Abstract
Plant has a large aquaporin family with more than 30 members which are divided into four subfamilies: plasma membrane intrinsic protein (PIP), tonoplast intrinsic protein (TIP), nodulin 26-like intrinsic proteins (NIP), and small and basic intrinsic proteins (SIP). Their primary structure, transport substrate, functional regulation, gene expression profile, protein amount, and intracellular localization are diversified. The SIP members have short N-terminal tails. Most aquaporins have two sets of common Asn–Pro–Ala (NPA) motif; however, the first motif of SIP1;1, SIP1;2, and SIP2;1 is changed to NPT, NPC, and NPL, respectively. SIP1;1 and SIP1;2, but not SIP2;1, have water transport activity. A recent study revealed that all three members of SIP are localized to the ER membrane and expressed in a cell specific manner in Arabidopsis thaliana. An overview is given on the main features of the SIP members in terms of their primary structure, ER membrane retention, homologues in mammals, and physiological function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction of plant aquaporins
Plant aquaporins comprise a large protein family. Arabidopsis thaliana (hereafter referred to as Arabidopsis), Zea mays, and rice have more than 30 aquaporin-encoding genes, respectively [6, 25, 33]. Plant aquaporins have been classified into four major subfamilies: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin 26-like intrinsic proteins (NIPs), and small and basic intrinsic proteins (SIPs; Fig. 1). The PIP2 group in the PIP subfamily has been demonstrated to play a key role in water flow across the plasma membrane, and the members have been identified in the purified plasma membranes [34, 40]. On the other hand, the physiological roles of the PIP1 group are still unclear. Several members of the PIP1 group did not show water transport activity [4, 37, 39] and showed gene expression profiles different from the PIP2 members [35, 36]. Recently, a few members of the PIP subfamily have been shown to transport carbon dioxide and stimulate photosynthesis [9, 12, 41].
Phylogenetic tree of aquaporins of Arabidopsis thaliana and human aquaporin-11 (AQP11) and -12 (AQP12). Relationships of A. thaliana SIPs with mammalian aquaporins was previously shown in the Phylip rooted phylogenetic tree by Gorelick et al. [11]
There are three major groups in the TIP subfamily, TIP1 (previous name, γ-TIP), TIP2 (δ-TIP), and TIP3 (α-TIP), in addition to TIP4 and TIP5 (Fig. 1). The members of TIP1, TIP2, and TIP4 groups have been demonstrated to be localized to the membranes of central vacuoles including pigment-containing vacuoles, and the TIP3 members are localized to the protein body, which is a vacuolar-derivative organelle [19, 27, 28] (Fig. 2). TIP members have their own characteristic substrates, including water, ammonia (TIP2 members), urea (members of TIP1, TIP2, and TIP4 groups), and glycerol (tobacco TIP) [18, 24, 29]. Recently, Arabidopsis TIP1;1 and TIP1;2 have been reported to mediate transport of hydrogen peroxide as well as human AQP8 [3]. Several reviews are devoted to plant aquaporins, especially physiological roles, gene expression profiles, transport substrates, and functional regulation of PIPs and TIPs [6, 13, 22, 25].
Schematic diagram of plant aquaporins localized at various organelle membranes. PIPs are localized to the plasma membrane, TIPs to the vacuolar membrane and the membrane of protein body, SIPs to the ER membrane, and NIPs to the plasma and ER membranes. In addition to water molecule, several small molecules are reported as transport substrates for these aquaporins as following. PIPs: carbon dioxide and urea; TIPs: urea, ammonia, glycerol, and hydrogen peroxide; and NIPs: ammonia, urea, glycerol, boron, silicon, lactate, and arsenite. Most of them were evidenced, and a few were suggested as candidates of substrate. See the text for the details
Arabidopsis has nine NIP members, which consist of 274–305 amino acid residues. The NIPs, except for NIP6;1, show extremely low expression [1]. NIPs are divided into two groups according to the architecture of their selectivity filter: group I (NIP1;1, NIP1;2, NIP2;1, NIP3;1, NIP4;1 and NIP4;2) and group II (NIP5;1, NIP6;1 and NIP7;1) [42]. The predicted pore structure of group I is similar to that of Glycine max nodulin 26 (GmNOD26), which transports both water and glycerol [8]. Arabidopsis NIP1 members have glycerol transport activity [43]. Arabidopsis NIP5;1, a member of group II, is localized to the plasma membrane and functions as a boric acid [probably a neutral form, B(OH)3] channel to uptake boron in the roots [38]. Rice NIP2;1, whose nearest homologue in Arabidopsis is AtNIP7;1 and different from AtNIP2;1, has been demonstrated to have silicon transport activity [26].
