Abstract
Benzalacetone synthase (BAS) is a member of the plant-specific type III PKS superfamily that catalyzes a one-step decarboxylative condensation of 4-coumaroyl-CoA with malonyl-CoA to produce p-hydroxybenzalacetone. In our recent work (Ma et al. in Planta 229(3):457–469, 2008), a three-intron type III PKS gene (PcPKS2) was isolated from Polygonum cuspidatum Sieb. et Zucc. Phylogenetic and functional analyses revealed this recombinant PcPKS2 to be a BAS. In this study, another three-intron type III PKS gene (PcPKS1) and its corresponding cDNA were isolated from P. cuspidatum. Sequence and phylogenetic analyses demonstrated that PcPKS1 is a chalcone sythase (CHS). However, functional and enzymatic analyses showed that recombinant PcPKS1 is a bifunctional enzyme with both, CHS and BAS activity. DNA gel blot analysis indicated that there are two to four CHS copies in the P. cuspidatum genome. RNA gel blot analysis revealed that PcPKS1 is highly expressed in the rhizomes and in young leaves, but not in the roots of the plant. PcPKS1 transcripts in leaves were inducible by pathogen infection and wounding. BAS is thought to play a crucial role in the construction of the C6–C4 moiety found in a variety of phenylbutanoids, yet so far phenylbutanoids have not been isolated from P. cuspidatum. However, since PcPKS1 and PcPKS2 (Ma et al. in Planta 229(3):457–469, 2008) have been identified in P. cuspidatum, it is possible that such compounds are also produced in that plant, albeit in low concentrations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polygonum cuspidatum (Japanese knotweed, Polygonaceae) is a medicinal plant rich in aromatic polyketides (Yi et al. 2007). Biosynthesis of these polyketides in P. cuspidatum is poorly understood, although type III polyketide synthases (PKSs) are thought to play a crucial role for construction of the basic skeleton of a variety of the aromatic polyketides. So far 14 plant-specific type III PKS enzymes have been identified. The most well-studied type III PKS is chalcone sythase (CHS), which provides the first committed step in flavonoid biosynthesis (Fig. 1; Schröder 1997). Benzalacetone synthase (BAS) is another member of the type III PKS superfamily that catalyzes a one-step decarboxylative condensation of 4-coumaroyl-CoA with malonyl-CoA to produce p-hydroxybenzalacetone (Fig. 1; Borejsza-Wysocki and Hrazdina 1996). BAS is thought to play a crucial role in the construction of the C6–C4 moiety found in a variety of medicinally important phenylbutanoids (Abe et al. 2001).
The reactions for the conversion of 4-coumaroyl-CoA and malonyl-CoA to p-hydroxybenzalacetone by the benzalacetone synthase (BAS), and to naringenin chalcone by chalcone synthase (CHS). Polyketide pyrones, bis-noryangonin (BNY), and 4-coumaroyltriacetic acid lactone (CTAL) are derailment side products of the type III PKS reactions in vitro when the reaction mixtures are acidified before extraction
A conserved gene structure is a common feature of the type III PKS genes found in flowering plants. Besides an early reported Antirrhinum majus CHS which has two introns (Sommer and Saedler 1986), all type III PKS genes contain a single intron at a conserved site (Schröder 1997; Durbin et al. 2000). Interestingly, two three-intron type III PKS genes, PcPKS1 and PcPKS2 (Ma et al. 2008), were isolated from P. cuspidatum, and the recombinant PcPKS2 was found to be a BAS. Interestingly, the catalytic efficiency (K cat/K m) of BAS activity of PcPKS1 was 70-fold higher than that of the PcPKS2 (Ma et al. 2008).
The results of a number of studies have shown that production of p-hydroxybenzalacetone is not an early released side product of CHS but that this compound is the specific product of BAS (Abe et al. 2001, 2003; Borejsza-Wysocki and Hrazdina 1994, 1996). CHSs have been cloned and sequenced from more than 40 plant species. Most of them have been expressed functionally in Escherichia coli and their catalytic properties have been investigated. However, almost all the CHSs do not have BAS activity in vitro under the standard assay conditions (without reducing agent). In this work a further type III PKS gene was isolated and characterized by functional and phylogenetic analyses. Since so far phenylbutanoids and their derivatives have not been isolated from P. cuspidatum, but the responsive genes have been identified, possible phenylbutanoid products have been studied at various stages of the plants development.
