Abstract
Plant nonspecific lipid transfer proteins (nsLTPs) are widely distributed through plant kingdom and are characterized by the presence of a central hydrophobic cavity, suitable for binding various hydrophobic molecules. Despite extensive research on nsLTP in different plant species, mostly angiosperm, and the great diversity of physiological processes in which they seem to be involved, their exact functions still remain unclear. Also, very limited experimental data are available on nsLTP in gymnosperm. In this study, we report for the first time on the molecular cloning of nsLTP, from Pinus sylvestris L. (PsLTP1, GenBank accession JN980402.1) and the expression pattern of PsLTP1 during ontogenesis and in response to environmental stress conditions. Total RNA from roots of 7-day old pine seedlings was used to isolate the cDNA clone, corresponding to Scots pine lipid transfer protein. The open reading frame of PsLTP1 consists of 372 bp encoding a protein of 123 amino acids. Amino acid sequence alignment revealed that mature PsLTP1 shares high level of similarity with nsLTP from other conifers and with well-studied nsLTPs from angiosperms. The PsLTP1 contains a 27-amino-acid N-terminal signal sequence and presents all the features of a plant nsLTP. Amino acid comparison analysis and 3D structure prediction showed that PsLTP1 is a type 1 nsLTP. The results of the expression analysis of Scots pine PsLTP1 gene revealed that its transcripts accumulate in actively growing tissues. Furthermore, transcription of PsLTP1 was upregulated in response to cold and salt treatments, and downregulated during acidic, osmotic and water stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Conifers are one of the oldest seed plants that have survived into the present day and are thus called living fossils (Hensiruk 2002). Scots pine (Pinus sylvestris L.) is an important tree species in northern Eurasia. In Ukraine, Scots pine covers 23% of the total forest area, and in Polissya, that area increases to 64.5%, making this tree species very important for Ukrainian forestry (Hensiruk 2002). Scots pine is economically valuable for its soft timber and derivative products that are widely used for pharmaceutical purposes. Besides the great economic benefit, boreal forests with Scots pine provide high biodiversity and oxygen. Conifers, like other plant species, are constantly exposed to a great variety of environmental conditions at every stage of their life cycle, including biotic (such as pests and pathogenic microorganisms) and abiotic stresses (drought, salt, alkali, flood, high and low temperature, oxidative stress and heavy metal toxicity). All these factors modulate plant vitality, negatively affecting plant growth and development and thereby decreasing productivity. To decrease the negative effects, conifers developed a set of defense mechanisms that include structural barriers, antimicrobial chemicals, hypersensitive response and pathogenesis-related (PR) protein genes and their products with strong antibacterial and antifungal activities (Adomas et al. 2008; Veluthakkal and Dasgupta 2010). Studies from various laboratories have revealed that some PR-genes are expressed constitutively while the expression of others is triggered only under certain conditions, such as infection or exposure to abiotic stresses. In recent years, considerable interest has developed in using specific genes and nature-friendly products to reduce damaging effects of different environmental stimuli and to enhance stress tolerance in plants. Genes encoding pathogenesis-related (PR) proteins are attractive candidates for that role (Goyala and Mattoo 2014; Sels et al. 2008). The best-known products of PR-genes in plants are chitinases, peroxidases, defensins, thaumatins, thionins and lipid transfer proteins (Goyala and Mattoo 2014).
Plant nonspecific lipid transfer proteins (nsLTPs) belong to the multigene PR-14 family with antimicrobial and antifungal activity in vitro (García-Olmedo et al. 1995). Research on nsLTPs has focused on their structure, subcellular localization, classification, expression patterns and evolution. The first nsLTP was identified nearly 40 years ago, and since then, they have been isolated from more than 50 plant species, and that number is still growing (José-Estanyol et al. 2004; Kader 1996). NsLTPs are low molecular weight basic proteins, ranging in size from 6.5 to 10.5 kDa, and typically are synthesized from a precursor that has a signal sequence, varying from 21 to 27 amino acids. Their tertiary structure is characterized by a conservative eight-cysteine motif (8 CM), where cysteine residues form four disulfide bonds, necessary to stabilize the hydrophobic cavity. In vitro studies showed that the cavity binds a diverse range of lipid molecules, including fatty acids, fatty acyl-CoA, phospholipids, glycolipids, hydroxylated fatty acids and prostaglandin B2 (Carvalho and Gomes 2007; Douliez et al. 2000, 2001; Tassin-Moindrot et al. 2000). The compact structure of nsLTPs is stabilized by the disulfide bonds, conferring stability to heat and resistance to proteolysis (Scheurer et al. 2004).