In addition to PIP, TIP, and NIP subfamilies, plants have the SIP subfamily, which is the smallest group of plant aquaporins [21]. Although there was a little information about SIPs for a long time, recent studies have revealed that SIPs are localized to the ER membrane and that SIPs are related to mammalian AQP11 and AQP12 in their intracellular localization and function [16]. In this study, the aim was to give a brief overview on the structural characteristics and the physiological functions of SIPs in comparison with mammalian aquaporins and the ER functions.
Structural characteristics of SIPs
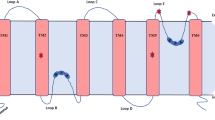
Arabidopsis has three members in the SIP subfamily (SIP1;1, SIP1;2, and SIP2;1) [20], maize has three [5], and rice has two (OsSIP1;1 and OsSIP2;1) [33]. In Arabidopsis, the mean molecular size of SIPs, 25.9 kDa, is the smallest among the four subfamilies (PIP, 30.6 kDa; TIP 26.2 kDa; and NIP, 30.7 kDa). Figure 3 shows the amino acid sequences of Arabidopsis SIPs. AtSIP1;1 and SIP1;2 share high sequence identity of 70%. There is only 26% amino acid sequence identity between SIP2;1 and SIP1;1 or SIP1;2.
Amino acid sequence alignment of three SIP members. Six transmembrane domains (TM1 to TM6) estimated from hydropathy plots are overlined, and two NPA motifs are boxed. Five loops (A to E) are marked by dashed lines. Identical (*) and conservative (′) residues among three sequences are marked. Amino acid sequence identities between SIPs, AQP11, and AQP12 are shown at the bottom
SIP members have the following structural characteristics. (1) The first NPA motifs are changed to NPT, NPC, and NPL in SIP1;1, SIP1;2, and SIP2;1, respectively. Human aquaporins AQP11 and AQP12 have NPC and NPT, respectively, as the first NPA motif [16]. Needless to say, a pair of the Asn–Pro–Ala (NPA) motifs is involved in the selection of substrate through the hydrogen bond between a water molecule and asparagine residue. The variation of the NPA motif might directly reflect the substrate specificity and/or velocity of the water transport as discussed previously [16]. (2) The N-terminal tail is short compared with other plant aquaporins, although the C-terminal tail is the same length as that of PIPs and TIPs. There is a possibility that the N-terminal part is related to the intracellular destination as discussed later. (3) The SIP members are relatively rich in basic residues such as lysine (Fig. 3), and their isoelectric points are higher than the other subfamilies [21]. (4) The loop C (length, 14~19 residues) between TM3 and TM4 is shorter than that of PIPs and TIPs (22~26 residues). The short loop C might affect the tertiary structure of SIPs and suggest a possible unique higher order structure of SIPs. Indeed, it is hard to estimate the structures of SIPs by the computer homology modeling.
The other structural characteristics of SIPs have been described and discussed in an excellent study [21]. By comparisons of sequences and estimation of higher order structure with well-characterized aquaporins AQP1 and GlpF, the aspartic acid residue in the TM1 (Asp11 of SIP1) conserved among three SIPs has been estimated to be important in fixing loop B in the right position by forming a hydrogen bond to the residues in the loop [21].