Materials and methods
Plant material and chemicals
Polygonum cuspidatum Sieb. and Zucc. plants were maintained in the medicinal plant garden and in a greenhouse at the Institute of Botany, The Chinese Academy of Sciences, Beijing, China. Details of culture and chemicals used were as previously described (Ma et al. 2008).
PCR-based cloning
Genomic DNA was isolated from young leaves using a modified CTAB method. The core DNA fragment was amplified using degenerate primers (Fig. 2; Ma et al. 2008). To obtain the full-length gene, TAIL-PCR was performed using six gene-specific primers, designed according to the terminal sequences of the core fragment, and performed as described (Liu et al. 1995).
Sequence organization of the PcPKS1 gene. Putative regulatory motifs are shown within the promoter region of the gene. Blue arrows indicate the primer sites used in this study for cloning a core fragment of the PcPKS1. The red arrow indicates the position of the insertion site of the second intron in the Antirrhinum majus CHS gene. The 5′ and 3′ splice sites of introns are boxed. The active site residues conserved in the type III PKSs (Cys164, His303, M. sativa CHS2 numbering) are marked with #
The ORF of the cDNA was produced using N-terminal and C-terminal PCR primers that were designed on the basis of the full-lengh PcPKS1: sense, the NdeI site is underlined 5′-TATACATATGGCACCATCGGTCCAGGAGATC-3′ and antisense, the SalI site is underlined 5′-TATAGTCGACGTGATGAGCAACTGGTACACTGTG-3′. The amplified DNA was digested with NdeI/SalI, and cloned into the NdeI/SalI site of pET-30b (+) (Novagen, Darmstadt, Germany). The recombinant enzyme contained a hexahistidine tag at the C terminus.
Heterologous expression in Escherichia coli and recombinant enzyme purification
Heterologous expression and purification of recombinant PcPKS1 were performed according to the method as described (Ma et al. 2008).
Enzyme reaction and product analysis
The standard assay (250 μl) contained 25 μM starter CoA, 65 μM malonyl-CoA, 0.1 M potassium phosphate (pH 7.0), and 2.0 μg of protein, and was incubated at 30°C for 60 min, then extracted twice with 250 μl of ethyl acetate and centrifuged at 10,000g for 10 min. Acetic acid (5% final concentration) was added before the extraction step to detect any side product. After drying under vacuum, the residue was dissolved in 50 μl of 50% (v/v) methanol. The product analysis of recombinant PcPKS1 were as described (Ma et al. 2008).
Enzyme kinetics
Kinetic constants were determined according to the method as described (Ma et al. 2008).
Enzyme native molecular mass determination
The relative molecular mass of PcPKS1 was determined on a calibrated gel filtration column as described (Liu et al. 2003).
DNA and RNA gel blot analysis
Ten microgram genomic DNA was digested with restriction enzymes EcoRI, EcoRV, and HindIII. Hybridization was performed using an [α-32P]dCTP-labeled PcPKS1 fragment (364 bp) including both the coding and promoter regions as probe. The primers used to amplify the probe were: forward, 5′-GTACACAACAGAGAGATAACTGTC-3′; and reverse, 5′-GGTCATGTGC TCGCTGTTGGTG-3′.
RNA gel blot analysis of PcPKS1 was performed according to the method as described (Ma et al. 2008). Hybridization was performed using an [α-32P]dCTP-labeled PcPKS1 fragment (295 bp) including both the coding and 3′ untranslated regions as probe. The primers used to amplify the probe were: forward, 5′-GATGAGATGAGGAAGAAGTCG-3′; and reverse, 5′-CCGTATATCACATGCTAGCTAGC-3′.
Phylogenetic tree construction
A neighbor-joining tree was generated as described (Ma et al. 2008).
Results
Isolation of the PcPKS1 gene and cDNA
Using primers Pc1 and Pc2 (Fig. 2; Ma et al. 2008), a 715 bp core fragment was cloned and its deduced amino acid sequence exhibited over 90% identity with CHSs of other plants in the Polygonaceae family. Interestingly, the Pc1/Pc2 PCR fragment contains an additional intron in the 3′ region of the gene sequence (Fig. 2). Flanking sequences were obtained by thermal asymmetric interlaced (TAIL)-PCR (Liu et al. 1995), and the resulting gene was designated as PcPKS1 (Accession no. EF090266; Fig. 2). The cDNA (Accession no. EF090604) of PcPKS1 was amplified with gene specific primers for full-length ORF and the reverse transcription products as template.
