Abstract
Purpose
To automatically detect and classify geographic atrophy (GA) in fundus autofluorescence (FAF) images using a deep learning algorithm.
Methods
In this study, FAF images of patients with GA, a healthy comparable group and a comparable group with other retinal diseases (ORDs) were used to train a multi-layer deep convolutional neural network (DCNN) (1) to detect GA and (2) to differentiate in GA between a diffuse-trickling pattern (dt-GA) and other GA FAF patterns (ndt-GA) in FAF images.
-
1.
For the automated detection of GA in FAF images, two classifiers were built (GA vs. healthy/GA vs. ORD). The DCNN was trained and validated with 400 FAF images in each case (GA 200, healthy 200, or ORD 200). For the subsequent testing, the built classifiers were then tested with 60 untrained FAF images in each case (AMD 30, healthy 30, or ORD 30). Hereby, both classifiers automatically determined a GA probability score and a normal FAF probability score or an ORD probability score.
-
2.
To automatically differentiate between dt-GA and ndt-GA, the DCNN was trained and validated with 200 FAF images (dt-GA 72; ndt-GA 138). Afterwards, the built classifier was tested with 20 untrained FAF images (dt-GA 10; ndt-GA 10) and a dt-GA probability score and an ndt-GA probability score was calculated.
For both classifiers, the performance of the training and validation procedure after 500 training steps was measured by determining training accuracy, validation accuracy, and cross entropy.
Results
For the GA classifiers (GA vs. healthy/GA vs. ORD), the achieved training accuracy was 99/98%, the validation accuracy 96/91%, and the cross entropy 0.062/0.100. For the dt-GA classifier, the training accuracy was 99%, the validation accuracy 77%, and the cross entropy 0.166.
The mean GA probability score was 0.981 ± 0.048 (GA vs. healthy)/0.972 ± 0.439 (GA vs. ORD) in the GA image group and 0.01 ± 0.016 (healthy)/0.061 ± 0.072 (ORD) in the comparison groups (p < 0.001). The mean dt-GA probability score was 0.807 ± 0.116 in the dt-GA image group and 0.180 ± 0.100 in the ndt-GA image group (p < 0.001).
Conclusion
For the first time, this study describes the use of a deep learning-based algorithm to automatically detect and classify GA in FAF. Hereby, the created classifiers showed excellent results. With further developments, this model may be a tool to predict the individual progression risk of GA and give relevant information for future therapeutic approaches.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In Western countries, age-related macular degeneration (AMD) is the leading cause of visual impairment and blindness in elderly patients [1,2,3]. Its prevalence increases with age and affects more than 10% of the > 65-year-old people [1].
In accordance with the fundoscopic findings, the classification of the Beckman Initiative for Macular Research Classification Committee distinguishes between early AMD, intermediate AMD, and late AMD. Late AMD is defined with the appearance of any geographic atrophy (GA) or choroidal neovascularizations (CNV) [4].
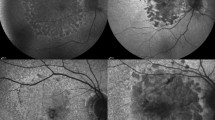
Nowadays, many different imaging modalities are used in the diagnosis of AMD. Most of these imaging techniques, e.g., optical coherence tomography (OCT), OCT angiography, and fluorescein angiography preferably, have their advantage in detecting exudative AMD [5,6,7]. By contrast, fundus autofluorescence (FAF) imaging is particularly useful in the diagnosis of GA [8,9,10]. It provides useful information about the function of the retinal pigment epithelium (RPE) by displaying ocular fluorophores, mainly lipofuscin [3, 9, 10]. In areas affected by GA, there is a progressive loss of the retinal pigment epithelium (RPE), the corresponding choriocapillary layer, and the photoreceptor layer [2, 10]. Due to the absence of the RPE cells and its fluorophores, GA areas appear hypoautofluorescent in the FAF [8,9,10]. However, the junction area of the GA can also show different hyperautofluorescent patterns [3, 9, 10]. In accordance with these different fundus autofluorescence patterns, GA can be subdivided in different phenotypes with differing characteristics [3]. Within these classes, the diffuse-trickling pattern (dt-GA) is shown to have an extremely rapid progression [10, 11].