ER localization of SIPs
Recently, all SIP members have been demonstrated to be localized to the ER membrane by expression of green fluorescent protein (GFP)–SIP fusion proteins in plant cells [14]. Green fluorescence of GFP/SIP fusion proteins was detected on the ER membrane as the same as that of an ER membrane marker Sec22 (Fig. 4). These long, reticular, and sheet-shaped structures are typical in the suspension-cultured cells. Green fluorescence was not detected on the plasma or vacuolar membranes. Free GFP expressed in plant cells was distributed to the cytoplasm and nucleus. If the fusion protein linked with GFP was cleaved at the linkage site, green fluorescence would be detected in the cytoplasm. In both constructs in which GFP was linked to the N or C terminus of SIP, all the SIP members were localized to the ER membrane [15]. These results clearly indicate the ER membrane localization of SIPs. However, there is a possibility that addition of GFP changes the intracellular localization because fusion with GFP, which is composed of 238 amino acids, generates artificial proteins.
Expression of SIP-GFP fusion proteins in protoplasts. Constructs GFP::SIP1;1, SIP1;1::GFP, GFP::SIP2;1, SIP2;1::GFP, GFP::AtSec22, and free GFP were transiently expressed in Arabidopsis suspension-cultured cells. The green fluorescence of GFP-tagged proteins was viewed by confocal laser scanning microscopy (left photograph in each set). Nomarski images were also recorded (middle photographs) and then merged with fluorescence images (right photographs). Bar 10 μm
Ishikawa et al. [15] demonstrated the ER membrane localization of SIPs in plant tissues by a combination of biochemical subcellular fractionation of tissue homogenate and immunochemical detection with antibodies specific to SIP1;1 and SIP2;1. After sucrose density gradient centrifugation, SIPs were recovered in the fractions together with the ER luminal protein BiP not in the Golgi, plasma, or vacuolar membranes. Further, SIPs and BiP were shifted to lighter fractions when the fractionation was carried out in the presence of magnesium chelator ethylenediaminetetraacetic acid, which causes release of ribosomes from rough ER membranes. This means that the SIP proteins are mainly localized to the rough ER. Like SIPs, mouse AQP11 and AQP12 were demonstrated to be localized to the ER membrane by heterologous expression of AQP linked with GFP [17, 31].
What is the ER retention signal for SIPs?
At present, there is no universal model of targeting of the membrane intrinsic proteins. In other words, we cannot estimate the membrane destination of transmembrane proteins from the primary sequences. In yeast and mammals, however, several classes of ER export signals have been identified in the cytosolic domains of transmembrane proteins [2]. Especially, the diacidic motif, such as Glu–His–Asp and Asp–Leu–Glu, is an effective signal for export from ER. The diacidic motifs in transmembrane domains in the cytosolic domains interact with protein components in the coat protein complex II (COPII). Hanton et al. [14] demonstrated that diacidic motif located at the cytoplasmic domains functioned as ER export signal in plant cells for a transmembrane protein GDP-mannose transporter. Plasma membrane K+ channel KAT1 of Arabidopsis has also been demonstrated to require diacidic motifs for correct targeting in plant cells [30]. When the diacidic motifs were deleted or substituted with other amino acids, the transmembrane proteins were localized to the ER. Like other transmembrane proteins, aquaporins are synthesized on the rough ER and then transferred to their destinations. Therefore, the ER retention or ER export signal in aquaporins might be key information for targeting in cells. A KDEL (Lys–Asp–Glu–Leu) sequence is known to be the ER retention signal of ER lumen proteins, but no signal has been identified for the transmembrane proteins.
The PIP members have one or plural diacidic motifs such as Asp–Val–Glu in their N-terminal regions, which face the cytoplasm. On the other hand, all SIP members had no diacidic motif at the N-terminal tails and loops B and D, which face the cytosol. There is an exception, a Glu–Glu–Gln–Glu sequence at the C-terminal tail of SIP2;1, which does not function as the ER export signal. This may be due to the position or steric effect in the tertiary structure as discussed for other transmembrane proteins [30]. At least, the diacidic motifs must be located at the surface of the molecule to interact with COPII proteins. These structural characteristics also theoretically support the ER localization of SIP members.