Sequence analysis
The full-length PcPKS1 is 2,424 bp, contains a promoter region of 720 bp, 1,182 bp of coding region, and 209 bp of 3′ untranslated region. Interestingly, the coding region of the PcPKS1 is interrupted by three intervening sequences of 106, 103, and 104 bp (Fig. 2). In comparison with PcPKS2 (Ma et al. 2008), the insertion sites of the three introns are completely identical in PcPKS1 (Fig. 2). No significant homologies were found between the three introns sequences of PcPKS1 and three introns sequences of PcPKS2 (Ma et al. 2008). Sequences of the introns 1, 2, and 3 of PcPKS1 revealed 22, 23, and 49% identity with the corresponding three intron sequences of PcPKS2. PcPKS1 intron 1 and intron 2 conform to the GT-AG rule; while the 5′ splice site and the 3′ splice site of the intron 3 follow a rare GC-AG rule (Brown et al. 1996; Buchanan et al. 2002; Fig. 2). Various conserved regulatory motifs were identified within the promoter region (Fig. 2).
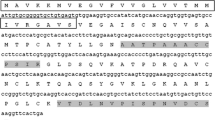
The PcPKS1 cDNA is 1,182 bp in length and encodes a deduced protein of 393 amino acid residues with a predicted molecular mass of 43.2 kDa and a pI of 5.88 (data not shown). The deduced amino acid sequence showed 57–97% identity with those of other type III PKSs of plant origin (Fig. 3): 97% identity with Rheum palmatum CHS2, 83% identity with A. majus CHS (Sommer and Saedler 1986), 80% identity with M. sativa CHS2 (Ferrer et al. 1999), 72% identity with R. idaeus RiPKS4 (Zheng and Hrazdina 2008), 72% identity with R. palmatum BAS (Abe et al. 2001), 70% identity with P. cuspidatum PcPKS2 (Ma et al. 2008) and 57% identity with Aloe arborescens octaketide synthase (OKS) (Abe et al. 2005).
Comparison of the primary sequences of P. cuspidatum PcPKS1 and other plant-specific type III PKSs. Multiple sequence alignment was calculated with the DNAMAN package. Black shading shows amino acid identities. The active site residues conserved in the type III PKSs (Cys164, His303, and Asn336, M. sativa CHS2 numbering) are marked with #, and residues for CoA binding are marked with +. Amino acid residues are conserved in PcPKS1, R. palmatum BAS and PcPKS2 (Ma et al. 2008), but are absent from other CHSs (marked with filled inverted triangle). Amino acid residues that are conserved in the CHS enzymes but do not appear in P. cuspidatum PcPKS1 are marked with filled triangle. The abbreviations for species and accession numbers are: PcPKS1 (Polygonum cuspidatum ABK92282), MsCHS2 (Medicago sativa P30074), AmCHS (Antirrhinum majus BAE80511), RiPKS4 (Rubus idaeus ABV54602), RpBAS (Rheum palmatum AAK82824), and PcPKS2 (Polygonum cuspidatum ABY47640)
Polygonum cuspidatum PcPKS1 contains the catalytic triad of Cys164, His303, and Asn336, and all identical CoA binding sites (numbering of M. sativa CHS2) (Fig. 3). Furthermore, most of the active site residues, including Val98, Thr132, Ser133, Met137, Gly163, Thr194, Gly211, Gly216, Ile254, Ser338, and Pro375, along with the CHS “gatekeeper” Phe215 and Phe265 (Austin and Noel 2003), are well conserved in PcPKS1. However, the CHS active site residues Val196 and Thr197 are uniquely replaced with Ile and Cys in PcPKS1, respectively (Fig. 3). In addition, 18 amino acid residues are conserved in PcPKS1, R. palmatum BAS (Abe et al. 2001) and PcPKS2 (Ma et al. 2008), but are different from other CHSs, and ten amino acid residues that are conserved in the CHS enzymes but do not appear in P. cuspidatum PcPKS1 as indicated in Fig. 3.
