Abstract
Purpose
Strenuous exercise induces a rapid and transient elevation of cell free DNA (cfDNA) concentration in blood plasma. The detection of cfDNA in the presence of plasma nucleases could indicate an association of cfDNA with protective vesicular structures. Several cell types release extracellular vesicles (EVs), including exosomes and shedding microvesicles, which are known to mediate the exchange of proteins and nucleic acids (largely RNA) between cells. Here, we assessed whether EVs play a role in the exercise-dependent release of cfDNA in blood plasma.
Methods
Venous blood collected from healthy volunteers before and after incremental treadmill exercise was separated into vesicular (EV) and soluble fractions. Nuclear and mitochondrial DNA content in plasma supernatants and EV fractions was determined by quantitative real-time PCR (qPCR).
Results
We show that the majority of cfDNA is located in the plasma supernatants. Only minute amounts of DNA were observed in the EV-associated fractions including microvesicles and exosomes. Nuclear and mitochondrial DNA species differ in terms of their quantities in the several plasma fractions.
Conclusions
Our results indicate that cfDNA liberated in response to acute physical exercise is not released by vesicular means and circulates in a soluble form in blood plasma which could indicate different biological functions exerted by cfDNA and EVs. The different nature of DNA species in plasma has major implications for the preparation of plasma and other bodily fluids prior to analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The analysis of cell free DNA (cfDNA) in blood plasma is becoming increasingly important in biomedical disciplines. Physiologically low concentrations of cfDNA increase due to a variety of acute and chronic pathological conditions such as cancer, autoimmune diseases, sepsis, stroke, myocardial infarction and trauma (Holdenrieder and Stieber 2009; Schwarzenbach et al. 2011; Swarup and Rajeswari 2007; Ziegler et al. 2002). Although studies have already proved the diagnostic and prognostic value of cfDNA concentrations for several of these disorders (Yi et al. 2014), the nature and release mechanisms of cfDNA are not fully understood. Depending on the physiological condition, there could be different mechanisms of releasing cfDNA into the blood stream. The main source of disease-associated cfDNA elevations is thought to be apoptosis and necrosis, thereby reflecting the degree of cellular damage (Yi et al. 2014). In cancer patients, additional active secretory mechanisms are proposed (Schwarzenbach et al. 2011). However, elevated concentrations of cfDNA in blood plasma have also been reported after acute bouts of physical exercise (Atamaniuk et al. 2004; Beiter et al. 2011). Since the appearance of DNA fragments in the bloodstream in response to cell death events would take several hours, different mechanisms of release must be evident in the case of short exercise settings that are characterised by already rising cfDNA levels during the first steps of an incremental exercise test (Breitbach et al. 2014). Recently, the elevation of cfDNA concentration observed under physical exercise was attributed to netting neutrophils (Beiter et al. 2014). In this process, the formation of neutrophil extracellular traps (NETs) occurs via a vesicular mechanism in which vesicles, filled with nuclear DNA, were extruded in the extracellular space (Pilsczek et al. 2010).
The existence of cfDNA in the bloodstream indicates a protection of the released DNA from nucleases, which could be mediated by the inclusion of cfDNA within vesicles (Lee et al. 2014). Numerous cell types release different classes of vesicles into their environment. Consequently, a mixture of these extracellular vesicles (EVs) is present in body fluids such as blood (Raposo and Stoorvogel 2013). EVs comprise apoptotic bodies released from dying cells as well as shedding microvesicles (MVs) and exosomes secreted by healthy cells (Raposo and Stoorvogel 2013; Thery 2011). EVs differ in size, cargo and release mechanisms, but notably, exhibit overlapping features that challenge nomenclature (Gould and Raposo 2013; Kowal et al. 2014). MVs with a size of 100–1000 nm in diameter directly pinch off from the plasma membrane, whereas smaller exosomes (50–100 nm) are generated in the endosomal system and released by fusion of multivesicular bodies (MVBs) with the plasma membrane (Bobrie et al. 2011). One of the first discovered functions of exosomes was their role in protein disposal during reticulocyte maturation (Harding et al. 2013). Moreover, platelets release MV and exosomes in the process of their activation (Aatonen et al. 2012). In the past decade, it has become evident that EVs carry distinct RNA species (Valadi et al. 2007) and are implicated in intercellular communication (Fruhbeis et al. 2013; Robbins and Morelli 2014; Simons and Raposo 2009), e.g. by participating in immune responses (Thery et al. 2009). Furthermore, EVs have been associated with disease progression including neurodegenerative, cardiovascular and cancerous diseases (Bellingham et al. 2012; Hergenreider et al. 2012; Luga et al. 2012; Peinado et al. 2012). Since EVs carry characteristic lipids, proteins and RNA species reflecting the cell type of origin (Kalra et al. 2012), they may serve as disease biomarkers and helpful tools for point of care diagnostics (Revenfeld et al. 2014). Notably, in several pathological conditions, including cancer and autoimmune diseases, which are characterised by increased cfDNA concentrations, elevated EV levels were also detected in plasma or serum (Holdenrieder and Stieber 2009; Horstman et al. 2007; Lee et al. 2014; Pisetsky 2009; Schwarzenbach et al. 2011).