It should be noted that many transmembrane proteins without diacidic motifs are efficiently exported from the ER. Thus, the presence of the diacidic motif is not the only ticket for export from the ER. As other types of transport signals, a pair of bulky hydrophobic residues, such as Phe–Phe, Leu–Leu, Leu–Phe, Phe–Tyr, and Tyr–Tyr, has been identified in membrane proteins [2]. These di-aromatic or di-hydrophobic motifs are found in the N-terminal tails of PIPs, but not in SIPs. In SIPs, there are di-hydrophobic motifs (Leu–Leu or Phe–Phe stretches) in the loop D. If these stretch functions as ER export signal, the loop D should face the cytoplasm to interact with some protein components for protein trafficking. However, the loop D is very short in most aquaporins and is not accessible for other proteins. In any case, it should be examined whether or not the absence of the ER export motifs determined the ER retention of SIP proteins. Plant aquaporins in various organelles might provide a good system to investigate targeting mechanism of the transmembrane proteins.
SIP homologues in mammalians
Among mammalian aquaporins, only AQP11 and AQP12 have less conserved NPA motifs. Their first NPA motifs are varied to NPC and NPT, respectively. Therefore, the nearest homologues of plant SIP in mammalian are AQP11 and AQP12, as proposed by Ishibashi [16]. Aquaporins with these unique NPA motifs are known only in multicellular organisms. In relation to the similarity to SIPs, the most characteristic point of AQP11 and AQP12 is the ER membrane localization [17, 31]. Gene disruption study was carried out, and AQP11-knockout mice have been reported to be born normally but have vacuolated proximal tubules at birth [32]. These tubules formed cysts to develop polycystic kidneys, which are fatal. Vacuolation means swelling of the ER lumen in the cells and suggests involvement of AQP11 in water transport in ER.
Tissue-specific expression of SIPs
Determination of the cells expressing genes for SIPs may provide information to understand their physiological role. The accumulation of SIP proteins was examined by an immunochemical approach with antibodies specific to individual SIP proteins [15]. A relatively high amount of SIP1;1 protein was detected in young roots and flower buds. SIP2;1 protein was accumulated in young roots and open flowers. Immunoblot analysis of the membrane fractions prepared from plant tissues gives quantitative information on protein accumulation, but not on its cell specificity. On the other hand, morphological analysis of expression profiles of promoter-GUS (β-glucuronidase) provides good information on cell-specific gene expression. Ishikawa et al. [15] revealed characteristic expression patterns of each SIP members. SIP1;1 was expressed in the roots, rosette leaves, and flowers, especially in stamens, and pollens, trichomes in rosette leaves. SIP1;2 was expressed in the cotyledon and hydathode tissue of rosette leaves. SIP2;1 was expressed in the vascular tissue of roots and the leaf veins. SIP1;1 in trichomes, SIP1;2 in hydathodes, and SIP2;1 in leaf veins are typical examples of cell-specific gene expression. Thus, each SIP member may play a role specific to each cell. It should be noted that the SIP proteins were accumulated in the suspension-cultured cells [15], although the most PIPs and TIPs were not accumulated in the suspension-cultured cells [23]. Disappearance of PIPs and TIPs, which are abundant in growing plant tissues, from the suspension cells might be related to weakness of water stress. The accumulation of SIP proteins in the suspension-cultured cells suggests that SIPs have other functions than water channel.
The database of DNA microchip analysis provides comprehensive reliable information on the gene expression in plants. The database shows that the expression level of SIP1;1 is the highest among the three SIP genes (http://www.genevestigator.ethz.ch/at/) [46]. Expression of SIP1;2 is extremely low except for siliques. Pollen shows the highest levels of transcripts of SIP1;1 and SIP2;1. As electron microscopic observation revealed highly developed structures of the ER in pollen cells, the cells may require a large quantity of ER components. There may be two reasons for the cell-specific expression and accumulation of SIPs. (1) Expression of SIP genes is related to the content of the ER in the cells. (2) Expression of SIP genes reflects the specific role of individual SIP in differentiated cells in plants. This point is discussed in the next section.
