Abstract
The introgressed small-chromosome segment of Agropyron elongatum (Host.) Neviski (Thinopyrum ponticum Podp.) in F5 line II-1-3 of somatic hybrid between common wheat (Triticum aestivum L.) and A. elongatum was localized by sequential fluorescence in situ hybridization (FISH), genomic in situ hybridization (GISH) and karyotype data. Karyotype analysis offered basic data of arm ratios and relative lengths of 21 pairs of chromosomes in parent wheat Jinan177 and hybrid II-1–3. Using special high repetitive sequences pSc119.2 and pAs1 for FISH, the entire B- and D-genome chromosomes were detected. The FISH pattern of hybrid II-1-3 was the same as that of parent wheat. GISH using whole genomic DNA from A. elongatum as probe determined the alien chromatin. Sequential GISH and FISH, in combination with some of the karyotype data, localized the small chromosome segments of A. elongatum on the specific sites of wheat chromosomes 2AL, 1BL, 5BS, 1DL, 2DL and 6DS. FISH with probe OPF-031296 from randomly amplified polymorphic DNA (RAPD) detected E-genome chromatin of A. elongatum, which existed in all of the small chromosome segments introgressed. Microsatellite primers characteristic for the chromosome arms above were used to check the localization and reveal the genetic identity. These methods are complementary and provide comprehensive information about the genomic constitution of the hybrid. The relationship between hybrid traits and alien chromatin was discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Relatives of wheat have some important traits beneficial for its quality and resistance improvement. Somatic hybridization provides a novel way for crop breeding (Waara and Glimelius 1995). It offers the possibility of getting over the barriers of sexual crossing and allows the transfer of superior genes from nuclear and cytoplasmic genome separately or simultaneously (Bijoya et al. 1999; Yue et al. 2001). UV irradiation could induce introgression of DNA or chromosome segments during protoplast fusion (Forsberg et al. 1998; Xiang et al. 2003). A small portion of DNA segments from donor integrated into the genome of receptor has some advantages over adding a complete set of chromosomes or intact chromosome(s), which could increase the regeneration ability, inherited stability and fertility of the hybrid (Xiang et al. 2003, 2004; Wang et al. 2003; Chen et al. 2004). With the development of molecular biology, the identification of alien chromosome and chromatin in somatic hybrid has greatly extended from traditional analysis of morphology, cytology and biochemistry (Xia et al. 1996) to verification with molecular markers, including randomly amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), restriction fragment length polymorphism (RFLP), simple sequence repeat (SSR) and genomic in situ hybridization (GISH) etc. (Forsberg et al. 1998; Zhou et al. 2001; Xiang et al. 2003; Wang et al. 2003; Xu et al. 2003).
In wheat breeding programs, chromosome identification (Xu et al. 1994) and alien chromatin detection in wheat background is important (Gill 1987; Forsström et al. 2002; Ko et al. 2002). The localization of alien chromatin is particularly useful in connection with the physical mapping of other DNA sequences to chromosomes (Pedersen and Langridge 1997), which can offer the feasibility for target-gene cloning and marker-assistant breeding. We have described the detection of alien chromosomes and chromatin in several wheat somatic hybrids (Zhou et al. 2001; Xia et al. 2003; Xiang et al. 2003; Xu et al. 2003). However, there were no reports about the precise localization of alien chromatin in wheat somatic hybrids (also any other somatic hybrids). This limited the application and development of somatic-hybridization technology.
The fluorescence in situ hybridization (FISH) patterns of wheat chromosomes are beneficial for rapid and intuitive physical localization of heterogeneric chromatin. Using high repetitive sequences pSc119.2 from rye and pAs1 including the inserted repetitive sequences of Aegilops tauschii (Cross.) Schmal for two-color FISH, Mukai et al. (1993) established the idiogram of B- and D-genome chromosomes of Chinese Spring wheat. Pedersen and Langridge (1997) presented a detailed complete wheat molecular karyotype based on pAs1 sequence and GAA-satellite sequence (pHvG38), permitting identification of all 21 wheat chromosomes by two-color FISH. In addition, the identification of Aegilops elongatum chromatin in wheat background has been reported using special repeat sequences of E-genome from RAPD as probes (Zhang et al. 1998). GISH and two-color FISH in combination with the use of genetically mapped barley SSR have been developed for the identification of wheat–barley translocations (Nagy et al. 2002). It was also reported that several pairs of wheat SSR primers from 1A and 1D were used to locate the recombined chromosomes and intercalary 1D chromosome segments in a durum wheat background (Blanco et al. 2002). This research provides important information for alien chromatin localization of wheat somatic hybrid.