The two main types of nsLTP in flowering plants, termed nsLTP1 and nsLTP2, differ in molecular size (LTP1 consists of 90 amino acids; LTP2 has 70 amino acids.) and by disulfide bond arrangement (In the LTP1 type, cysteine 1 forms a disulfide bond with cysteine 6, and cysteine 5 bonds with cysteine 8; in the LTP2 type, cysteine 1 is paired with cysteine 5, while cysteine 6 is linked with cysteine 8) (Kalla et al. 1994). In addition, both LTPs have different internal cavities for binding hydrophobic ligands: LTP1 is characterized by a long tunnel-like cavity, suitable for binding fatty acids, while LTP2 has two adjacent hydrophobic cavities allowing to accommodate sterols, which is not possible for LTP1 (Cheng et al. 2004; Samuel et al. 2002). Beside LTP1 and 2, more isoforms have been discovered in recent years (Boutrot et al. 2008; Wang et al. 2012). Some are restricted to a single plant species, while others are apparently present in all plants, except for algae (Edstam et al. 2011). The absence of any nsLTP in algae indicates that nsLTPs arose during the conquest of the land by plants, thus, making them key elements in adaptation to harsh environment. Indeed, they are abundant in plants and constitute about 4% of the total soluble proteins (Kader 1996).
Plant lipid transfer proteins display complex expression patterns. They are present in different tissues and are upregulated at different stages of development. NsLTPs have been found in seeds, leaves, stems, roots, flowers and fruits (Pyee et al. 1994; Thoma et al. 1994; Yubero-Serrano et al. 2003). Members of this family are responsive to abiotic stimuli such as drought, cold, salt stress, and their expression is also induced by bacterial and fungal pathogens (Carvalho et al. 2006; Edstam et al. 2014; Guo et al. 2013; Jung et al. 2003).
Despite recent progress, the function of nsLTPs in plants is not well understood. Initially, nsLTPs were thought to be involved in lipid trafficking, due to their ability to bind and transfer lipids in vitro (Kader 1996). Later, LTPs were found to be secreted, but their function in the extracellular matrix was not defined (Sterk et al. 1991; Thoma et al. 1994). Studies of Arabidopsis to knockdown, knockout or overexpress members of the nsLTP family have uncovered the involvement of nsLTP in key physiological processes such as long-distance signaling, defense against pathogens and pests, pollen tube adhesion, cuticle formation, suberin biosynthesis, embryogenesis, seed germination, fruit ripening and allergen action (DeBono et al. 2009; Edstam et al. 2013; Eklund 2003; Maldonado et al. 2002; Park et al. 2007). Recent studies further support the role of nsLTPs in lipid trafficking based on their dynamic distribution (Ambrose et al. 2013; Edstam et al. 2013; Pagnussat et al. 2012).
To date, the research has mainly focused on nsLTP functions in angiosperms rather than gymnosperms. Members of nsLTP family identified in Ginkgo biloba were shown to function as proteinase inhibitors. In Norway spruce, nsLTPs are involved in somatic embryogenesis (Sabala et al. 2000; Sawano et al. 2008). Notably, nsLTPs in pines have not been studied.
In this paper, we describe for the first time the identification of a cDNA encoding Scots pine nsLTP (PsLTP1) and analyze the predicted amino acid sequence and the three-dimensional (3D) structure of the PsLTP1 protein. Furthermore, the PsLTP1 expression pattern was studied in different organs from Scots pine during ontogenesis and under different abiotic stresses.
Materials and methods
Plant material and treatment
Seeds of Scots pine from Lviv Forestry Breeding Seed Center (Lviv region, Ukraine) were washed with distilled water three times and placed on sterile filter paper soaked with distilled water on a Petri dish. They were then left to germinate in a growth chamber in a 16 h-photoperiod at 24 °C. Seedlings were collected at 3 and 7 days after sowing and organs excised separated (roots, hypocotyls and cotyledons). Needles, roots, buds, bark, female and pollen cones were also separately collected from 15-year-old Scots pine from the Ukrainian National Forestry University (UNFU) Dendrobotanical garden. Seeds were harvested from 2-year-old female cones. Endosperms and embryos were collected from mature seeds. All samples were immediately frozen in liquid nitrogen and used for RNA extraction.
For stress treatments in a growth chamber, twenty 7-day-old seedlings were used per experiment at same conditions as mentioned above. For acidulation and NaCl treatments, seedlings were transferred onto sterile filter paper in a Petri dish, soaked with 10 mM HCl or 250 mM NaCl, respectively. For osmotic and oxidative stresses, seedlings were placed onto sterile filter paper that had been moistened with 100 mM mannitol or 10 mM hydrogen peroxide, respectively. Solutions were prepared in a final volume of 5 mL of distilled water and sterilized using a 0.2 µm filter. For these treatments, seedlings were later kept at 24 °C, for 24 h. For cold treatment, plates with seedlings were placed at 4 °C for 24 h. For heat-shock stress, plates with seedlings were incubated at 37 °C for 24 h. For water stress, 7-day-old seedlings were submerged in distilled water on plates, kept at 24 °C and collected after 24 h. Non treated seedlings were grown at 24 °C as the control. After treatments, samples were stored at − 80 °C until total RNA isolation. For each treatment, three replicates were used and treated independently.