Water channel activity and physiological roles in ER
SIP1;1 and SIP1;2 have been demonstrated to have water channel activity by stopped-flow light scattering assay of membrane vesicles prepared from yeast cells expressing SIP [15]. In contrast, SIP2;1 gave no activity in the same assay condition. Recently, mouse AQP11 has been shown to have water channel activity with a stopped-flow spectrophotometer using proteoliposomes reconstituted with purified recombinant AQP11 [45]. Thus, SIP1;1 and SIP1;2, but not SIP2;1, have functions and structures relatively similar to those of mammalian AQP11. Transport substrate for SIP2;1 remains to be determined. In addition, the actual or alternative substrates for SIP1;1 and SIP1;2 should be surveyed. As mentioned above, it may be possible to transfer SIP proteins to the plasma membrane by adding the ER export signals in an artificial system. If realized in a yeast heterologous expression system, transport assay using small molecules as substrates of SIP2;1 can be performed easily.
The ER membrane has the widest surface area in plant cells and the diversity in morphology and functionality. In most cases, the ER membranes form a complex meshwork of tubular/reticular/sheet shapes. Biochemical fractionation by sucrose density gradient centrifugation revealed that three SIP proteins were located to the rough ER as mentioned in the previous section. The rough ER is the site of synthesis of membrane and secretary proteins and initial protein glycosylation, quality control of proteins, and refolding of misfolded proteins. The smooth ER is the site of elongation of fatty acids and synthesis of phospholipids and triacylglycerols. The ER also contains enzymes that catalyze reactions to detoxify lipid-soluble drugs and harmful products of metabolism. In Arabidopsis, an ethylene receptor ETR1 that lacks a diacidic motif [44] is located in the ER membrane [7]. Thus, the ER is a site of signal transduction of gaseous plant hormone ethylene. At present, it is unclear whether or not SIPs are related to these ER functions.
We have to answer the question why plants possess water channels at the ER membrane. If SIPs facilitate membrane diffusion of other small molecules, the actual substrate for SIPs should be determined. Other functions, such as membrane adhesion demonstrated in mammalian AQP0 [10], also remain to be examined. We should also examine the possibility that SIPs maintain the tubular/reticular/sheet shapes of the ER. Vacuolation or swelling of the ER in AQP11-knockout mice [32] suggests the involvement of aquaporin in the maintenance of tubular/reticular/sheet shapes of the ER structure. However, it is unclear which function of water channel or membrane adhesion is related to this phenotype in mice.
Mutant analysis of knockout and knockdown of SIP genes will provide much information on their physiological meaning in plants. T-DNA insertion mutants of SIP1;1 and SIP2;1 are available. In the preliminary experiments, the mutant lines did not show any phenotypic properties of growth, morphology, and sensitivities to physiological and physical stresses (data not shown, Maeshima). Structural change of the ER in the mutant plants remains to be examined. Double or triple knockout mutant lines and knockdown mutant lines are needed to understand the physiological roles of SIPs in plants.
This short review has highlighted all SIP members, which have incomplete NPA motifs and have been localized to the ER in plant cells. The ER-localized aquaporins have been identified only in multicellular organisms including higher plants and animals. Thus, these ER membrane aquaporins may be tightly related to the unique functions or structures of the ER in multicellular systems. The ER is heterogenic in function, structure, and physical linkage/interaction with other organelles, such as Golgi apparatus, nuclear envelope, vacuoles, oil bodies, plasma membrane, small vesicles, and actin filaments. Further investigation including visualization of the ER in living cells and survey of the actual transport substrate for SIPs is needed to clarify the physiological roles of the ER membrane aquaporins. We should also determine higher order structures of SIPs because their primary structures are considerably different from the other normal aquaporins, including plant PIP and human AQP0. Further studies are needed to define the physiological roles of the ER membrane aquaporins in living organisms.