Fertile hybrids and progenies have already been created via somatic hybridization between Triticum aestivum (cv. Jinan 177) and Agropyron elongatum treated by UV (Xia et al. 2003). Cytogenetical analysis of hybrids showed that the chromosome numbers varied in the range of 38~44 (Wang et al. 2004). GISH analysis revealed the different hybrid lines with different sites of translocation or insertion of chromosome segments (Xia et al. 2003; Wang et al. 2004). Some F1~F5 hybrid lines expressed valuable traits, such as high quality, short stalk, salt tolerance and disease resistance. These materials may have potential for wheat improvement. Of these, a hybrid line II-1-3 possessed salt tolerance (Chen et al. 2004) and high quality (Zhao et al. 2003). The stable phenotype from F2 to F5 and the normal PMC MI chromosome pairing in F5 (Wang et al. 2004) imply that II-1-3 is an introgressed small-chromosome-fragment line. The goal of this study was to localize the alien chromatin of II-1-3 using cytological and molecular approaches, including identification of wheat chromosomes involved and determination of the relative distance of A. elongatum chromatin introgressed.
Materials and methods
Plant materials
Seeds of T. aestivum L (cv. Jinan 177), A. elongatum (Host.) Neviski (synonym, Thinopyrum ponticum Podp.) and the F5 hybrid line II-1-3 between T. aestivum and A. elongatum were saved in our laboratory. Seeds of the 4E/4D substitution line of wheat, T. urartu Thum, Aegilops speltoides Tausch and Aegilops tauschii (Coss) were kindly provided by Quality and Resource Institute of Agriculture Science Academy of China.
Root tip and chromosome preparation
Seeds of T. aestivum, A. elongatum, II-1-3 and 4E/4D substitution line were germinated on moist filter paper for a few days at 25°C in the dark. The root tips of 0.5–1 cm in length were excised and placed in ice water for 24 h, and then fixed in 1:3 acetic acid–ethanol for 2 days. For chromosome counting, the root-tip meristem was squashed under a cover slip in Carbol Fuchsin solution. For GISH and FISH analysis, the root-tip meristem was squashed in 45% acetic acid.
Karyotype analysis
More than 500 cells were counted for the parent wheat and hybrid, respectively. Karyotype classification followed the method of Sear (1969) and consulted the data of Chinese Spring from Gill (1987). The parameters of the karyotypes were based on ten metaphase cells spread. Student’s t-test (Table 2) via the Statistical Package for the Social Sciences (SPSS) software was used to compare the karyotypes of Jinan177 with II-1-3.
DNA probes of repetitive sequence and genome
Genomic DNA of common wheat Jinan177, A. elongatum, T. urartu, Aegilops tauschii, Aegilops speltoides and II-1-3 were isolated by CTAB method according to Doyle and Doyle (1990). Total genomic DNA of A. elongatum was labeled as a probe for GISH.
pSc119.2 and pAs1 containing particular repetitive sequences of B and D genome, respectively, were offered by Dr. Zhang Xueyong (Quality and Resource Institute of Agriculture Science Academy of China). They were labeled as probes for FISH.
Three RAPD primers (Operon Technology, USA) were used to derive amplicons following Zhang et al. (1998). Annealing temperatures were 38°C for OPF-03, 41°C for OPB-08 and 48°C for OPN-01, respectively. The 1,296-bp segment of A. elongatum amplified with OPF-03 was retrieved by BioDev glassmilk kit (Boda Biocompany, Beijing) and labeled as a probe for E genome.
All probes were labeled with digoxigenin-11-dUTP using a nick translation kit following the manufacturer’s instructions (976776 Boehringer Mannheim).
GISH and FISH
GISH and FISH were carried out following the method described by Xiang et al. (2003). The combinations and ratios of probes and blocking DNA are listed in Table 1.
Some combinations of GISH with FISH were tested on the same sides: I. GISH+B; II. GISH+D; III. GISH+B+D. For I and II, pSc119.2 and pAs1 were used for FISH, respectively, after the preparation of GISH was rinsed. For III, FISH/ pAs1 followed the FISH/ pSc119.2, which was performed after the GISH preparation was rinsed.