In this study, we assess the role of EVs in the exercise-dependent release of cfDNA. Whole blood was collected from healthy volunteers before and after an incremental treadmill exercise test. The nuclear and mitochondrial DNA content was investigated in plasma supernatants and EV fractions. We show that the vast majority of cfDNA emitted during physical exercise is not associated with EVs.
Materials and methods
Ethical approval
All experimental procedures were approved by the Human Ethics Committee Rhineland-Palatine and conformed to the standards of the Declaration of Helsinki of the World Medical Association. All subjects were informed orally and in writing about the procedures and the aim of the study and gave written consent to participate.
Subjects and exercise setting
Five healthy, physically active (physical activity >3 h per week), non-smoking men volunteered to participate in this study. Subjects were excluded if they had any signs of infection or other diseases, injuries or took prescribed medications including anticoagulant treatment (e.g. aspirin). The participants were asked to abstain from physical exercise 24 h before the test since physical exercise influences physiological pathways, which is reflected by rising cfDNA concentrations, changes in blood cells and other laboratory values (Breitbach et al. 2014). We wanted to ensure all values at baseline when starting the test. All tests were conducted in the morning at 9:00 AM after taking a standardised breakfast.
The subjects performed an incremental exercise test on a motorised treadmill (HP Cosmos, Traunstein, Germany) with starting speed at 6 km/h and increasing velocity by 2 km/h every 3 min with a constant incline of 1.5 % until exhaustion. During the test heart rate, ventilatory und metabolic responses were monitored. After each exercise step, subjects were inquired for Borg scale value. For lactate measurement, 20 µl of capillary blood was collected from the earlobe before the run, at the end of each exercise step, immediately after the run and after 3, 5, 10, 30 and 90 min of recovery. The samples were measured with the lactate analyser Biosen 5130 (EKF Diagnostics, Magdeburg, Germany). 30 ml of blood was taken from the medial cubital vein with a Safety-Multifly® needle (0.8 × 19 mm) (Sarstedt, Nümbrecht, Germany) and collected in tripotassium-EDTA covered Monovettes® (Sarstedt, Nümbrecht, Germany) before (pre), immediately after (post), 10 min (10+), 30 min (30+) and 90 min after (90+) the treadmill test. Venous blood was analysed for complete blood counts before, immediately after and 90 min after the exercise.
Plasma collection and fractionation of EVs
Isolation of EVs was performed by differential centrifugation with modifications as described before (Krämer-Albers et al. 2007). Immediately after draw (maximum 5 min delay) blood was centrifuged at 1600×g for 10 min at 4 °C in an Eppendorf FA-45-6-30 rotor (Eppendorf, Hamburg, Germany) (“total plasma”). The plasma fraction was transferred carefully and 4 ml of the supernatant were centrifuged at 10,000×g for 30 min at 4 °C (“10,000×g SN”). The remaining pellet was washed two times with PBS to remove soluble proteins (“10,000×g pellet”). 3 ml of the 10,000×g supernatant was filtered through 0.2 µm syringe filters (Millex-GP, Millipore, Merck, Darmstadt, Germany) and 2 × 1.4 ml of the filtrate was centrifuged for 2 h at 47,000 rpm (100,000×g) and 4 °C in a Beckman TLA55 rotor (k factor at maximum speed: 66) (Beckman Coulter, Krefeld, Germany) to pellet EVs including smaller MVs and exosomes. The supernatant was retained (“100,000×g SN”). Both membrane pellets were resuspended in 20 µl DNase buffer. The samples were treated with 5 units DNase I (Roche Diagnostics, Mannheim, Germany) or in the absence of DNase I for 20 min at 37 °C (“100,000×g pellet +DNase I” and “100,000×g pellet”, respectively) and diluted in 1 ml PBS before final pelleting the membrane particles for 2 h at 100.000×g. The samples were stored at −20 °C and DNA was extracted within a maximum of 1 week.
DNA extraction
DNA was extracted from the different plasma fractions using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations. Prior to DNA extraction, the pellets were diluted in the kit’s ACL lysis buffer adjusted to 1/10 of the initial plasma volume (equivalent to 10 µl of plasma per µl lysate). 700 µl of the supernatants and 70 µl of the pellets were processed with the QIAamp Circulating Nucleic Acid Kit and DNA was eluted in 70 µl of the kit’s AVE elution buffer (final concentration equivalent to 10 µl of plasma per µl of extract). DNA samples were stored at −20 °C and concentrations were measured within a maximum of 1 week.
Quantification of nuclear and mitochondrial cfDNA
Quantification of total nuclear cfDNA was based on the amplification of long terminal repeat (LTR) sequences of the human endogenous retrovirus group K (HERVK) family LTR5 Hs. Sequence information for primer design was obtained from the UCSC Genome Browser on the February 2009 human reference sequence (GRCh37) (Rhead et al. 2010). The online software Primer3 was used to design multilocus primers 5′-ACC GAG ACA TTC CAT TGC C-3′ and 5′-GCC TCT TGC AGT TGA GAC AAG-3′ targeting a 70-bp fragment of an LTR5 sequence with 195 matches in the human haploid genome. The binding frequency was assessed by aligning the primers using the in silico PCR tool of the UCSC Genome Browser. For the quantification of mitochondrial cfDNA, we used primers described by Beiter et al. (2011).