References
Alexandersson E, Fraysse L, Sjovall-Larsen S, Gustavsson S, Fellert M, Karlsson M, Johanson U, Kjellbom P (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Mol Biol 59:469–484
Barlowe C (2003) Signals for COPII-dependent export from the ER: what’s the ticket out? Trends Cell Biol 13:295–300
Bienert GP, Møller ALB, Kristiansen KA, Schulz A, Møller IM, Schjoerring JK, Jahn TP (2007) Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J Biol Chem 282:1183–1192
Chaumont F, Barrieu F, Jung R, Chrispeels MJ (2000) Plasma membrane intrinsic proteins from maize cluster in two sequence subgroups with differential aquaporin activity. Plant Physiol 122:1025–1034
Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R (2001) Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol 125:1206–1215
Chaumont F, Moshelion M, Daniels MK (2005) Regulation of plant aquaporin activity. Biol Cell 97:749–764
Chen Y-F, Randlett MD, Findell JL, Schaller GE (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem 277:19861–19866
Dean RM, Rivers RL, Zeidel ML, Roberts DM (1999) Purification and functional reconstitution of soybean nodulin 26: an aquaporin with water and glycerol transport properties. Biochemistry 38:347–353
Flexas J, Ribas-Carbó M, Hanson DT, Bota J, Otto B, Cifre J, McDowell N, Medrano H, Kaldenhoff R (2006) Tobacco aquaporin NtAQP1 is involved in mesophyll conductance to CO2 in vivo. Plant J 48:427–439
Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T (2005) Lipid–protein interactions in double-layered two-dimensional AQP0 crystals. Nature 438:633–638
Gorelick DA, Praetorius J, Tsunenari T, Nielsen S, Agre P (2006) Aquaporin-11: a channel protein lacking apparent transport function expressed in brain. BMC Biochem 7:14
Hanba YT, Shibasaka M, Hayashi Y, Hayakawa T, Kasamo K, Terashima I, Katsuhara M (2004) Overexpression of the barley aquaporin HvPIP2;1 increases internal CO2 conductance and CO2 assimilation in the leaves of transgenic rice plants. Plant Cell Physiol 45:521–529
Hachez C, Zelazny E, Chaumont F (2006) Modulating the expression of aquaporin genes in planta: a key to understand their physiological functions? Biochim Biophys Acta 1758:1142–1156
Hanton SL, Renna L, Bortolotti LE, Chatre L, Stefano G, Brandizzi F (2005) Diacidic motifs influence the export of transmembrane proteins from the endoplasmic reticulum in plant cells. Plant Cell 17:3081–3093
Ishikawa F, Suga S, Uemura T, Sato MH, Maeshima M (2005) Three SIP aquaporins of Arabidopsis are localized in the ER membrane and expressed in a tissue- and cell-specific manner. FEBS Lett 579:5814–5820
Ishibashi K (2006) Aquaporin subfamily with unusual NPA boxes. Biochem Biophys Acta 1758:989–993
Itoh T, Rai T, Kuwahara M, Ko SBH, Uchida S, Sasaki S, Ishibashi K (2005) Identification of a novel aquaporin, AQP12, expressed in pancreatic acinar cells. Biochem Biophys Res Commun 330:832–838
Jahn TP, MØller ALB, Zeuthen T, Holm LM, Klaerke DA, Mohsin B, Kuhlbratndt WK, Schjoerring JK (2004) Aquaporin homologues in plants and mammals transport ammonia. FEBS Lett 574:31–36
Jiang L, Phillips TE, Hamm CA, Drozdowicz YM, Rea PA, Maeshima M, Rogers SW, Rogers JC (2001) The protein storage vacuole: a unique compound organelle. J Biol Chem 155:991–1002
Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjövall S, Fraysse L, Weig AR, Kjellbom P (2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for new nomenclature for major intrinsic proteins in plants. Plant Physiol 126:1358–1369
Johanson U, Gustavsson S (2002) A new subfamily of major intrinsic proteins in plants. Mol Biol Evol 19:456–461
Kaldenhoff R, Fischer M (2006) Functional aquaporin diversity in plants. Biochem Biophys Acta 1758:1134–1141
Kobae Y, Mizutani M, Segami S, Maeshima M (2006) Immunochemical analysis of aquaporin isoforms in Arabidopsis suspension cultured cells. Biosci Biotech Biochem 70:980–987
Loque D, Ludewig U, Yuan L, von Wiren N (2005) Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol 137:671–680
Luu DT, Maurel C (2005) Aquaporins in a challenging environment: molecular gears for adjusting plant water status. Plant Cell Environ 28:85–96