Phylogenetic analysis
The phylogenetic tree showed that PcPKS1 is located together with R. palmatum (Polygonaceae) CHS2 in a cluster containing other typical chalcone-forming enzymes (Fig. 4). For PcPKS2 (Ma et al. 2008), the phylogenetic analysis showed that it grouped together with R. palmatum (Polygonaceae) BAS (Abe et al. 2001) in a cluster containing all functionally divergent plant-specific type III PKSs (Fig. 4).
Neighbor-joining tree of type III PKSs. Numbers at the forks are bootstrap values from 100 replicates. Four bacterial type III PKSs were used to root the tree. The accession numbers used in the analysis were as previously described (Ma et al. 2008)
Characterization of PcPKS1
Polygonum cuspidatum PcPKS1 was heterologously expressed in E. coli with an additional hexahistidine tag at the C terminus. The purified enzyme gave a single band with a molecular mass of 43 kDa on SDS-PAGE (Fig. 5). On a calibrated gel-filtration column, the native molecular mass of the enzyme was about 90 kDa, indicating a homodimeric structure of PcPKS1.
SDS-PAGE analysis of recombinant PcPKS1 stained with Coomassie brilliant blue. Lane 1, molecular mass markers with masses indicated in kDa. Lane 2, total protein from uninduced E. coli strain BL21-Rosetta (DE3) harboring the His-tagged PcPKS1 construct. Lane 3, soluble extract from uninduced E. coli. strain BL21-Rosetta (DE3) harboring the His-tagged PcPKS1 construct. Lane 4, total protein from BL21-Rosetta (DE3) cells harboring the His-tagged PcPKS1 construct after induction with IPTG for 6 h at 30°C. Lane 5, soluble extract from BL21-Rosetta (DE3) cells harboring the His-tagged PcPKS1 construct after induction with IPTG for 6 h at 30°C. Lane 6, recombinant PcPKS1 after Ni2+-chelating chromatography. Lane 7, recombinant PcPKS1 (7.5 μg) after passage through a PD-10 column
Identification and quantification of the enzymatic products showed that recombinant PcPKS1 efficiently afforded naringenin as a single product at pH 7.0 (Fig. 6a). It effectively yielded p-hydroxybenzalacetone as almost the sole product at pH 9.0 (Fig. 6c). Both products are observed at pH 8.0, with a preference for naringenin formation (Fig. 6b). The enzymatic products were identified by HPLC and LC-MS in comparison with authentic samples. Moreover, no traces of bis-noryangonin (BNY) and 4-coumaroyltriacetic acid lactone (CTAL) (Fig. 1) could be detected in the enzyme reaction mixture at different pH values (Fig. 7).
HPLC elution profiles of enzyme reaction products of the chalcone-forming activity and the p-hydroxybenzalacetone-forming activity of the PcPKS1 at pH 7.0 (a), pH 8.0 (b), and pH 9.0 (c). HPLC separation conditions are described in “Materials and methods”. Note that naringenin chalcone is converted by acid treatment to racemic naringenin through a nonstereospecific ring C closure
For the chalcone-forming activity, PcPKS1 exhibited an optimum within a range of pH 7.0–8.0 in either Tris–HCl or potassium phosphate buffer (Fig. 7). Besides 4-coumaroyl-CoA, its derivatives were accepted as starter substrates; however, the relative activities were lower (Table 1). In contrast to CHS, which showed broad substrate specificity toward aliphatic CoA esters, PcPKS1 did not accept isobutyryl-CoA, isovaleryl-CoA, or acetyl-CoA as a substrate (Table 1). For the chalcone-forming reaction, we determined the kinetic parameters for 4-coumaroyl-CoA (K m = 12.9 μM, K cat = 6.48 min−1, K cat/K m = 8,388 M−1 s−1) and malonyl-CoA (K m = 57.5 μM, K cat = 6.48 min−1, K cat/K m = 1,878 M−1 s−1).