The 1,296-bp segment of E genome from A. elongatum was used for FISH on the chromosome plates of the hybrid II-1-3 and 4E/4D substitution line.
Microsatellite marker
DNA was extracted from leaves of the hybrid II-1-3 and parents by using the same method as above. Microsatellite loci mapping on particular wheat chromosomes (based on the locations from GISH/FISH/karyotype) were amplified using 52 pairs of primers identified according to the procedure described by Röder et al. (1998).
Results
Chromosome number and karyotype
Statistical data of chromosome numbers revealed that 77.25% of the cells in hybrid II-1-3 were 2n=42, near to the proportion of parent wheat Jinan 177. The karyotypes (Fig. 1) and basic data (Table 2) of arm ratios and relative lengths of 21 pairs of chromosomes from hybrid and parent wheat were similar at a global level. However, the arm ratios of 2A, 6B and 6D and the relative length of 3D in II-1-3 chromosomes were obviously greater than that of Jinan177(P<0.05) (Table 2). It was suggested that these differences were due to the introgression of A. elongatum. These data provide a reference for the location of A. elongatum chromatin.
Localization of A. elongatum chromatin on wheat chromosomes
FISH patterns of B- and D-genome chromosomes of hybrid and parent wheat
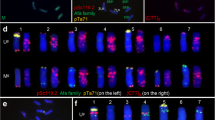
Using the clone pSc119.2 and pAs1 separately for FISH produced hybrid bands on all B- and D-genome chromosomes of II-1-3 and parent wheat (Fig. 2). According to the FISH patterns of Chinese Spring wheat (Mukai et al. 1993), seven pairs of chromosomes of B and D genome were paired and identified, respectively (Fig. 2). In addition, a pAs1 probe hybridized with a pair of 4A chromosomes, showing minor sites on the terminals of long arms (Fig. 2), the same as the result in Chinese Spring (Mukai et al. 1993). The FISH patterns of B and D genome of II-1-3 (Fig. 2a, c) were identical with parent wheat (Fig. 2b, d), and in general agreement with Mukai et al. (1993). The results indicate that pSc119.2 and pAs1 can be efficiently used for the detection of B- and D-genome chromosomes in wheat Jinan 177 and probably permit identification of related chromosomes in most wheat cultivars.
FISH karyotype of metaphase chromosomes of the hybrid II-1-3 and parent Jinan 177. The probes were labeled using digoxigenin and detected with FITC (yellow–green). The chromosomes counterstained with PI (red). a, b B-genome patterns of II-1-3 (a) and Jinan 177 (b) with rye repetitive DNA probe (pSc119.2) on metaphase plates of the root tips. c, d D-genome patterns of II-1-3 (c) and Jinan 177 (d) with Aegilops tauschii repetitive (pAsI) probe on metaphase plates of the root tips. Scale bars 10 μm
Distribution of A. elongatum chromatin on B-, D- and A-genome chromosomes
Sequential GISH and FISH were used to identify the chromatin of A. elongatum on the B-, D- and several A-genome chromosomes of wheat in the hybrids. The combinations of in situ hybridization include: GISH+B (I), GISH+D (II) and GISH+B+D (III).
GISH results from combination I, II and III detected the alien chromatin translocated or inserted into wheat chromosomes. Statistical data from 510 cells indicated that 67.73 % of them had six green hybridization signals (Figs. 3a, c, 4a), accounting for the highest proportion.
Sequencial GISH and FISH (a/b and c/d) on the same spread of metaphase chromosomes of root tip in hybrid II-1-3. The probes were labeled using digoxigenin and detected with FITC (yellow–green). The chromosomes were counterstained with PI (red). Right arrow Hybridized signal. a, c GISH patterns of II-1-3 with total genome DNA of A. elongatum. b, d B- and D-genome patterns of II-1-3 with pSc119.2 and pAsI probes on the same plates of the hybrid, respectively. Scale bars 10 μm
FISH results using pSc119.2 as a probe in combination I and III showed two GISH signals (Figs. 3a, 4a) on 1B and 5B chromosomes, respectively (Figs. 3b, 4b). This indicates that there are two alien segments in the B genome.
