00115
Komatsu A, Moriguchi T, Koyama K, Omura M, Akiham T (2002) Analysis of sucrose synthase genes in citrus suggests different roles and phylogenetic relationships. J Exp Bot 53:61–71
Langenkamper G, Fung RWM, Newcomb RD, Atkinson RG, Gardner RC, MacRae EA (2002) Sucrose phosphate synthase genes in plants belong to three different families. J Mol Evol 54:322–332
Liu W, Han X, Zhan G, Zhao Z, Feng Y, Wu C (2015) A novel sucrose regulatory MADS-Box transcription factor GmNMHC5 promotes root development and nodulation in soybean (Glycine max [L.] Merr.). Inter J Mol Sci 16:20657–20673
Lunn JE, MacRae E (2003) New complexities in the synthesis of sucrose. Curr Opin Plant Biol 6:208–214
Lutfiyya LL, Xu N, D’Ordine N, Morrell RL, Miller JA, Duff SMG (2007) Phylogenetic and expression analysis of sucrose phosphate synthase isoenzymes in plants. J Plant Physiol 164:923–993
Mohmed SG (2014) Determining the role of sucrose phosphate synthase in C/N metabolism in plants using genetic engineering tools. Ph. D. Dissertation, New Mexico State University
Nguyen-Quoc B, Foyer CH (2001) A role for ‘futile cycles’ involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. J Exp Bot 52:881–889
Nguyen-Quoc B, N’Tchobo H, Foyer CH, Yelle S (1999) Overexpression of sucrose phosphate synthase increases sucrose unloading in transformed tomato fruit. J Exp Bot 50:785–791
Okamura M, Aoki N, Hirose T, Yonekura M, Ohto C, Ohsugi R (2011) Tissue specificity and diurnal change in gene expression of the sucrose phosphate synthase gene family in rice. Plant Sci 181:159–166
Oldroyd GED, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45:119–144
Park JY, Canam T, Kang KY, Ellis DD, Mansfield SD (2008) Over-expression of an Arabidopsis family A sucrose phosphate synthase (SPS) gene alters plant growth and fiber development. Transgenic Res 17:181–192
Park JY, Canam T, Kang KY, Unda F, Mansfield SD (2009) Sucrose phosphate synthase expression influences poplar phenology. Tree Physiol 29:937–946
Privat I, Foucrier S, Prins A, Epalle T, Eychenne M, Kandalaft L, Caillet V, Lin C, Tanksley S, Foyer C, McCarthy J (2008) Differential regulation of grain sucrose accumulation and metabolism in Coffea arabica (Arabica) and Coffea canephora (Robusta) revealed through gene expression and enzyme activity analysis. New Phytol 178:781–797
Reimholz R, Geiger M, Haake V, Deiting U, Krause KP, Sonnewald U, Stitt M (1997) Potato plants contain multiple forms of sucrose phosphate synthase, which differ in their tissue distributions, their levels during development, and their response to low temperature. Plant Cell Environ 20:291–305
Roy Choudhury S, Roy S, Das R, Sengupta D (2008) Differential transcriptional regulation of banana sucrose phosphate synthase gene in response to ethylene, auxin, wounding, low temperature, and different photoperiods during fruit ripening and functional analysis of banana gene promoter. Planta 229:207–223
Ruan YL (2012) Signaling role of sucrose metabolism in development. Mol Plant 5:763–765
Seger M, Gebril S, Tabilona J, Peel A, Sengupta-Gopalan C (2015) Impact of concurrent overexpression of cytosolic glutamine synthetase (GS1) and sucrose phosphate synthase (SPS) on growth and development in transgenic tobacco. Planta 241:69–81
Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30:1126–1149
Stein O, Granot D (2019) An overview of sucrose synthases in plants. Front Plant Sci 10:95
Stitt M, Wilke I, Feil R, Heldt HW (1988) Coarse control of sucrose-phosphate synthase in leaves: alterations of the kinetic-properties in response to the rate of photosynthesis and the accumulation of sucrose. Planta 174:217–230
Stitt M, Lunn J, Usadel B (2010) Arabidopsis and primary photosynthetic metabolism–more than the icing on the cake. Plant J 61:1067–1091
Sun J, Zhang J, Larue CT, Huber SC (2011) Decrease in leaf sucrose synthesis leads to increased starch turnover and decreased RuBP regeneration-limited photosynthesis but not Rubisco-limited photosynthesis in Arabidopsis null mutants of SPSA1. Plant Cell Environ 34:592–604
Tian H, Ma L, Zhao C, Hao H, Gong B, Yu X, Wang X (2010) Antisense repression of sucrose phosphate synthase in transgenic muskmelon alters plant growth and fruit development. Biochem Biophys Res Comm 393:365–370
Verma AK, Upadhyay SK, Verma PC, Solomom S, Singh SB (2011) Functional analysis of sucrose phosphate synthase (SPS) and sucrose synthase (SS) in sugarcane (Saccharum) cultivars. Plant Biol 13:325–332
Volkert K, Debast S, Voll LM, Voll H, Schiebl I, Hofmann J, Schneider S, Bornke F (2014) Loss of the two major leaf isoforms of sucrose phosphate synthase in Arabidopsis thaliana limits sucrose synthesis and nocturnal starch degradation but does not alter carbon partitioning during photosynthesis. J Exp Bot 65:5217–5229
Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M (2013) Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339:704–707
Wind J, Smeekens S, Hanson J (2010) Sucrose: metabolite and signaling molecule. Phytochemistry 71:1610–1614
Winter H, Huber SC (2000) Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit Rev Biochem Mol Biol 35:253–289
Worrell AC, Bruneau JM, Summerfelt K, Boesig M, Voelker TA (1991) Expression of a maize sucrose phosphate synthase in tomato alters leaf carbohydrate partitioning. The Plant Cell 3:1121–1130
Yonekura M, Aoki N, Hirose T, Onai K, Ishiura M, Okamura M, Ohsugi R, Ohto C (2013) The promoter activities of sucrose phosphate synthase genes in rice, OsSPS1 and OsSPS11, are controlled by the light and circadian clock, but not by sucrose. Front Plant Sci 4:31
Acknowledgements
This work was supported by the Agricultural Experiment Station at New Mexico State University and by the National Institutes of Health (Research Initiative for Scientific Research Enhancement program). The support from LIFTED, LED grow lights (Rio Rancho, NM) is greatly appreciated.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Padhi, S., Grimes, M.M., Muro-Villanueva, F. et al. Distinct nodule and leaf functions of two different sucrose phosphate synthases in alfalfa. Planta 250, 1743–1755 (2019). https://doi.org/10.1007/s00425-019-03261-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-019-03261-9