For the p-hydroxybenzalacetone-forming activity, the pH optimum of PcPKS1 was 9.0 (Fig. 7). At this pH value, 4-coumaroyl-CoA and feruloyl-CoA were the only cinnamoyl-CoA derivatives accepted as starter substrates (Table 1). PcPKS1 did not accept isobutyryl-CoA, isovaleryl-CoA, or acetyl-CoA as a substrate (Table 1). For the p-hydroxybenzalacetone-forming reaction, we determined the kinetic parameters for 4-coumaroyl-CoA (K m = 30.5 μM, K cat = 8.64 min−1, K cat/K m = 4,721 M−1 s−1) and malonyl-CoA (K m = 131.7 μM, K cat = 8.64 min−1, K cat/K m = 1,094 M−1 s−1).
Gene organization of CHS in P. cuspidatum
Genomic DNA isolated from young leaves of P. cuspidatum was digested with EcoRI, EcoRV and HindIII. DNA gel blot analysis with a probe of 364 bp fragment (-211–153th nucleotide acid sequence) including both the coding and promoter regions of the PcPKS1 showed the presence of two to four bands differing in size (Fig. 8). Since there is no EcoRI, EcoRV, and HindIII restriction site in the PcPKS1 gene, the DNA gel blot results indicate that 2–4 copies of CHS exist in the genome of P. cuspidatum.
DNA gel blot analysis of PcPKS1. 10 μg genomic DNA of P. cuspidatum was digested with EcoRI, EcoRV and HindIII. Hybridizations were performed for 16 h at 65°C with an [α-32P] dCTP labeled PcPKS1 fragment (364 bp) including the coding and promoter regions as probe. Washing of the membrane and detection were as described in “Materials and methods”
Expression of PcPKS1
RNA gel analysis performed using a PcPKS1 fragment (nucleotides 1,348–1,643) including both the coding and 3′ untranslated regions as probe, in this region nucleotide homology with PcPKS2 (Ma et al. 2008) was only about 53%. RNA gel blot analysis revealed that PcPKS1 expressed differentially in various tissues (Fig. 9a) and in response to different types of environmental stimuli, such as treatment with a pathogen elicitor, or wounding (Fig. 9b, c).
RNA gel blot analysis of PcPKS1 tissue-specific expression (a) and induction (b, c). For tissue-specific expression (a), total RNA was prepared from young leaves (1), mature leaves (2), petioles (3), stems (4), rhizomes (5), and roots (6). For pathogen treatment (b), mature leaves were collected from the seedlings inoculated with Agrobacterium tumefaciens (EHA105) after 0, 2, 4, 8, 12, 24, and 48 h. For wounding (c), attached mature leaves were cut. Samples from all treatments were harvested after 0, 2, 4, 8, 12, 24, and 48 h. Hybridization was performed using an [α-32P]dCTP-labeled PcPKS1 fragment (295 bp) including both the coding and 3’ untranslated regions as probe. Washing of the membrane and detection were as described above for DNA gel blotting analysis
Discussion
It is a common phenomenon that type III PKSs produce some early released derailment side product such as BNY and CTAL (Fig. 1; Akiyama et al. 1999; Austin and Noel 2003). The results of a number of studies have proven that production of p-hydroxybenzalacetone is not an early released side product of CHS but instead it is a specific product of BAS (Fig. 1; Abe et al. 2001, 2003; Borejsza-Wysocki and Hrazdina 1994, 1996). In this study, we described the enzymatic properties of a bifunctional PKS that possesses both CHS and BAS activities. The following evidence suggests that the formation of p-hydroxybenzalacetone is an intrinsic property of recombinant PcPKS1: (i) for the enzymatic property, the pH optimum of the p-hydroxybenzalacetone-forming activity of PcPKS1 is 9.0 (Fig. 7), which is similar to R. palmatum BAS (8.0–8.8) (Abe et al. 2001) and identical with PcPKS2 (Ma et al. 2008), and PcPKS1 effectively yielded p-hydroxybenzalacetone as almost the sole product at this pH optimum (Fig. 6c); (ii) for catalytic capability, PcPKS1 exhibited the highest level of activity for p-hydroxybenzalacetone production. The catalytic efficiency (K cat/K m) of BAS activity of PcPKS1 is 4,721 M−1 s−1 which 1.5- and 70-fold higher than that of the R. palmatum BAS (Abe et al. 2001) and PcPKS2 (Ma et al. 2008) at pH 9.0; (iii) for substrate specificity, at the pH 9.0, like the PcPKS2 (Ma et al. 2008), 4-coumaroyl-CoA and feruloyl-CoA were the only cinnamoyl-CoA derivatives accepted as starter substrates (Table 1). In contrast to typical CHS (Morita et al. 2000; Abe et al. 2002), such as the R. palmatum BAS (Abe et al. 2001) and PcPKS2 (Ma et al. 2008), PcPKS1 did not accept isobutyryl-CoA, isovaleryl-CoA, or acetyl-CoA as a substrate at different pH values (Table 1).