Total RNA extraction and cDNA synthesis
Total RNA was extracted from freshly frozen seedlings using lithium chloride precipitation with minor modifications (Chang et al. 1993). The quality of total RNA samples was verified using 1% agarose gel electrophoresis and the quantity measured using the WPA UV 1101 Biotech Photometer UV1101 (Biochrom, Holliston, MA, USA) at OD260 with ratio OD260/OD280 determination. cDNA was synthesized from 1 µg of total RNA using reverse transcriptase RevertAid Premium (Fermentas, Vilnius, Lithuania), according to the manufacturer protocol.
Cloning Scots pine lipid transfer protein gene (PsLTP1 gene)
To clone the cDNA of lipid transfer protein-1 gene from P. sylvestris, we used a PCR-based approach. Primers for Scots pine LTP cloning were designed using the known LTP cDNA sequence of Pinus resinosa Aiton (accession number AF223405.1, NCBI database). Both primers contained sites for restriction enzymes: NcoI in the forward primer and EcoRI in the reverse primer (listed in Table 1 as F-PsLTPc and R-PsLTPc, respectively; restriction sites are underlined). cDNA from roots of 7-day-old seedlings was used as a template. PCR was run under the following conditions (95 °C for 5 min; 30 cycles of 95 °C for 30 s, 54 °C for 1 min, 72 °C for 1 min; final extension at 72 °C for 5 min). PCR products were separated by gel electrophoresis and excised from the gel. For obtaining the pET23d-PsLTP1 expression plasmid, PCR products were digested with both NcoI and EcoRI and subcloned into the bacterial cloning/expression vector pET23d (+) (Novagen, Merck KGaA, Darmstadt, Germany) digested with the same enzymes. The resulting construct was used to transform E. coli strain DH5α, and recombinant clones were grown on selective Luria–Bertani (LB) medium containing ampicillin 100 µg/mL. The presence of a DNA insert carrying the complete coding region for PsLTP1 was verified by PCR analysis. Inserts from positive colonies were purified and sequenced using the automatic DNA sequencer ABI 73 TM (SEQGEN, Torrance, CA, USA).
Sequence analysis and three-dimensional (3D) structure prediction of PsLTP
The amplified and cloned sequence of the PsLTP1 cDNA was analyzed using the Expasy Translate tool (Gasteiger et al. 2003). The presence of the signal peptide was verified using the online program SignalP, version 3.0 (Bendtsen et al. 2004). BLAST was used for searches of sequence databases (https://blast.ncbi.nlm.nih.gov/). Multiple sequence alignments (detailed later) were created using Clustal Omega, version 1.2.1 (Sievers et al. 2011). Default parameters were used. The phylogenetic tree was constructed through comparison with sequences from the BLAST databases using the Clustal W method in LaserGene software (DNASTAR, Madison, WI, USA). LTP sequences used for multiple sequence alignment and phylogenetic tree construction were from Monterey pine (Pinus radiata D. Don), GO448405; loblolly pine (Pinus taeda L.), U10432; shore pine (Pinus contorta Douglas ex Loudon), GT250957; red pine (Pinus resinosa Aiton), AF223405; Japanese black pine (Pinus thunbergii Parl.), FY841227; Jack pine (Pinus banksiana Lamb.), GW765141; Japanese red pine (Pinus densiflora Siebold and Zucc.), FG617230; maritime pine (Pinus pinaster Aiton), CT576220; Scots pine (Pinus sylvestris L.), JN980402; white spruce [Picea glauca (Moench) Voss], EX413050; Norway spruce [Picea abies (L.) H. Karst], GT886696; black spruce [Picea mariana (Mill.) Britton, Sterns and Poggenb.], JZ079655; Sitka spruce [Picea sitchensis (Bong.) Carrière], GW719531; ginkgo (G. biloba L.), DQ836633; barley (Hordeum vulgare L.), M15207; Arabidopsis [Arabidopsis thaliana (L.) Heynh.], AF159798; tree tobacco (Nicotiana glauca Graham), AY621631; maize (Zea mays L.), Uniprot P19656; strawberry [Fragaria ×ananassa (Duchesne ex Weston) Duchesne ex Rozier], AJ315844. The 3D structure of PsLTP1 was predicted by submitting the primary sequence to I-TASSER and Phyre2 servers (Kelley et al. 2015; Yang et al. 2015). The signal peptide of PsLTP1 was removed from the sequence submitted to I-TASSER and Phyre2 since it is absent in the mature protein. Both 3D structures were determined using the automated modeling mode. The protein structures were visualized using ChimeraX version 0.1 (Pettersen et al. 2004).