Samples were analysed by qPCR with a CFX384 Touch™ Real-Time PCR detection system (Bio-Rad, München, Germany) using the following protocol for the amplification of nuclear DNA: 5 min incubation at 95 °C, followed by 40 cycles of denaturation at 94 °C for 15 s, annealing at 61 °C for 30 s and extension at 80 °C for 30 s. Amplification of mitochondrial DNA was performed under the following conditions: 5 min incubation at 95 °C, 40 cycles of denaturation at 94 °C for 15 s, annealing at 59 °C for 30 s, and extension at 72 °C for 10 s. The experiments were performed in triplicate with a final volume of 5 µl per single reaction including 1.6 µl of template DNA, 2.6 µl PCR master mix containing 0.1 U/µl HotStarTaq Plus Polymerase (Qiagen, Hilden, Germany), 2× PCR buffer (Qiagen, Hilden, Germany), 1 mM MgCl2 (Qiagen, Hilden, Germany), 0.4 mM dNTPs (Carl Roth, Karlsruhe, Germany), 0.28× SYBR green (Sigma–Aldrich, Taufkirchen, Germany), 5 nM FITC (Sigma-Aldrich, Taufkirchen, Germany) and 0.8 µl primer mix of a final concentration of 312 nM. Nontemplate controls and positive controls for interplate calibration were also analysed in triplicate in each PCR run. Formation of the expected PCR product was confirmed by melting curve analysis.
The quantification of DNA concentrations was based on serially diluted genomic reference standards of known concentrations including the target sequences for the respective primers. The standards were generated from human genomic DNA (Novagen, Merck, Darmstadt, Germany) by PCR using primers 5′ TTC TCA AAG AGG GGG ATG TG 3′ and 5′ GTG GGA AGG GAA AGA CCT GA 3′ to amplify a 400-bp fragment of an LTR-sequence and using primers as described by Beiter et al. (2011) to amplify an 85-bp fragment of the mitochondrial genome. The amplifications were performed on a Mini Thermal Cycler (Bio-Rad, München, Germany) with the following conditions for the LTR standard: initial denaturation at 95 °C for 5 min and 40 cycles of denaturation at 94 °C for 15 s, annealing at 59 °C for 30 s, and extension at 72 °C for 30 s. The protocol for the amplification of the mitochondrial sequence differed in terms of annealing at 55 °C for 15 s. The PCR products were TA cloned in a pCR® 2.1 vector (Invitrogen, Carlsbad, CA, USA) and the sequences were confirmed by Sanger sequencing (StarSEQ®, Mainz, Germany). For standard preparation, the PCR products were cut out of the vector, and stock solutions of genomic reference standards were determined fluorophotometrically (NanoDrop 3300, Thermo Fisher Scientific Inc., Waltham, MA). DNA concentrations were converted into copy numbers by using the online program Finnzymes DNA copy number calculation. Standards were prepared in defined copy numbers, ranging from 2 × 105 to 50 copies/µl for the LTR5 assay and from 2 × 105 to 25 copies/µl for the mitochondrial qPCR assay and subjected to qPCR analysis. Standard curves were constructed by plotting the quantification cycle (Cq) value against the logarithm of calibrator copy number in each dilution. The copy number calculation was based on the molecular weight of a human haploid genome of 3.3 pg with one copy being equivalent to one molecule of double-stranded DNA. Mitochondrial cfDNA concentrations were given as copies/ml plasma, based on the equation by Chiu et al. (2003). Nuclear cfDNA results were converted from copy numbers to ng/ml plasma.
We assessed the lower limit of quantification (LOQ), defined as the minimal concentration that could be quantified with 80 % accuracy (Beiter et al. 2011). The LOQ was determined with each genomic reference standard measured in seven replicates. The LOQ of the LTR5 assay was set to 50 copies/µl, corresponding to a DNA concentration of 0.78 pg/µl eluate. The LOQ of the mitochondrial qPCR assay was determined at 25 copies/µl eluate. Since the eluate was tenfold concentrated with the DNA extraction method used here, samples with a nuclear DNA concentration below 0.078 ng/ml plasma and with a mitochondrial DNA concentration below 2500 copies/ml plasma were not quantifiable.
Western blot analysis
The 10,000×g and 100,000×g pellets were resuspended in sample buffer and subjected to SDS-PAGE (4–12 % Bis–Tris gel) and Western blotting (NuPAGE, Life Technologies, Darmstadt, Germany). Proteins were blotted onto a PVDF membrane. Next, the membrane was blocked with 4 % milk powder and 0.1 % Tween in PBS and incubated sequentially with primary and HRP-coupled secondary antibodies and chemiluminescence reagents.
The following antibodies were used: mouse anti-Hsp70 (SC-24; Santa Cruz, Heidelberg, Germany, 1:1000), rabbit anti-Flotillin-1 (F1180, Sigma–Aldrich, Taufkirchen, Germany, 1:1000) and HRP-coupled secondary antibodies (Goat-anti-Mouse-HRP, 115-035-003, 1:10,000; Goat-anti-Rabbit-HRP, 111-035-003, 1:10,000, Dianova, Hamburg, Germany).