Sequencial GISH and FISH (a/b/c) on the same spread of metaphase chromosomes of the hybrid II-1-3 root tip. The probes were labeled using digoxigenin and detected with FITC (yellow–green). The chromosomes were counterstained with PI (red). Right arrow Hybridized signal. a GISH patterns of II-1-3 with total genome DNA of A. elongatum. b, c FISH patterns of B- and D-genome chromosomes of II-1-3 with pSc119.2 or pAsI probes on the same plates, respectively. Scale bars 10 μm
Using pAs1 for FISH in combination II and III showed three GISH signals (Figs. 3c, 4a) on 1D, 2D and 6D chromosomes, respectively (Figs. 3d, 4c). Therefore, the D genome contains three A. elongatum segments.
Integrating B-genome and D-genome signals of FISH in the same chromosome spread from combination III can differentiate parts of A-genome chromosomes including 1A, 4A and 5A (Mukai et al. 1993). In combination with karyotype data, 2A (the longest) and 6A (the shortest) could also be identified. Thus, a small A. elongatum chromosome segment has been located on the 2A chromosome (Fig 4c).
In sum, all A. elongatum small chromosome segments were localized on 2AL, 1BL, 5BS, 1DL, 2DL and 6DS of wheat chromosomes of the hybrid (Table 3; Fig.4). In addition, the arm ratios of the chromosomes involved and the relative distances from centromeres to the breaking points were analyzed, based on the data from GISH. The relative distances from centromeres to the breaking points were 72.45±11.12, 75.54±10.12, 50.89±12.45, 69.23±15.33, 72.88±14.02 and 20.63±13.57, respectively (Table 3).
Origin of A. elongatum chromosome segments
RAPD profiles of wheat, A. elongatum and II-1-3 showed specific segments of 525, 1,296 and 817 bp in A. elongatum. There was no evidence for any introgressed segment homologous with 525 and 817 bp in II-1-3. In contrast, II-1-3 displayed an allele of A. elongatum 1,296-bp sequence amplified by prime OPF-03(CCTGATCACC)(Fig. 5) (Zhang et al. 1998). It implies the introgression of A. elongatum DNA via asymmetric somatic hybridization.
The OPF-031296 segment was hybridized with II-1-3 and 4E/4D substitution line which was used as control. According to expectations, typical hybridized signals localized on a pair of chromosome terminals of 4E/4D substitution in the FISH pattern(Fig. 6a). Among 77 cells in hybrid II-1-3 observed, 48 (62.33%) have six green hybridized sites(Fig. 6b), coinciding with the result from GISH. This indicated that E-genome DNA was involved in A. elongatum chromatin in hybrid II-1-3.
FISH to metaphase chromosomes of hybrid II-1-3 and 4E/4D substitution line. The 1,296-bp segment of E genome amplified from A. elongatum with OPF-03 primer was labeled using digoxigenin and detected with FITC (yellow–green). The chromosomes were counterstained with PI (red). Right arrow Hybridized signal. a FISH patterns of 4E/4D substitution line with 1,296-bp segment probe on metaphase plate of the root tips. b FISH patterns of II-1-3 with 1296-bp segment probe on metaphase plate of the root tip. Scale bars 10 μm
SSR analysis of special sites on hybrid chromosomes
To test the result from GISH/ FISH, a total of 52 pairs of microsatellite primers located on 2AL, 1BL, 5BS, 1DL, 2DL and 6DS of wheat were used to amplify the genomic DNA of the hybrid and parents. Sixteen markers (Table 4; Fig. 7) showed clear polymorphism among different genotypes. The SSR profiles of II-1-3 (Table 4; Fig. 7) carried (1) the fragments originated from both parents (P); (2) the fragments derived from A. elongatum(A); (3) the fragments from A. elongatum/novel fragment(s) (A, N) and (4) the biparental/novel fragment(s) (P, N). Markers P, A, N (Fig. 7) indicated the integration and rearrangement between biparental DNA. The relative distance (%) from the centromeres to SSR loci checked were counted. Several SSR bands were located on 2AL, 1BL and 2DL and the relative distances [relative distance = (distance from centromere to SSR locus checked/length of the arm involved) × 100%] of most fragments ranged from 63.79% to 77.59% (Table 4), in agreement with the results of sequential GISH and FISH (Table 3). The alien segments, 2AL, 1BL, 5BS, 2DL and 6DS, have also been primarily positioned on the macrosatellite genetic map of wheat with centiMorgans according to the results of Röder et al. (1998) (Fig. 8).