Gene expression analysis by semiquantitative multiplex RT-PCR
After first-strand cDNA synthesis, the cDNA samples were used as a template for semiquantitative multiplex RT-PCR (mxPCR) using a Proteus thermocycler (Helena Bioscience, Gateshead, UK) (95 °C for 5 min; 35 cycles at 95 °C for 30 s, 54 °C for 30 s, 72 °C for 30 s; final extension at 72 °C for 7 min). The housekeeping gene 60S ribosomal protein L44 (RPL44; GenBank EL342388) was used as a standard control for the mxPCR. The primer sequences used in the mxPCR are listed in Table 1. All mxPCR products were separated by electrophoresis in 1.5% (w/v) agarose gel.
Statistical analyses
The data were analyzed statistically using ANOVA. Results are represented as the mean ± SE. Densitometrical analysis of the electrophoregrams was performed using GelProAnalyzer 4.0 (MediaCybernetics, Rockville, MD, USA). The values for expression level of PsLTP1 were calculated relative to 60S rpl44. The experiments were run in triplicate.
Results
Molecular cloning and sequence analysis of PsLTP cDNA
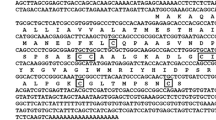
To obtain the full-length cDNA encoding Scots pine lipid transfer protein (PsLTP1), RT-PCR was performed using specific primers and mRNA extracted from seedling roots (GenBank JN980402.1). The PsLTP1 ORF is predicted to encode a polypeptide of 123 amino acid residues containing a signal peptide of 27 amino acids at the N-terminus (Fig. 1). After the signal sequence was removed, the predicted molecular mass of the mature form of PsLTP1 is 9.83 kDa with a theoretical pI of 8.12.
By multiple sequence alignment, PsLTP1 was shown to share identities with other known nsLTPs. Comparison analyses with other nsLTPs from angiosperms and gymnosperms showed amino acid identity ranging from 39% for Arabidopsis to 100% for three Pinus species in the database: Jack pine, Maritime pine and Japanese red pine (Fig. 2a). LTPs from other pine species also showed high identity with Scots pine PsLTP1 (99% with black pine nsLTP, 98% with red pine and shore pine nsLTPs, 97 and 96% with loblolly and Monterey pine, respectively). An identity of 86% was found for PsLTP1 from Sitka and 91% for nsLTPs from white spruce, respectively, from genus Picea. Despite high similarity between nsLTPs from gymnosperms, Gb-nsLTP1 from ginkgo showed only 37% sequence identity with Scots pine PsLTP1, while barley nsLTP and 41% identity. Furthermore, 51% identity was found with tree tobacco and 47% identity with strawberry FaLTP. nsLTP from maize, which also was used for 3D modeling, had 45% identity with Scots pine PsLTP1. To further investigate the relationship between PsLTP1 and nsLTPs, we constructed a phylogenetic tree (Fig. 2b), which showed that PsLTP1 formed one monophyletic group with other pine members and had a closer relationship with nsLTPs from the gymnosperms than with angiosperms, except for Gb-nsLTP1 from ginkgo.
Sequence analysis of PsLTP1 with 18 plant nsLTPs. a Sequence alignment of mature PsLTP1 from seedling roots with mature nsLTPs from gymnosperms and angiosperms. All N-terminal signal sequences were removed from the alignment using SignalP. The eight strictly conserved cysteine residues are shaded in red. Asterisks below the sequences indicate identical amino acids; colon indicates strong amino acid similarity, and a period indicates weak similarity. Percentage of identity between PsLTP and each amino acid sequence used in the alignment is reported for each sequence. LTPs sequences were: Monterey pine, loblolly pine, shore pine, red pine, Japanese black pine, Jack pine, Japanese red pine, maritime pine, Scots pine, white spruce, Norway spruce, black spruce, Sitka spruce, ginkgo, barley, Arabidopsis, tree tobacco, maize, strawberry. b Phylogenetic tree of PsLTP1 and other plant nsLTPs constructed by Clustal W program using neighbor-joining method. The length of each pair of branches displays the distance between sequence pairs, and units at the bottom of the tree indicate the number of substitution events
Scots pine PsLTP1 was further analyzed using 3D structure modeling based on templates of the lipid transfer protein from Zea mays L. (PDB codes 1fk5 and 1ahf), where both PDB codes correspond to the same protein sequence (Uniprot P19656), but the structure of 1ahf was experimentally determined by NMR spectroscopy and the structure of 1fk5 was established by X-ray crystallography. For building the 3D model using the I-TASSER server, 1ahf was used as a template, while coordinates from 1fk5 were applied using the Phyre2 server. In both cases PsLTP1 had 45% amino acid sequence identity with Z. mays. The predicted model of PsLTP1 using I-TASSER consisted of 54% α-helices (Fig. 3a). Phyre2 predicted a model with 100% confidence and 96% coverage. From 96 amino acids, which codify the PsLTP1, 54% consisted of α-helices, and 38% of the query was found to be disordered (Fig. 3b). The 3D structural model of PsLTP1 shows that the protein consists of four α-helices, where the secondary structures are stabilized by four disulfide bridges and an unstructured C-terminal tail. The four pairings are formed between eight cysteines: C7–C55 (cysteine pair 1–6), C17–C32 (cysteine pair 2–3), C33–C77 (cysteine pair 4–7) and C53–C92 (cysteine pair 5–8).