Statistical analysis
The qPCR data were captured with the CFX Manager Software, Version 3.0 (Bio-Rad, München, Germany). Microsoft® Excel 2007 was used for data analyses. We considered p values <0.05 to be statistically significant and performed statistical analysis with IBM SPSS statistics 22 (IBM, Ehningen, Germany). Unless otherwise indicated, all data were presented as mean (±SD). Quantitative changes in variables at the various times or differences in cfDNA concentrations between the plasma fractions were compared by multivariate analysis of variance (MANOVA) with Bonferroni correction. Data were excluded from statistical analyses if DNA concentrations were outside the quantification range (LOQ) of the respective qPCR assay.
Results
Exercise test
Five healthy, physically active men performed an incremental treadmill exercise test until volitional exhaustion. To determine the individual exhaustion, the values of VO2max, lactate and perceived exertion expressed by Borg values were analysed immediately after the exercise test (Table 1). The individual anaerobic threshold (IAT) was determined on the basis of lactate accumulations according to the Dickhuth model (Dickhuth et al. 1999).
Physical exercise induces changes in blood values
To investigate if changes in cfDNA are accompanied by changes in blood values during physical exercise, we analysed full blood counts and lactate before and after the treadmill test (Table 2). Lactate and numbers of erythrocytes, platelets, lymphocytes and monocytes were significantly elevated immediately after exercise. After 90 min of recovery, erythrocytes, platelets, lymphocytes and monocytes reached baseline levels. Neutrophil counts increased after exercise and during recovery, but did not reach statistical significance.
Most of the cfDNA released after exercise is not associated with EVs
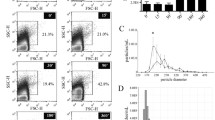
To analyse if the increase in cfDNA concentrations observed after physical exercise is associated with EV release, we collected venous blood before and after an incremental treadmill test and separated blood plasma by differential centrifugation into distinct soluble and EV fractions. Larger MVs, apoptotic bodies and cell debris were pelleted at 10,000×g. Subsequently, smaller MVs and exosomes were isolated by filtration through a 200-nm filter and centrifugation at 100,000×g. Both EV fractions were analysed by Western blotting for marker proteins. Flotillin 1 and Hsc/Hsp70 were detected in 10,000×g (Fig. 1a) and 100,000×g pellets (Fig. 1b) confirming the presence of EVs including MVs and exosomes. The markers indicated decreased levels of MVs in plasma immediately after exercise before returning to baseline, while levels of smaller MVs and exosomes appeared slightly increased in response to exercise (Frühbeis et al. 2015).
Next, we investigated the nuclear and mitochondrial DNA content of all plasma fractions. The amounts of DNA were quantified by two different qPCR assays, targeting genomic LTR and mitochondrial loci. To investigate if DNA detected in association with EVs was located inside EVs or on the surface, 100,000×g pellets were resuspended and treated with DNase to digest DNA bound to the outer surface of the vesicles.
Upon exhaustive exercise, nuclear cfDNA increased in each plasma fraction around tenfold, followed by a decline to baseline levels within 90 min of recovery (Fig. 2a; Table 3). A 20-fold increase of cfDNA in response to exercise was observed in untreated 100,000×g pellets. After DNase treatment, DNA concentrations reached fold change levels comparable to the other fractions, indicating that simply more cfDNA was bound to the vesicle surface.
Kinetics of mean (a) nuclear and (b) mitochondrial cfDNA concentrations in five healthy subjects before and after an incremental treadmill exercise. LOQ indicates the lower limit for the quantification of DNA concentrations. Error bars indicate ± SD. SN supernatant. Total plasma, 1600×g supernatant; 10,000×g SN, 10,000×g supernatant; 100,000×g SN, 100,000×g supernatant. For further details please see Table 3
The highest amounts of nuclear DNA were observed in the supernatants, whereas nuclear DNA in the EV fractions was scarce. Across all time points, we detected mean (±SD) 4.67 (4.22) % of total plasma DNA (represented by the 1600×g supernatant) in the 10,000×g pellet, 3.94 (3.16) % in the untreated 100,000×g pellets and 0.94 (0.86) % in DNase-treated 100,000×g pellets. In addition, the amount of nuclear DNA in the remaining supernatants after subsequent centrifugation at 1600×g, 10,000×g and 100,000×g was nearly similar, demonstrating that centrifugation and filtration had no critical influence on the cfDNA levels in supernatants. Thus, the major part of the released nuclear DNA exists in a non-vesicle-bound form in blood plasma. Moreover, considering the low amounts of nuclear DNA in 10,000×g and 100,000×g pellets, the contribution of EV-associated DNA to the total nuclear cfDNA pool is marginal in healthy individuals. These findings are not restricted to eccentric running exercise, since concentric bicycle exercise revealed similar results (Supplemental Figure S1).