To date, two PKSs, RiCHS (Beekwilder et al. 2007) and RiPKS4 (Zheng and Hrazdina 2008) were described as bifunctional PKSs exhibiting both CHS and BAS activity. BAS activity of RiCHS was found in the engineered E. coli and yeast cells, there was no exact sequence information for RiCHS (Beekwilder et al. 2007). Zheng and Hrazdina (2008) postulate that BAS activity of RiPKS4 was created by the sequence variation in the C terminus due to DNA recombination at the 3′ region of its coding sequence (Fig. 3). However, this speculation could not be proven by the PcPKS1 sequence, no notable similarity was found in the 3′ region between PcPKS1 and RiPKS4 (Fig. 3). In R. palmatum BAS, one of the characteristic features is that the active-site Phe215 is uniquely replaced by Leu. This exchange was essential for R. palmatum BAS activity as confirmed by mutagenesis, it interrupted polyketide chain elongation at the diketide stage (Abe et al. 2003). Interestingly, sequence analysis showed that Phe215 and Phe265 are uniquely replaced by Leu and Cys in PcPKS2 (Fig. 3; Ma et al. 2008). This mechanism does not apply to PcPKS1 as well because F215 in PcPKS1 is conserved (Fig. 3). It is hard to explain why recombinant PcPKS1 possesses both CHS and BAS activities. Austin and Noel (2003) proposed that BAS utilizes a second active-site Cys for the decarboxylation reaction of a diketide intermediate to produce benzalacetone. We speculate that the PcPKS1 uses an alternative pocket to hold the coumaroyl moiety for the diketide formation. It is tempting to speculate that PcPKS1 may be an intermediate in the evolution from CHS to BAS, and it may have a peculiar subtle structure as compared with typical CHS. It is also possible is that the residues surrounding the F215 are responsible for multiple product formation. Detailed sequence analysis showed that 18 amino acid residues are conserved in PcPKS1, R. palmatum BAS (Abe et al. 2001) and PcPKS2 (Ma et al. 2008), but are different from other CHSs, and ten amino acid residues that are conserved in the CHS enzymes but do not appear in P. cuspidatum PcPKS1 (Fig. 3). These amino acids may be important for the bifunctional characteristics and pH dependence of PcPKS1. In order to test the function of these amino acids, site-directed mutagenesis experiments of the PcPKS1 are in progress in our laboratory. Furthermore, efforts of homology modeling failed to find an explanation for special characteristics of PcPKS1, but the good solubility of overexpressed PcPKS1 might allow to carry out protein crystallization (Fig. 5).
The presence of putative regulatory motifs such as ACE (Arias et al. 1993) and E-box (Hartmann et al. 2005) in the promoter region suggested that the expression of PcPKS1 may respond to different environmental stimuli and tissue-specific activation (Fig. 2). Indeed, PcPKS1 is highly expressed in the rhizomes and in young leaves, but not in the roots of the plant, and its transcripts in leaves were inducible by pathogen infection and wounding. Similar expression patterns of PcPKS1 (Fig. 9) and PcPKS2 (Ma et al. 2008) most likely suggested that PcPKS1 and PcPKS2 have similar functions in this plant. Phenylbutanoid and its derivatives have been found in several plants, such as anti-inflammatory glucoside lindleyin in R. palmatum, gingerol, and curcumin in ginger plants (Abe et al. 2001), and raspberry ketone, the characteristic aroma of the raspberry fruit (Borejsza-Wysocki and Hrazdina 1996). Besides several aromatic polyketides including anthraquinones, such as emodin, physcion and chrysophanol, and stilbenes, such as resveratrol, piceid and 2,3,5,4′-tetrahydroxy stilbene-2-O-d-glucoside (Xiao et al. 2002; Hegde et al. 2004; Yi et al. 2007), the phenylbutanoids have not been isolated from P. cuspidatum so far. However, the similar evolutionary history between P. cuspidatum and R. palmatum and the fact that PcPKS1 and PcPKS2 (Ma et al. 2008) were both isolated from P. cuspidatum may indicate that such compounds are produced, albeit in low concentrations. Therefore, further studies are needed to address the physiological role of PcPKS1 and PcPKS2 (Ma et al. 2008) and elucidate whether PcPKS1 is a true bifunctional enzyme with both CHS activity and BAS activity in P. cuspidatum.