Three-dimensional model predicted for PsLTP1. The hypothetical 3D structural modeling was performed based on a template of lipid transfer protein from Zea mays (PDB codes 1fk5 and 1ahf). To build the 3D model, the I-TASSER program used 1ahf (a) as a template, while the Phyre2 used 1fk5 (b). In both cases, PsLTP had 45% amino acid sequence identity with Z. mays. The 3D structural model of PsLTP shows that the protein consists of four α-helices, where the secondary structures are stabilized by four disulfide bridges and unstructured C-terminal tail. The four pairings are formed between eight cysteines: C7–C55, C17–C32, C33–C77 and C53–C92
Expression profile of PsLTP1 gene
In the expression analysis of PsLTP1 in different tissues during ontogenesis using mxPCR, PsLTP1 transcripts were expressed in all organs of the seedlings and an adult tree. The relative transcript levels in young seedlings were higher than in the adult tree. In seedlings, the highest level of PsLTP1 transcripts accumulated in apical parts of the seedlings and gradually decreased in lower parts (Fig. 4).
Expression analysis of PsLTP1 gene in different organs of Scots pine. Top panels: a electrophoresis of mxPCR products using RNA from roots (1), hypocotyls (2), cotyledons (3) of 7-day-old seedlings; bark (4), needles (5), buds (6), roots (7), female cones (8), pollen cones (9) of 15-years-old tree; embryos (10), endosperms (11) and 3-day-old seedlings (12). Bottom panel, b transcript levels calculated relative to housekeeping gene rpl44 from Scots pine as an internal control
In organs of an adult tree, the quantity of PsLTP1 transcripts was relatively low compared to the level in seedlings. As shown in Fig. 4, PsLTP1 transcripts are present in all investigated organs, and were relatively high in female and pollen cones and buds. PsLTP1 transcripts were not detected in the endosperm, but levels were high in embryos collected from dry seeds, indicating that PsLTP1 gene expression is tissue- and organ-specific and time-dependent.
The analysis of PsLTP1 expression in response to several abiotic stresses (Fig. 5) showed that cold and salinity led to an upregulation of PsLTP1 expression. Treatment with hydrogen peroxide and high temperature did not substantially affect expression in pine seedlings, while HCl and mannitol induced a downregulation that was more pronounced than that caused by water stress (32% when compared to the control).
Expression of Scots pine PsLTP1 under various environmental stresses. a Electrophoresis of mxPCR products using RNA extracted from seedlings treated with water as the control (1), exposed to HCl (2), cold (3), or salt (4), submersed in water (5), exposed to H2O2 (6), heat shock (7) or osmotic stress (8). b Expression level of PsLTP1 relative to rpl44 from Scots pine as the internal control. Expression of the control, grown under typical conditions, was set to zero. Values below zero indicate downregulation above zero indicate upregulation of PsLTP1
Discussion
In this study, we cloned a cDNA carrying the entire coding sequence for an nsLTP (PsLTP1 gene) from P. sylvestris. The deduced amino acid sequence of PsLTP1 shares most of the characteristics present in other plant nsLTPs, including the presence of eight cysteines in defined positions, crucial for correct disulfide bridge formation. PsLTP1 is a low molecular weight protein with basic pI. Signal P predicted an N-terminal signal peptide of 27 amino acids that engages the protein into the secretory pathway (Carvalho and Gomes 2007). The predicted mature PsLTP1 is a type 1 nsLTP with a calculated molecular mass of approximately 10 kDa and a positive net charge. Using I-TASSER and Phyre2 programs generated the hypothetical 3D structure of PsLTP1, consisting of 4 α-helices and a C-terminal tail. The disulfide bond arrangements C7–C55 (cysteine pair 1–6) and C53–C92 (cysteine pair 5–8) corroborate that PsLTP1 belongs to type 1 in the nsLTP family. The predicted structure with internal cavity allows the protein to accommodate hydrophobic compounds such as fatty acids, but further in vitro and in vivo studies are required to validate this prediction. Multiple sequence alignment with nsLTP from other plant species revealed that PsLTP1 shares a high degree of identity with nsLTPs from other conifers, and they clustered together in the phylogenetic tree. Thus, the mature PsLTP1 protein shares 100% identity of its amino acid sequence with three other members of the same genus: Jack pine, maritime pine and Japanese red pine. Notably, all have different geographical locations, very distant from each other. Scots pine is native to northern Eurasia, maritime pine is native to the Mediterranean region, Jack pine is an eastern North American pine, and Japanese red pine is native to the northeastern part of China and the Korean peninsula, Japan and the far eastern part of Russia. From these data, we can conclude that PsLTP1-like sequences are highly conserved inside this genus.