In contrast to nuclear cfDNA, we could not determine a significant increase in mitochondrial DNA in any fraction (Fig. 2b; Table 3). The majority [mean (±SD) 48.39 (21.25) %] of mitochondrial DNA was observed in the 10,000×g pellet, which is expected to include mitochondrial fragments pelleting together with MVs. Trace amounts of mitochondrial DNA were identified in the 100,000×g supernatants and pellets. We detected only (mean ± SD) 0.12 (0.11) % of total mitochondrial DNA in the untreated 100,000×g pellets and 0.04 (0.03) % in the DNase-treated 100,000×g pellets. In summary, most of the cfDNA liberated in the blood stream during physical exercise is released independent of EVs into the soluble plasma fraction.
The results also permit some conclusions concerning pre-analytical considerations. It was recommended to perform a first centrifugation step at 1200–1600×g and a second step at 16,000×g to remove cellular debris which could influence cfDNA quantification (El Messaoudi et al. 2013). Our results indicate that this second step is not required when analysing nuclear cfDNA. Moreover, longer cfDNA fragments (≥272 bp) quantified with the LTR-based qPCR assay described here tend to decrease with further centrifugation and filtration steps (data not shown). Recently, it was shown that the relative amount of longer cfDNA fragments in the total cfDNA pool is increased after physical exercise, which could reflect the degree of cellular damage or inflammatory reactions (Breitbach et al. 2014). Therefore, losing longer fragments after high-speed centrifugation could imply the loss of additional information concerning athletic performance. In the case of mitochondrial DNA, different centrifugation protocols would considerably affect DNA concentrations. Filtration and ultracentrifugation of the blood plasma decrease mitochondrial cfDNA concentration. It was suggested that particle-associated mitochondrial DNA, represented in the pellets, and a free form of mitochondrial DNA, represented in the 100,000×g supernatant, coexist in plasma (Chiu et al. 2003). This should be taken into account when analysing mitochondrial DNA in blood plasma.
Discussion
The potential of cfDNA as biomarker for exercise and clinical diagnostics is challenged by open questions concerning the cellular source and release mechanisms. Our study addresses primarily the role of EVs in cfDNA release. While the dynamics of cfDNA und EVs with regard to their levels detected in plasma coincides under several conditions, we find that they are not linked.
Our study may allow limited interpretation concerning the cellular source of cfDNA, considering the observed changes in full blood counts during physical exercise. Acute physical exercise exerts numerous effects on, e.g. metabolic and immune functions, as reflected by transient hyperlactatemia, lymphocytosis, neutrophilia, monocytosis and activation of leucocytes and platelets (Adeva-Andany et al. 2014; Chaar et al. 2011; Walsh et al. 2011). We observed parallel accumulations of nuclear cfDNA and lymphocyte counts. T cell activation could be driven by acute exercise and in response to mitogen and antigen stimulation (Walsh et al. 2011). Interestingly, it was reported that lymphocytes secrete DNA in vitro after mitogen stimulation or in the presence of antigens (Galeazzi et al. 2003). In a few cases, T cell-specific but no endothelial cell-specific DNA could be detected in the plasma of cancer patients (Jahr et al. 2001). We also observed parallel kinetics of erythrocyte counts and cfDNA. However, mature erythrocytes do not contain DNA, but enucleation of erythroblasts during erythropoiesis could contribute to the cfDNA pool in blood plasma (Nagata et al. 2010). Although it was shown that cfDNA detected in blood plasma originates predominantly from the hematopoietic cell lineage (Lui et al. 2002), the contribution of distinct cell subsets is currently unknown. The identification of cell-specific methylation patterns in cfDNA could help solve this question.
According to other reports (Beiter et al. 2011; Breitbach et al. 2014), we showed that short-term treadmill exercise induces the release of nuclear cfDNA. This phenomenon was also observed in different exercise settings, e.g. endurance treadmill exercise (Fatouros et al. 2010), half-marathon (Atamaniuk et al. 2004), ultra-marathon (Atamaniuk et al. 2008) and weightlifting (Atamaniuk et al. 2010). An early study on cfDNA kinetics during physical exercise attributed elevated cfDNA concentrations after a 6-h ultra-marathon to muscle damage and apoptosis of skeletal and immune cells (Atamaniuk et al. 2008). Later, it was shown that short-term exercise is also sufficient to induce a cfDNA response, with increasing levels in the first steps of incremental exercise (Beiter et al. 2011; Breitbach et al. 2014). Rapidly increasing concentrations of nuclear cfDNA during exercise could not be attributed to cell death events which take several hours (Blagosklonny 2000). Moreover, the kinetics of cfDNA and muscle damage markers like myoglobin and creatine kinase in response to endurance exercise revealed completely different patterns (Beiter et al. 2014; Fatouros et al. 2010). Concentrations of plasma creatine kinase peaked 24 h after finishing a 90-min treadmill run when cfDNA concentrations were already at baseline (Fatouros et al. 2010). These results were confirmed by the lack of correlation between cfDNA and the cytoplasmic muscle components myoglobin and fatty acid-binding protein (FASB), which also showed a delayed response to 60-min high-intense cycling compared to cfDNA (Beiter et al. 2014). Furthermore, acute short-term bicycle exercise was not sufficient to cause DNA damage or to activate apoptotic pathways in mononuclear blood cells of trained and untrained individuals (Ceci et al. 2013). The amount of mitochondrial DNA, maybe mainly represented by particles containing large quantities of mitochondrial DNA, like e.g. mitochondria or apoptotic vesicles (Chiu et al. 2003; Thery et al. 2009), did not increase in response to exercise. The steady mitochondrial DNA level in plasma, also shown by others (Beiter et al. 2011), most likely excludes apoptosis as main mechanism of DNA release following exhaustive exercise. The different nature and release mechanisms of cell free mtDNA and nuclear DNA could be also reflected by their lack of correlation in some diseases (Budnik et al. 2013). It was suggested that cfDNA concentrations increase by unknown active or passive mechanisms in response to acute stress during exercise (Breitbach et al. 2012). Beiter et al. hypothesised that an increased production of reactive oxygen species during exercise is associated with lactate accumulation kinetics and furthermore could trigger cfDNA release via netting neutrophils (Beiter et al. 2011). However, it was noted that NETs do not increase in physically active people due to exercise (Breitbach et al. 2014). Therefore, the absolute contribution of NETs to the cfDNA pool during physical exercise remains to be elucidated.