In addition to PcPKS1 and PcPKS2, there are more type III PKS genes with 1–2 intron(s) in P. cuspidatum (Ma et al. 2008). It is still unclear whether multi-intron type III PKS genes exist in other members of the Polygonaceae. This phenomenon was found in bryophytes; among the 19 putative type III PKS genes in Physcomitrella patens, six contained no introns, four contained one intron, and others contained two introns (Jiang et al. 2006). Plant β-ketoacyl synthases (KS) are considered as ancestors of type III PKSs. Most of 3-ketoacyl-CoA synthase (KAS) genes do not have introns in the coding region (James et al. 1995; Todd et al. 1999; Kajikawa et al. 2003a; Kajikawa et al. 2003b; Blacklock and Jaworski 2006). There are two “intron evolutionary” models concerning the original form of genes that today are interrupted. The “introns early” model supposes that introns have always been an integral part of the gene, and genes without introns have lost them in the course of evolution. The “introns late” model supposes that the ancestral protein-coding units consisted of uninterrupted sequences of DNA, and that introns were subsequently inserted into them (Lewin 2000). The fact that some type III PKS gene do not have introns in bryophytes and most of the KAS genes do not have introns in the coding region seems to support the “introns late” hypothesis. But the example of the evolution of terpene synthases seems to support the “introns early” model (Trapp and Croteau 2001). Clues obtained from studies of bifunctional enzyme activity combined with the unusual gene structure of P. cuspidatum PcPKS1, provide an interesting example for the molecular evolution and structure-function studies on type III PKS enzymes.
Abbreviations
- ALS:
-
Aloesone synthase
- BAS:
-
Benzalacetone synthase
- CHS:
-
Chalcone synthase
- CoA:
-
Coenzyme A
- LC-ESIMS:
-
Liquid chromatography-electron spray ionization mass spectrometry
- OKS:
-
Octaketide synthase
- PKS:
-
Polyketide synthase
- STS:
-
Stilbene synthase
References
Abe I, Takahashi Y, Morita H, Noguchi H (2001) Benzalacetone synthase. A novel polyketide synthase that plays a crucial role in the biosynthesis of phenylbutanones in Rheum palmatum. Eur J Biochem 268:3354–3359
Abe I, Takahashi Y, Noguchi H (2002) Enzymatic formation of an unnatural C(6)-C(5) aromatic polyketide by plant type III polyketide synthases. Org Lett 4:3623–3626
Abe I, Sano Y, Takahashi Y, Noguchi H (2003) Site-directed mutagenesis of benzalacetone synthase. The role of the Phe215 in plant type III polyketide synthases. J Biol Chem 278:25218–25226
Abe I, Oguro S, Utsumi Y, Sano Y, Noguchi H (2005) Engineered biosynthesis of plant polyketides: chain length control in an octaketide-producing plant type III polyketide synthase. J Am Chem Soc 127:12709–12716
Akiyama T, Shibuya M, Liu HM, Ebizuka Y (1999) p-Coumaroyltriacetic acid synthase, a new homologue of chalcone synthase, from Hydrangea macrophylla var. thunbergii. Eur J Biochem 263:834–839
Arias JA, Dixon RA, Lamb CJ (1993) Dissection of the functional architecture of a plant defense gene promoter using a homologous in vitro transcription initiation system. Plant Cell 5:485–496
Austin MB, Noel JP (2003) The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep 20:79–110
Beekwilder J, van der Meer IM, Sibbesen O, Broekgaarden M, Qvist I, Mikkelsen JD, Hall RD (2007) Microbial production of natural raspberry ketone. Biotechnol J 2:1270–1279
Blacklock BJ, Jaworski JG (2006) Substrate specificity of Arabidopsis 3-ketoacyl-CoA synthases. Biochem Biophys Res Commun 346:583–590
Borejsza-Wysocki W, Hrazdina G (1994) Biosynthesis of p-hydroxyphenylbutan-2-one in raspberry fruits and tissue culture. Phytochemistry 35:623–628