PsLTP1 showed was expressed at the highest level in young tissues, including roots, hypocotyls and cotyledons of pine seedlings and in buds, female and pollen cones of the adult tree. Moreover, PsLTP1 transcripts were abundant in embryos and 3-day-old seedlings but were absent in the endosperm. A thorough search of the EST database revealed that nucleotide sequences of similar nsLTPs have been isolated mostly from young seedlings. Similar results were also obtained in studies of other species in which expression of nsLTPs tends to be higher in aerial plant organs and in actively growing tissues. In Betula plathyphilla Sukaczev, expression of BplLTP1 was much higher in young organs than in adults (Guan et al. 2013). In Arabidopsis, nsLTPs are expressed in vascular tissues and epidermal cells during development and in broccoli in the cuticular waxy layer (Pyee et al. 1994; Thoma et al. 1994). The expression of nsLTP genes is necessary for normal embryo development in both Arabidopsis and Norway spruce (Sabala et al. 2000; Vroemen et al. 1996). Interestingly, nsLTPs decrease during plant aging and in fully expanded tissues in angiosperms (Pyee et al. 1994; Thoma et al. 1994).
Some abiotic stresses caused downregulation of the PsLTP1 gene, but low temperature and high salinity led to the upregulation of PsLTP1 in Scots pine seedlings. A thorough search of EST libraries of P. sylvestris revealed that several other sequences identical to PsLTP1 have been isolated from young pine plants after treatment with ozone. Similar expression patterns were described for other plant species for which several stress-induced LTP genes have been isolated and characterized. For example, TSW12 in tomato was upregulated after NaCl treatment (Torres-Schumann et al. 1992; Trevino and O’Connell 1998). Two pepper genes (CALTPI and CALTPIII) were upregulated during drought, in a high salt concentration, at low temperature and after wounding (Jung et al. 2003). In strawberry, FaLTP is upregulated during fruit development, and FaLTP transcripts increase after ABA, SA and wounding treatments but decrease slightly after cold treatment; they do not accumulate after salt treatment (Yubero-Serrano et al. 2003). Three LTP transcripts (PpLTPG3, PpLTPG8 and PpLTPG9) in the moss Physcomitrella patens (Hedw.) Bruch and Schimp. accumulate to high levels during cold treatment, and the expression of four LTP genes (PpLTPG2, PpLTPG3, PpLTPG6, PpLTPG9) associated with the synthesis and/or deposition of cutin and cuticular wax is upregulated during dehydration (Edstam et al. 2014).
Because members of the nsLTP gene family vary widely in their expression patterns during development, in different tissues and in response to environmental stresses, clarifying the exact physiological role of each member of this gene family is a complex task. Taking into account that PsLTP1 transcripts were detected mostly from actively growing young tissues, the homology among members from different pine species is very high, and their expression profiles also seems to be similar, it is not unreasonable to think that the identified PsLTP1 could be involved in pine development and response to abiotic stress.
Previously, we isolated a nsLTP (p1 protein) from Scots pine seedlings and confirmed its identity by mass spectrometry (Kovaleva et al. 2009). Two peptides were obtained by tryptic digestion RP-HPLC of this endogenous p1 protein (VTDLNVPISPNVDCSK and AATPAAACCPSIR), then identified using MS. A search of different databases revealed that both peptides are similar to the nonspecific lipid-transfer protein precursor (nsLTP) from P. resinosa (GenBank AF22340) (Kovaleva et al. 2009). Further studies allowed us to isolate a cDNA clone, encoding P. sylvestris LTP, which we termed PsLTP1. Sequence analysis of the coding region and the predicted translated protein revealed that PsLTP1 contains the same two peptide sequences obtained by tryptic digestion of the endogenous nsLTP (p1 protein) from Scots pine seedlings. Taken together, our studies suggest that PsLTP1 could have a similar structure and/or role in defense as the p1 protein previously described. However, the biological function of PsLTP1 needs to be further investigated.