The release of cfDNA via EVs during exercise has not been examined to date. We studied the association of cfDNA with EVs by fractionation of blood derived from healthy individuals and demonstrated that the bulk of cfDNA detected in plasma before and after exercise is not present in EVs. While the presence of RNA species in EVs derived from both tumour and non-tumour cells is established (Huang et al. 2013; Valadi et al. 2007), the association of DNA with EVs has only recently been documented and appears characteristic of tumour-derived exosomes. It was shown that EVs from normal skin fibroblasts, murine MC/9 mast cells and urine contain less or no DNA (Balaj et al. 2011; Miranda et al. 2010; Valadi et al. 2007). In contrast, EVs derived from different cancer cell lines and tumour-bearing mice were found to contain single- (Balaj et al. 2011) and double-stranded DNA, thereby representing the mutational status of the parental cell lines (Thakur et al. 2014). Studies on cancer patients suggested that extracellular DNA is released by exosomal-like vesicles (Lazaro-Ibanez et al. 2014) and, moreover, the majority of serum cfDNA found in the circulation of tumour patients is probably not present in a free floating form (Kahlert et al. 2014). The DNA sequences identified in cancer cell-derived EVs were found to be representative for the entire host genome, including LINE and Alu elements (Balaj et al. 2011; Lee et al. 2014; Thakur et al. 2014). Thus, EV-associated DNA was mainly detected in tumour-derived cell lines and cancer patients. Most of the studies did not investigate the amount of DNA in EVs compared to total culture medium or serum. Exosomal DNA was detected in low quantity and in one case even preamplified prior to analysis (Lazaro-Ibanez et al. 2014). In principle, our data indicated the presence of EV-associated DNA under physiological conditions representing genomic, as verified by the LTR-based qPCR assay, and trace amounts of mitochondrial DNA. The absolute amount of EV-derived DNA is very low with the majority located on the surface of EVs as indicated by DNase treatment. We cannot exclude at present that the digest of surface DNA is incomplete. However, it is beyond the scope of this study to determine whether the DNA still present after the digest is protected inside EVs.
In summary, our results provide evidence that the majority of cfDNA is not released via EVs in response to acute physical exercise, but rather circulates in a free, non-vesicle-associated form in blood plasma. It seems that the release of cfDNA under physical exercise occurs independently of EVs which could hint at different physiological functions exerted by these molecules. The origin and biological significance of cfDNA in response to physical exercise warrant further study to estimate the potential of cfDNA as biomarker for exercise diagnostics and performance. Likewise, the physiological relevance of EV-mediated communication during exercise should be addressed.