The SSR genetic map of chromosomes 2AL, 1BL, 5BS, 2DL and 6DS and genetic distances (cM) from Röder et al. (1998). Shadows represent the special SSRs on relative chromosomes of II-1-3
Discussion
It was clear that UV induced both donor chromosome elimination and fragmentation (Hall et al. 1992; Xia et al. 2003; Wang et al. 2003; Xiang et al. 2003, 2004). Novel chromosomes, including intercalary translocations (Figs. 3, 4; Tables 2, 3), could produce when the instant break points of receptor DNA linked with donor DNA segments. Such a chromotype is particularly interesting in the context of alien introgression (Xia et al. 2003), as it is superior to big-segment translocation. e.g. it can largely exclude the interference of disadvantage genes; it benefits heredity and localization of alien target genes. Therefore, it is very important to both genetic theory research and genetic breeding of wheat.
The amount of chromatin introgressed in translocated wheat lines can be evaluated using GISH (Blanco et al. 2002). In combination with other cytogenetic and molecular genetic approaches, the translocation breakpoint and the physical size of introgressed chromosome segment in recombined wheat could be determined. For example, Jiang et al. (1993) detected wheat–rye substitution using GISH/C-banding/N-banding for the first time. Zhang et al. (2001) localized the Yellow dwarf disease resistant gene of Thinopyrum intermedium (Host) to 7DL using GISH/RFLP/RAPD. Malysheva et al. (2003) identified barley chromosomes and chromosome segments in wheat–barley hybrids via GISH/SSR. Wei et al. (1999) detected rye chromatin in the new wheat germplasm 10-A with FISH/RFLP/A-PAGE. However, most of the translocation lines of wheat reported contained alien whole arm or big segment created by sexual cross/chromosome engineering (Ren and Zhang (1997). Only a few small-segment-translocation lines were produced (Blanco et al. 2002; Malysheva et al. 2003).
Small segment translocation was more complex in the somatic hybrid line than in sexual hybrid of wheat. The karyotype analysis offered an assistant means for identification of A-genome chromosome in the hybrid (Fig. 4) and some information on the hybrid genome variation (Table 2, Fig. 1). However, the chromosomes with distinct differences in the karyotype between the hybrid and parent wheat are not all in correspondence with the chromosomes involved in the introgression. Out of four chromosomes detected by karyotype analysis (Table 2), only 2A and 6D related to the introgression of alien chromosome segments detected by GISH. So, the karyotype change was likely derived from not only alien chromatin introgression but also somaclonal variation. Through single-color FISH with pSc119.2 and pAs1, consulting some of the karyotype data, we detected the chromosomes of B, D, 1A, 4A and 5A (Figs. 2, 3b, d, 4b, c), 2A and 6A (Table 2, Fig. 4a, b). In combination with GISH, we have successfully localized the small chromosome segments of A. elongatum on wheat chromosomes (Figs. 2, 3, 4; Table 3). The reliability of the location was confirmed further by using SSR markers mapped on the specific sites of wheat chromosomes (Figs. 7, 8; Table 4). It is noted from SSR data that Xgwm311, Xgwm265 and Xgwm382 far from the centromere were located on the long arm of 2A. They represent the small fragment on the 2AL (Fig. 8; Table 4). But the other locus Xgwm448 close to the centromere is also determined. This information indicates that chromosome 2A has two small interstitial translocations and one of them is too small to be visible with GISH. Therefore, more SSR data are needed to identify the size and break points of the hybrid in further studies.
It is reported that most of the substitution and translocation occurred on D-genome chromosomes of wheat in the sexual hybrids of wheat and Th. intermedium (Zhang et al. 1991). Another experiment indicated that the Th. intermedium chromosome segment controlling blue-grain character was translocated to D-genome chromosomes of wheat (Ying et al. 2001). In our experiment, four of six A. elongatum chromosome segments were localized on the similar sites of group 1 and 2 chromosomes, and three of six hybridization signals were positioned on D-genome chromosomes (Figs. 3, 4). So, maybe the introgression of the small-chromosome fragment of A. elongatum also prefers some special chromosomes and loci in wheat somatic hybridization.