Borejsza-Wysocki W, Hrazdina G (1996) Aromatic polyketide synthases (purification, characterization, and antibody development to benzalacetone synthase from raspberry fruits). Plant Physiol 110:791–799
Brown JW, Smith P, Simpson CG (1996) Arabidopsis consensus intron sequences. Plant Mol Biol 32:531–535
Buchanan BB, Gruissem W, Jones RL (2002) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Waldorf
Durbin ML, McCaig B, Clegg MT (2000) Molecular evolution of the chalcone synthase multigene family in the morning glory genome. Plant Mol Biol 42:79–92
Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP (1999) Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Biol 6:775–784
Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B (2005) Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol 57:155–171
Hegde VR, Pu H, Patel M, Black T, Soriano A, Zhao W, Gullo VP, Chan TM (2004) Two new bacterial DNA primase inhibitors from the plant Polygonum cuspidatum. Bioorg Med Chem Lett 14:2275–2277
James DW Jr, Lim E, Keller J, Plooy I, Ralston E, Dooner HK (1995) Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon Activator. Plant Cell 7:309–319
Jiang C, Schommer CK, Kim SY, Suh DY (2006) Cloning and characterization of chalcone synthase from the moss, Physcomitrella patens. Phytochemistry 67:2531–2540
Kajikawa M, Yamaoka S, Yamato KT, Kanamaru H, Sakuradani E, Shimizu S, Fukuzawa H, Ohyama K (2003a) Functional analysis of a beta-ketoacyl-CoA synthase gene, MpFAE2, by gene silencing in the liverwort Marchantia polymorpha L. Biosci Biotechnol Biochem 67:605–612
Kajikawa M, Yamato KT, Kanamaru H, Sakuradani E, Shimizu S, Fukuzawa H, Sakai Y, Ohyama K (2003b) MpFAE3 a beta-ketoacyl-CoA synthase gene in the liverwort Marchantia polymorpha L., is preferentially involved in elongation of palmitic acid to stearic acid. Biosci Biotechnol Biochem 67:1667–1674
Lewin B (2000) Gene VII. Oxford University Press Inc., New York
Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8:457–463
Liu B, Falkenstein-Paul H, Schmidt W, Beerhues L (2003) Benzophenone synthase and chalcone synthase from Hypericum androsaemum cell cultures: cDNA cloning, functional expression, and site-directed mutagenesis of two polyketide synthases. Plant J 34:847–855
Ma LQ, Pang XB, Shen HY, Pu GB, Wang HH, Lei CY, Wang H, Li GF, Liu BY, Ye HC (2008) A novel type III polyketide synthase encoded by a three-intron gene from Polygonum cuspidatum. Planta 229(3):457–469
Morita H, Takahashi Y, Noguchi H, Abe I (2000) Enzymatic formation of unnatural aromatic polyketides by chalcone synthase. Biochem Biophys Res Commun 279:190–195
Schröder J (1997) A family of plant-specific polyketide synthases: facts and predictions. Trends Plant Sci 2:373–378
Sommer H, Saedler H (1986) Structure of the chalcone synthase gene of Antirrhinum majus. Mol Gen Genet 202:429–434
Todd J, Post-Beittenmiller D, Jaworski JG (1999) KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J 17:119–130
Trapp SC, Croteau RB (2001) Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158:811–832
Xiao K, Xuan L, Xu Y, Bai D, Zhong D (2002) Constituents from Polygonum cuspidatum. Chem Pharm Bull (Tokyo) 50:605–608
Yi T, Zhang H, Cai Z (2007) Analysis of rhizoma Polygoni cuspidati by HPLC and HPLC-ESI/MS. Phytochem Anal 18:387–392
Zheng D, Hrazdina G (2008) Molecular and biochemical characterization of benzalacetone synthase and chalcone synthase genes and their proteins from raspberry (Rubus idaeus L.). Arch Biochem Biophys 470:139–145
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Ma, LQ., Guo, YW., Gao, DY. et al. Identification of a Polygonum cuspidatum three-intron gene encoding a type III polyketide synthase producing both naringenin and p-hydroxybenzalacetone. Planta 229, 1077–1086 (2009). https://doi.org/10.1007/s00425-009-0899-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-009-0899-1