Abbreviations
- cfDNA:
-
Cell free DNA
- EV:
-
Extracellular vesicles
- Flot-1:
-
Flotillin-1
- IAT:
-
Individual anaerobic threshold
- LOQ:
-
Limit of quantification
- MV:
-
Microvesicles
- MVB:
-
Multivesicular bodies
- NET:
-
Neutrophil extracellular trap
- qPCR:
-
Quantitative real-time PCR
- SN:
-
Supernatant
- VO2max :
-
Maximal oxygen consumption
References
Aatonen M, Gronholm M, Siljander PR (2012) Platelet-derived microvesicles: multitalented participants in intercellular communication. Semin Thromb Hemost 38:102–113. doi:10.1055/s-0031-1300956
Adeva-Andany M, Lopez-Ojen M, Funcasta-Calderon R, Ameneiros-Rodriguez E, Donapetry-Garcia C, Vila-Altesor M, Rodriguez-Seijas J (2014) Comprehensive review on lactate metabolism in human health. Mitochondrion 17C:76–100. doi:10.1016/j.mito.2014.05.007
Atamaniuk J, Vidotto C, Tschan H, Bachl N, Stuhlmeier KM, Muller MM (2004) Increased concentrations of cell-free plasma DNA after exhaustive exercise. Clin Chem 50:1668–1670. doi:10.1373/clinchem.2004.034553
Atamaniuk J, Stuhlmeier KM, Vidotto C, Tschan H, Dossenbach-Glaninger A, Mueller MM (2008) Effects of ultra-marathon on circulating DNA and mRNA expression of pro- and anti-apoptotic genes in mononuclear cells. Eur J Appl Physiol 104:711–717. doi:10.1007/s00421-008-0827-2
Atamaniuk J, Vidotto C, Kinzlbauer M, Bachl N, Tiran B, Tschan H (2010) Cell-free plasma DNA and purine nucleotide degradation markers following weightlifting exercise. Eur J Appl Physiol 110:695–701. doi:10.1007/s00421-010-1532-5
Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J (2011) Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nature Commun 2:180. doi:10.1038/ncomms1180
Beiter T, Fragasso A, Hudemann J, Niess AM, Simon P (2011) Short-term treadmill running as a model for studying cell-free DNA kinetics in vivo. Clin Chem 57:633–636. doi:10.1373/clinchem.2010.158030
Beiter T, Fragasso A, Hudemann J, Schild M, Steinacker J, Mooren FC, Niess AM (2014) Neutrophils release extracellular DNA traps in response to exercise. J Appl Physiol 117:325–333. doi:10.1152/japplphysiol.00173.2014
Bellingham SA, Guo BB, Coleman BM, Hill AF (2012) Exosomes: vehicles for the transfer of toxic proteins associated with neurodegenerative diseases? Front Physiol 3:124. doi:10.3389/fphys.2012.00124
Blagosklonny MV (2000) Cell death beyond apoptosis Leukemia 14:1502–1508
Bobrie A, Colombo M, Raposo G, Thery C (2011) Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12:1659–1668. doi:10.1111/j.1600-0854.2011.01225.x
Breitbach S, Tug S, Simon P (2012) Circulating cell-free DNA: an up-coming molecular marker in exercise physiology. Sports Med 42:565–586. doi:10.2165/11631380-000000000-00000
Breitbach S, Sterzing B, Magallanes C, Tug S, Simon P (2014) Direct measurement of cell-free DNA from serially collected capillary plasma during incremental exercise. J Appl Physiol 117:119–130. doi:10.1152/japplphysiol.00002.2014
Budnik LT, Kloth S, Baur X, Preisser AM, Schwarzenbach H (2013) Circulating mitochondrial DNA as biomarker linking environmental chemical exposure to early preclinical lesions elevation of mtDNA in human serum after exposure to carcinogenic halo-alkane-based pesticides. PLoS One 8:e64413. doi:10.1371/journal.pone.0064413
Ceci R et al (2013) Oxidative stress responses to a graded maximal exercise test in older adults following explosive-type resistance training. Redox Biol 2C:65–72. doi:10.1016/j.redox.2013.12.004
Chaar V et al (2011) Effect of strenuous physical exercise on circulating cell-derived microparticles. Clin Hemorheol Microcirc 47:15–25. doi:10.3233/CH-2010-1361
Chiu RW, Chan LY, Lam NY, Tsui NB, Ng EK, Rainer TH, Lo YM (2003) Quantitative analysis of circulating mitochondrial DNA in plasma. Clin Chem 49:719–726
Dickhuth HH, Yin L, Niess A, Rocker K, Mayer F, Heitkamp HC, Horstmann T (1999) Ventilatory, lactate-derived and catecholamine thresholds during incremental treadmill running: relationship and reproducibility. Int J Sports Med 20:122–127. doi:10.1055/s-2007-971105
El Messaoudi S, Rolet F, Mouliere F, Thierry AR (2013) Circulating cell free DNA: preanalytical considerations. Clin Chim Acta 424:222–230. doi:10.1016/j.cca.2013.05.022
Fatouros IG et al (2010) Time of sampling is crucial for measurement of cell-free plasma DNA following acute aseptic inflammation induced by exercise. Clin Biochem 43:1368–1370. doi:10.1016/j.clinbiochem.2010.08.020
Fruhbeis C, Frohlich D, Kuo WP, Kramer-Albers EM (2013) Extracellular vesicles as mediators of neuron-glia communication. Front Cell Neurosci 7:182. doi:10.3389/fncel.2013.00182
Frühbeis C, Helmig S, Tug S, Simon P, Krämer-Albers EM (2015) Physical exercise induces rapid release of small extracellular vesicles into the circulation. J Extracell Vesicles 4:28239. doi:10.3402/jev.v4.28239
Galeazzi M, Morozzi G, Piccini M, Chen J, Bellisai F, Fineschi S, Marcolongo R (2003) Dosage and characterization of circulating DNA: present usage and possible applications in systemic autoimmune disorders. Autoimmun Rev 2:50–55
Gould SJ, Raposo G (2013) As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles. doi:10.3402/jev.v2i0.20389
Harding CV, Heuser JE, Stahl PD (2013) Exosomes: looking back three decades and into the future. J Cell Biol 200:367–371. doi:10.1083/jcb.201212113