St, Ee and Eb, the three basic genomes of A. elongatum, were much related to wheat A-, B-, D-genome (Zhang et al. 1999). Thinopyrum bessarabicum (Savul. and Rayss), the donor of Eb genome, has a high salt-tolerant trait(Zhuang et al. 2003). In our experiment, the involvement of E-genome chromatin in the small chromosome segments of A. elongatum was proved by FISH using the peculiar repetitive sequence from E genome (Zhang et al. 1998) as a probe (Figs.5, 6). It is deduced that the salt tolerance of II-1-3 related to Eb chromatin from A. elongatum. In addition, II-1-3 contained special high molecular weight (HMW) glutenin subunits with the same mobility as A. elongatum and had a high quality (Zhao el al. 2003). It has been reported that HMW glutenin subunit genes localized on long arms of wheat group-1 chromosomes (Blasnco et al. 2002). It is worth studying whether the small chromosome segments and the SSR loci of A. elongatum on 1BL and 1DL (Figs.3, 4, 7, 8 and Tables 3, 4) are related with the novel subunits.
This study indicates that the introgressed alien small fragment in somatic hybrid of wheat can be localized with GHSH/FISH/SSR etc., which provides a new mode of practice for wheat marker-assistant breeding.
References
Bijoya B, Aniruddha PS, Hari SG (1999) Transfer of wild abortive cytoplasmic male sterility through protoplast fusion in rice. Mol Breed 5:319–327
Blanco A, Cenci A, Simeone R, Gadaleta A, Pignone D, Galasso I (2002) The cytogenetics and molecular characteristics of a translocated chromosome 1AS1AL-1DL with a GLU-D1 locus in durum wheat. Cell Mol Biol Lett 7:559–567
Chen SY, Xia GM, Quan T, Xiang F, Chen HM (2004) Studies on the salt-tolerance of F3–F6 hybrid Lines orginated from somatic hybridization between common wheat and Thinopyrum ponticum. Plant Science 167:773–779
Doyle JJ, Doyle JI (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Forsberg J, Dixelius C, Lagercrantz U, Glimelius K (1998) UV dose-dependent DNA elimination in asymmetric somatic hybrids between Brassica napus and Arabidopsis thaliana. Plant Sci 131:65–76
Forsström PO, Merker A, Schwarzacher T (2002) Characterization of mildew resistant wheat-rye substitution lines and identification of an inverted chromosome by fluorescent in situ hybridization. Heredity 88(5):349–355
Gill BS (1987) Chromosome banding methods standard chromosome nomenclature and applications in cytogenetic analysis. In: Heyne EG (eds) Wheat and wheat improvement, 2nd edn. American Society of Agronomy, Madison, pp 243–254
Hall RD, Rouwendal GJA, Krens FA (1992) Asymmetric somatic cell hybridization in plants II. Electrophoretic analysis of radiation-induced DNA damage and repair following exposure of sugarbeet (Beta vulgaris L.) protoplasts to UV and garmma rays. Mol Gen Genet 234:315–324
Jiang J, Chen P, Fribe B (1993) Alloplasmic wheat-Elymus ciliaris chromosome addition lines. Genome 37:327–333
Ko JM, Seo BB, Suh DY, Do GS, Park DS, Kwack YH (2002) Production of a new wheat line possessing the 1BL1RS wheat-rye translocation derived from Korean rye cultivar Paldanghomil. Theor Appl Genet 104:171–176
Malysheva L, Sjakste T, Matzk F, Roder M, Ganal M (2003) Molecular cytogenetic analysis of wheat–barley hybrids using genomic in situ hybridization and barley microsatellite markers. Genome 46(2):314–322
Mukai Y, Nakahara Y, Yamamoto M (1993) Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome 36:489–494
Nagy ED, Molnár-Láng M, Linc G, Láng L (2002) Identification of wheat-barley translocations by sequential GISH and two-colour FISH in combination with the use of genetically mapped barley SSR markers. Genome 45:1238–1247
Pedersen C, Langridge P (1997) Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome 40:589–593
Ren ZL, Zhang HQ (1997) Induction of small-segment-transposition between wheat and rye chromosomes. Sci China Ser C 40(3):323–331
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Sear ER (1969) Wheat cytologenetics. Annu Rev Genet 3:451–468