References
Adomas A et al (2008) Comparative analysis of transcript abundance in Pinus sylvestris after challenge with a saprotrophic, pathogenic or mutualistic fungus. Tree Physiol 28:885–897
Ambrose C, DeBono A, Wasteneys G (2013) Cell geometry guides the dynamic targeting of apoplastic GPI-linked lipid transfer protein to cell wall elements and cell borders in Arabidopsis thaliana. PLoS ONE 8(11):e81215
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Boutrot F, Guirao A, Alary R, Joudrier P, Gautier MF (2008) Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtpgenes by EST data mining. BMC Genom 9:86–105
Carvalho AO, Gomes VM (2007) Role of plant lipid transfer proteins in plant cell physiology—a concise review. Peptides 28:1144–1153
Carvalho AO, Souza-Filho GA, Ferreira BS, Branco AT, Araújo IS, Fernandes KV, Retamal CA, Gomes VM (2006) Cloning and characterization of a cowpea seed lipid transfer protein cDNA: expression analysis during seed development and under fungal and cold stresses in seedlings’ tissues. Plant Physiol Biochem 44:732–742
Chang SJ, Puryear J, Cairney J (1993) A simple and efficient method for isolating rna from pine trees. Plant Mol Biol Report 11:113–116
Cheng CS, Samuel D, Liu YJ, Shyu JC, Lai SM, Lin KF, Lyu PC (2004) Binding mechanism of nonspecific lipid transfer proteins and their role in plant defense. Biochemistry 43:13628–13636
DeBono A, Yeats TH, Rose JKC, Bird D, Jetter R, Kunst L, Samuels L (2009) Arabidopsis LTPG is a GPI anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21:1230–1238
Douliez JP, Michon T, Elmorjani K, Marion D (2000) Mini review: structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. J Cereal Sci 32:1–20
Douliez JP, Jegou S, Pato C, Molle D, Tran V, Marion D (2001) Binding of two mono-acylated lipid monomers by the barley lipid transfer protein, LTP1, as viewed by fluorescence, isothermal titration calorimetry and molecular modelling. Eur J Biochem 268:384–388
Edstam MM, Viitanen L, Salminen TA, Edqvist J (2011) Evolutionary history of the non-specific lipid transfer proteins. Mol Plant 4:947–964
Edstam MM, Blomqvist K, Eklof A, Wennergren U, Edqvist J (2013) Coexpression patterns indicate that GPI-anchored non-specific lipid transfer proteins are involved in accumulation of cuticular wax, suberin and sporopollenin. Plant Mol Biol 83:625–649
Edstam MM, Laurila M, Höglund A, Raman A, Dahlström KM, Salminen TA, Edqvist J, Blomqvist K (2014) Characterization of the GPI-anchored lipid transfer proteins in the moss Physcomitrella patens. Plant Physiol Biochem 75:55–69
Eklund DM (2003) Localization of nonspecific lipid transfer proteins correlate with programmed cell death responses during endosperm degradation in Euphorbia lagascae seedlings. Plant Physiol 132:1249–1259
García-Olmedo F, Molina A, Segura A, Moreno M (1995) The defensive role of nonspecific lipid-transfer proteins in plants. Trends Microbiol 3:72–74
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Goyala RK, Mattoo AK (2014) Multitasking antimicrobial peptides in plant development and host defense against biotic/abiotic stress. Plant Sci 228:135–149
Guan MX, Chai RH, Kong X, Liu XM (2013) Isolation and characterization of a lipid transfer protein gene (BplLTP1) from Betula platyphylla. Plant Mol Biol Report 31:991–1001
Guo L, Yang HB, Zhang XY, Yang SH (2013) Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J Exp Bot 64:1755–1767
Hensiruk SA (2002) Forests of Ukraine. Shevchenko Scientific Society, Ukrainian State university of Forestry and Wood Technology, Lviv
José-Estanyol M, Gomis-Ruth FX, Puigdomenech P (2004) The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol Biochem 42:355–365
Jung HW, Kim W, Hwang BK (2003) Three pathogen-inducible genes encoding lipid transfer protein from pepper are differentially activated by pathogens, abiotic, and environmental stresses. Plant, Cell Environ 26:915–928
Kader JC (1996) Lipid transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 47:627–654
Kalla R, Shimamoto K, Potter R, Nielsen PS, Linnestad C, Olsen OA (1994) The promoter of the barley aleurone-specific gene encoding a putative 7 kDa lipid transfer protein confers eleurone cell-specific expression in transgenic rice. Plant J 6:849–856
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858
Kovaleva V, Kiyamova R, Cramer R, Krynytskyy H, Gout I, Filonenko V, Roman G (2009) Purification and molecular cloning of antimicrobial peptides from Scots pine seedlings. Peptides 30:2136–2143
Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK (2002) A putative lipid transfer protein involved in systemic resistance signaling in Arabidopsis. Nature 419:399–403
Pagnussat L, Burbach C, Baluška F, de la Canal L (2012) An extracellular lipid transfer protein is relocalized intracellularly during seed germination. J Exp Bot 63:6555–6563
Park S, Lee J, Shin S, Park Y, Lee S, Hahm K (2007) Characterization of a heat-stable protein with antimicrobial activity from Arabidopsis thaliana. Biochem Biophys Res Commun 362:562–567
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Pyee J, Yu HS, Kolattukudy PE (1994) Identification of a lipid transfer protein as the major protein in the surface wax of broccoli (Brassica oleracea) leaves. Arch Biochem Biophys 311:460–468
Sabala I, Elfstrand M, Farbos I, Clapham D, von Arnold S (2000) Tissue-specific expression of Pa18, a putative lipid transfer protein gene, during embryo development in Norway spruce (Picea abies). Plant Mol Biol 42:461–478
Samuel D, Liu YJ, Cheng CS, Lyu PC (2002) Solution structure of plant nonspecific lipid transfer protein-2 from rice (Oryza sativa). J Biol Chem 277:35267–35273
Sawano Y, Hatano K-I, Miyakawa T, Komagata H, Miyauchi Y, Yamazaki H, Tanokura M (2008) Proteinase inhibitor from ginkgo seeds is a member of the plant nonspecific lipid transfer protein gene family. Plant Physiol 146:1909–1919
Scheurer S, Lauer I, Foetisch K, San Miguel Moncin M, Retzek M, Hartz C, Enrique E, Lidholm J, Cistero-Bahima A, Vieths S (2004) Strong allergenicity of Pru av 3, the lipid transfer protein from cherry, is related to high stability against thermal processing and digestion. J Allergy Clin Immunol 114:900–907
Sels J, Mathys J, De Coninck BMA, Cammue BPA, De Bolle MFC (2008) Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem 46:941–950
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Sterk P, Booij H, Schellekens GA, Van Kammen A, De Vries SC (1991) Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell 3:907–921
Tassin-Moindrot S, Caille A, Douliez JP, Marion D, Vovelle F (2000) The wide binding properties of a wheat nonspecific lipid transfer protein. Solution structure of a complex with prostaglandin B2. Eur J Biochem 267:1117–1124
Thoma S, Hecht U, Kippers A, Botella J, De Vries S, Somerville C (1994) Tissue-specific expression of a gene encoding a cell wall-localized lipid transfer protein from Arabidopsis. Plant Physiol 105:35–45
Torres-Schumann S, Godoy JA, Pintor-Toro JA (1992) A probable lipid transfer protein gene is induced by NaCl in stems of tomato plants. Plant Mol Biol Report 18:749–757
Trevino MB, O’Connell MA (1998) Three drought-responsive members of the nonspecific lipid-transfer protein gene family in Lycopersicon pennellii show different developmental patterns of expression. Plant Physiol 116:1461–1468
Veluthakkal R, Dasgupta MG (2010) Pathogenesis-related genes and proteins in forest tree species. Trees 24:993–1006
Vroemen CW, Langeveld S, Mayer U, Ripper G, Jurgens G, Van Kammen A, De Vries SC (1996) Pattern Formation in the Arabidopsis embryo revealed by position-specific lipid transfer protein gene expression. Plant Cell 8:783–791
Wang HW, Hwang SG, Karuppanapandian T, Liu A, Kim W, Jang CS (2012) Insight into the molecular evolution of non-specific lipid transfer proteins via comparative analysis between rice and sorghum. DNA Res 19:179–194
Yang JY, Yan RX, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8
Yubero-Serrano E-M, Moyano E, Medina-Escobar N, Muñoz-Blanco J, Caballero JL (2003) Identification of a strawberry gene encoding a non-specific lipid transfer protein that responds to ABA, wounding and cold stress. J Exp Bot 54:1865–1877
Author information
Authors and Affiliations
Corresponding author
Additional information
Project funding: The work was supported by grants from the Ministry of Education and Science of Ukraine (0116U003593) and grant from cieA3 (Campus de Excelencia Internacional Agroalimentario)-UCO, Spain.
The online version is available at http://www.springerlink.com
Corresponding editor: Tao Xu.
Rights and permissions
About this article
Cite this article
Hrunyk, N., Kovaleva, V., Krynytskyy, H. et al. Molecular cloning and characterization of a lipid transfer protein gene (PsLTP1) from Pinus sylvestris (L.). J. For. Res. 30, 1149–1158 (2019). https://doi.org/10.1007/s11676-018-0648-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-018-0648-z