Hergenreider E et al (2012) Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol 14:249–256. doi:10.1038/ncb2441
Holdenrieder S, Stieber P (2009) Clinical use of circulating nucleosomes. Crit Rev Clin Lab Sci 46:1–24. doi:10.1080/10408360802485875
Horstman LL, Jy W, Minagar A, Bidot CJ, Jimenez JJ, Alexander JS, Ahn YS (2007) Cell-derived microparticles and exosomes in neuroinflammatory disorders. Int Rev Neurobiol 79:227–268. doi:10.1016/S0074-7742(07)79010-4
Huang X et al (2013) Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom 14:319. doi:10.1186/1471-2164-14-319
Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R (2001) DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 61:1659–1665
Kahlert C et al (2014) Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 289:3869–3875. doi:10.1074/jbc.C113.532267
Kalra H et al (2012) Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol 10:e1001450. doi:10.1371/journal.pbio.1001450
Kowal J, Tkach M, Thery C (2014) Biogenesis and secretion of exosomes. Curr Opin Cell Biol 29C:116–125. doi:10.1016/j.ceb.2014.05.004
Krämer-Albers EM et al (2007) Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: trophic support for axons? Proteomics Clin Appl 1:1446–1461
Lazaro-Ibanez E, Sanz-Garcia A, Visakorpi T, Escobedo-Lucea C, Siljander P, Ayuso-Sacido A, Yliperttula M (2014) Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: apoptotic bodies, microvesicles, and exosomes. Prostate. doi:10.1002/pros.22853
Lee TH, Chennakrishnaiah S, Audemard E, Montermini L, Meehan B, Rak J (2014) Oncogenic ras-driven cancer cell vesiculation leads to emission of double-stranded DNA capable of interacting with target cells. Biochem Biophys Res Commun. doi:10.1016/j.bbrc.2014.07.109
Luga V et al (2012) Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151:1542–1556. doi:10.1016/j.cell.2012.11.024
Lui YY, Chik KW, Chiu RW, Ho CY, Lam CW, Lo YM (2002) Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin Chem 48:421–427
Miranda KC et al (2010) Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int 78:191–199. doi:10.1038/ki.2010.106
Nagata S, Hanayama R, Kawane K (2010) Autoimmunity and the clearance of dead cells. Cell 140:619–630. doi:10.1016/j.cell.2010.02.014
Peinado H et al (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 18:883–891. doi:10.1038/nm.2753
Pilsczek FH et al (2010) A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J Immunol 185:7413–7425. doi:10.4049/jimmunol.1000675
Pisetsky DS (2009) Microparticles as biomarkers in autoimmunity: from dust bin to center stage. Arthritis Res Ther 11:135. doi:10.1186/ar2856
Raposo G, Stoorvogel W (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200:373–383. doi:10.1083/jcb.201211138
Revenfeld AL, Baek R, Nielsen MH, Stensballe A, Varming K, Jorgensen M (2014) Diagnostic and prognostic potential of extracellular vesicles in peripheral blood. Clin Ther 36:830–846. doi:10.1016/j.clinthera.2014.05.008
Rhead B et al (2010) The UCSC Genome Browser database: update. Nucleic Acids Res 38:D613–D619. doi:10.1093/nar/gkp939
Robbins PD, Morelli AE (2014) Regulation of immune responses by extracellular vesicles. Nat Rev Immunol 14:195–208. doi:10.1038/nri3622
Schwarzenbach H, Hoon DS, Pantel K (2011) Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 11:426–437. doi:10.1038/nrc3066
Simons M, Raposo G (2009) Exosomes–vesicular carriers for intercellular communication. Curr Opin Cell Biol 21:575–581. doi:10.1016/j.ceb.2009.03.007
Swarup V, Rajeswari MR (2007) Circulating (cell-free) nucleic acids–a promising, non-invasive tool for early detection of several human diseases. FEBS Lett 581:795–799. doi:10.1016/j.febslet.2007.01.051
Thakur BK et al (2014) Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 24:766–769. doi:10.1038/cr.2014.44
Thery C (2011) Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep 3:15. doi:10.3410/B3-15
Thery C, Ostrowski M, Segura E (2009) Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9:581–593. doi:10.1038/nri2567
Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. doi:10.1038/ncb1596
Walsh NP et al (2011) Position statement. Part one: Immune function and exercise. Exerc Immunol Rev 17:6–63
Yi J et al (2014) Increased plasma cell-free DNA level during HTNV infection: correlation with disease severity and virus load. Viruses 6:2723–2734. doi:10.3390/v6072723
Ziegler A, Zangemeister-Wittke U, Stahel RA (2002) Circulating DNA: a new diagnostic gold mine? Cancer Treat Rev 28:255–271
Acknowledgments
The work of CF and EMKA was supported by Deutsche Forschungsgemeinschaft, DFG grant KR 3668/1-1. The work of SH, PS and ST was supported by an internal funding of the Johannes Gutenberg-University of Mainz. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest.
Additional information
Communicated by Fabio Fischetti.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Helmig, S., Frühbeis, C., Krämer-Albers, EM. et al. Release of bulk cell free DNA during physical exercise occurs independent of extracellular vesicles. Eur J Appl Physiol 115, 2271–2280 (2015). https://doi.org/10.1007/s00421-015-3207-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00421-015-3207-8