Waara S, Glimelius K (1995) The potential of somatic hybridization in crop breeding. Euphytica 85:217–223
Wang YP, Sonntag K, Rudloff E (2003) Development of rapeseed with high erucic acid content by asymmetric somatic hybridization between Brassica napus and Crambe abyssinica. Theor Appl Genet 106:1147–1155
Wang J, Xiang FN, Xia GM (2004) Transfer of small chromosome fragments of Agropyron elongatum to wheat chromosome via asymmetric somatic hybridization. Sci China 47(4):1–8
Wei YM, Zheng YL, Zhou RH (1999) Detection of the rye chromatin in multispikelet wheat germplasm 10-A background using fluorescence in situ hybridization FISH and RFLP markers. Acta Bot Sin 41(7):722–725
Xia GM, Chen HM (1996) Plant regeneration from intergeneric somatic hybridization between Triticum aestivum L. and Leymus chinensis Trin. Tzvel. Plant Sci 120:197–203
Xia GM, Xiang FN, Zhou AF, Wang H, Chen HM (2003) Asymmetric somatic hybridization between Wheat (Triticum aestivum L) and Agropyron elongatum Host Nevishi. Theor Appl Genet 107:299–305
Xiang FN, Xia GM, Chen HM (2003) Effect of UV dosage on somatic hybridization between common wheat (Triticum aestivum L.) and Avena sativa L. Plant Sci 164:697–707
Xiang FN, Xia GM, Zhi DY, Wang J, Nie H, Chen HM (2004) Hybrid plant regeneration in relation to the nuclear and cytoplasmic genomes of wheat and Setaria italica. Genome 47(4):680–688
Xu J, Conner RL, Laroche A (1994) C-banding and florescence in situ hybridization studies of the wheat-alien hybrid Agrotana. Genome 37:477–481
Xu CH, Xia GM, Zhi DY, Xiang FN, Chen HM (2003) Integration of maize nuclear and mitochondrial DNA into the wheat genome through somatic hybridization. Plant Sci 165:1001–1008
Ying J, Li B, Mu SM, Zhou HP, Liu JZ, Li ZS (2001) Identification of blue-grained wheat translocation lines using fluorescence in situ hybridization. Acta Bot Sin 43(2):164–168
Yue W, Xia GM, Zhi D Y, Chen HM (2001) Transfer of salt tolerance from Aeleuropus littoralis Sinensis to wheat (Triticum aestivum L.) via asymmetric somatic hybridization. Plant Sci 161:259–266
Zhang XY (1991) Production and utilization of alien translocation lines of common wheat. Heredita China 13(5):39–44
Zhang XY, Dong YC (1999) Genome composition of Thinopyrum and rules of new species formation. J Yunnan Univ (Nat Sci) 21:66–67
Zhang XY, Li DY (2000) Repetitive DNA sequences in wheat and its relatives. Sci Agric Sin 33 (5) :1–7
Zhang XY, Dong YC, Li P, Richard R-C Wang (1998) Distribution of E- and St- specific RAPD fragments in few genomes of Triticeae. Acta Genet Sin 25(2):131–141
Zhang ZY, Xin ZY, Larkin PJ (2001) Molecular characterization of a Thinopyrum intermedium Group 2 chromosome (2Ai-2) conferring resistance to barley yellow dwarf virus. Genome 44:1129–1135
Zhao TJ, Quan TY, Xia GM, Chen HM (2003) Glutenin and SDS sedimentation analysis of the F5 somatic hybrids between Triticum aestivum and Agropyron elongatum. J Shandong Univ 38(3):112–116
Zhou AF, Xia GM, Chen HM (2001) Comparative study of symmetric and asymmetric somatic hybridization between common wheat and Haynaldia villosa. Sci China Ser C 31(4):298–305
Zhuang LF, Qi ZJ, Ying J, Chen PD, Liu DJ (2003) Development and identification of a set of Triticum aestivum–Thinopyrum bessarabicum disomic alien addition lines. Acta Genet Sin 30(10):919–925
Acknowledgements
J. Wang and F. Xiang contributed equally to this work. The National Natural Science Foundation of China, No.30370857, Major Project of Ministry of Education in China and National 863 High Technology Research and Development Project No. 2001AA241032 supported this study. We are grateful to Dr. Zhang Xueyong (Chinese Academy of Agriculture Sciences) for providing repeat sequences of B and D genome and control materials.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, J., Xiang, F. & Xia, G. Agropyron elongatum chromatin localization on the wheat chromosomes in an introgression line. Planta 221, 277–286 (2005). https://doi.org/10.1007/s00425-004-1443-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-004-1443-y