Ma JF, Tamai K, Yamaji N, Mitani N, Konishi S, Ishiguro M, Katsuhara M, Murata Y, Yano M (2006) Silicon transporter in rice. Nature 440:688–691
Maeshima M, Hara-Nishimura I, Takeuchi Y, Nishimura M (1994) Accumulation of vacuolar H+-pyrophosphatase and H+-ATPase during reformation of the central vacuole in germinating pumpkin seeds. Plant Physiol 106:61–69
Maeshima M (2001) Tonoplast transporters: organization and function. Annu Rev Plant Physiol Plant Mol Biol 52:469–497
Maurel C (2007) Plant aquaporins: Novel functions and regulation properties. FEBS Lett 581:2227–2236
Mikosch M, Hurst AC, Hertel B, Homann U (2006) Diacidic motif is required for efficient transport of the K+ channel KAT1 to the plasma membrane. Plant Physiol 142:923–930
Morishita Y, Matsuzaki T, Hara-Chikuma M, Andoo A, Shimono M, Matsuki A, Kobayashi K, Ikeda M, Yamamoto T, Verkman A, Kusano E, Ookawara S, Takata K, Sasaki S, and Ishibashi K (2005) Disruption of aquaporin-11 produces polycystic kidneys following vacuolization of the proximal tubule. Mol Cell Biol 25:7770–7779
Mizutani M, Watanabe S, Nakagawa T, Maeshima M (2006) Aquaporin NIP2;1 is mainly localized to the ER membrane and shows root-specific accumulation in Arabidopsis thaliana. Plant Cell Physiol 47:1420–1426
Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M (2005) Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol 46:1568–1577
Santoni V, Verdoucq L, Sommerer N, Vinh J, Pflieger D, Maurel C (2006) Methylation of aquaporins in plant plasma membrane. Biochem J 400:189–197
Suga S, Imagawa S, Maeshima M (2001) Specificity of the accumulation of mRNAs and proteins of the plasma membrane and tonoplast aquaporins in radish organs. Planta 212:294–304
Suga S, Komatsu S, Maeshima M (2002) Aquaporin isoforms responsive to salt and water stresses and phytohormones in radish seedlings. Plant Cell Physiol 43:1229–1237
Suga S, Maeshima M (2004) Water channel activity of radish plasma membrane aquaporins heterologously expressed in yeast and their modification by site-directed mutagenesis. Plant Cell Physiol 45:823–830
Takano J, Wada M, Ludewig U, Schaaf G, Wirén N, Fujiwara T (2006) The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18:1498–1509
Tournaire-Roux C, Sutka M, Javot H, Gout E, Gerbeau P, Luu DFT, Bligny R, Maurel C (2003) Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 425:393–397
Törnroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, Neutze R, Kjellbom P (2006) Structural mechanism of plant aquaporin gating. Nature 439:688–694
Uehlein N, Lovisolo C, Siefritz F, Kalenhoff R (2003) The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 425:734–737
Wallace IS, Choi WG, Roberts DM (2006) The structure, function and regulation of the nodulin 26-like intrinsic protein family of plant aquaglyceroporins. Biochem Biophys Acta 1758:1165–1175
Weig AR, Jakob C (2000) Functional identification of the glycerol permease activity of Arabidopsis thaliana NLM1 and NLM2 proteins by heterologous expression in Saccharomyces cerevisiae. FEBS Lett 481:293–298
Wilkinson JQ, Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ (1995) An ethylene-inducible component of signal transduction encoded by Never-ripe. Science 270:1809–1813
Yakata K, Hiroaki Y, Ishibashi K, Sohara E, Sasaki S, Mitsuoka K, Fujiyoshi Y (2007) Aquaporin-11 containing a divergent NPA motif has normal water channel activity. Biochem Biophys Acta 1768:688–693
Zimmermann P, Hennig L, Gruissem W (2005) Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci 10:407–409
Acknowledgments
We thank Sei Sasaki for the chance to write this review. Work in our laboratory was supported by the Program for Promotion of Basic Research Activities of Innovative Biosciences (PROBRAIN), RITE, Grants-in-Aid for Scientific Research (18380064 and 16085204) from the Ministry of Education, Sports, Culture, Science and Technology of Japan, and the Global Research Program of the Ministry of Science and Technology of Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Maeshima, M., Ishikawa, F. ER membrane aquaporins in plants. Pflugers Arch - Eur J Physiol 456, 709–716 (2008). https://doi.org/10.1007/s00424-007-0363-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-007-0363-7