Deep learning is an interesting research field that gains importance in many different medical areas, especially those dealing with imaging issues. Among others, it enables the automated detection of different structures by self-learning algorithms working with a deep convolutional neural network (DCNN) [12]. Ophthalmology with a lot of different imaging modalities is a potential field of application. Recently, there have been some publications dealing with the use of machine learning in AMD [13,14,15,16,17,18,19,20,21,22,23]. In these studies, machine learning was only used to interpret fundus photography or OCT images. To the best of our knowledge, there exists no study that uses a DCNN in the automated evaluation of fundus autofluorescence images.
Therefore, the aim of our study was to create a deep learning-based classifier for the evaluation of fundus autofluorescence to (1) automatically detect GA and (2) identify eyes exhibiting rapidly progressing dt-GA.
Methods
Deep learning process
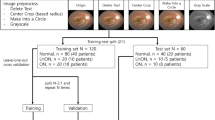
For this study, 30° FAF images of GA patients, healthy patients, and patients with other retinal diseases (ORDs), all obtained by the same FAF device (Spectralis, Heidelberg Engineering, Heidelberg, Germany), were used to train and test a DCNN classifier (Fig. 1). Only images with an image quality that was adequate for manual diagnosis were used. The selection of the images as well as the following assignment towards the training and testing set was performed randomly. There was a strict separation of the training data from the test data to prevent inter-eye and intra-eye correlations. Images of patients that were used for training the DCNN classifier were not used to test it. Furthermore, only one image of a single patient was used for testing. A DCNN is a self-learning algorithm that can perform deep learning by processing input data (e.g., images) within many different hierarchal layers from simple (e.g., lines) and to more complex forms [14, 24].
For this study, the fast working deep learning framework TensorFlow™ (Google Inc., Mountain view, USA) was used to provide deep learning with a multi-layer DCNN. The first layers of this DCNN had been pretrained with millions of already classified everyday life images (e.g., dog, cat, house and car) from the image database ImageNet [25]. In order to obtain a classifier being able to detect GA and GA patterns in FAF images, the last layer of the used DCNN was subsequently trained with above-mentioned FAF images [12, 14, 25,26,27,28].
Detection of GA
In order to differentiate not just between healthy and pathological FAF images, but also to detect GA in FAF images, two classifiers were built with the DCNN: one to differentiate between GA FAF images and healthy FAF images and another to differentiate between GA and ORD (e.g., AMD without GA, adult-onset foveamacular vitelliform dystrophy, central serous chorioretinopathy, and epiretinal membranes). Therefore, the DCNN was trained with 400 FAF images in 500 training steps in both cases (GA: n = 200; healthy or ORD: n = 200).
For both classifiers, the quality of the training process was assessed by determining the training accuracy (performance to correctly classify already trained images), the validation accuracy (performance to correctly classify not so far trained images), and the cross entropy (a function that gives information about the training progress and decreases in a successful training process) [12, 14].
Finally, the two DCNN classifiers were tested with 60 untrained FAF images in each case (GA: n = 30; healthy or ORD: n = 30). For every image probability scores both for GA FAF (GA probability score) and healthy FAF (normal FAF probability score) or ORD FAF (ORD probability score) were automatically calculated by the DCNN.
Detection of diffuse-trickling pattern
In a second step, the DCNN was trained to discriminate GA with dt-GA in FAF from other GA FAF patterns (ndt-GA). For this training process, 72 FAF images with dt-GA and 138 FAF images with ndt-GA were used. As described above, training accuracy, validation accuracy, and cross entropy were determined.
Finally, this classifier was tested with 10 FAF images with dt-GA and 10 images with ndt-GA. By analogy with the GA probability score and the normal FAF probability score described for the GA classifier above, a dt-GA probability score and an ndt-GA probability score were automatically calculated.
Statistics
Statistical analysis was performed with the software SPSS (IBM SPSS Statistics 23.0; IBM, Armonk, NY, USA). The nonparametric Mann-Whitney-U test for independent samples was used to compare the automatically calculated probability scores of the two classifiers. The level of significance was defined p < 0.05. Descriptive statistics were performed with Excel® (Microsoft® Excel® for Mac 2011, 14.6.2; Microsoft®, Redmond, USA).
In order to get information about the precision and repeatability of the two created classifiers, the whole deep learning procedure including the final testing was repeated for a second time. Therefore, the mean absolute probability score difference and the coefficient of variation were calculated. In addition, a Bland-Altman plot was constructed to visualize information about the repeatability of the testing results.
Results
GA classifiers (GA vs. healthy/GA vs. ORD)
Performance of the training process
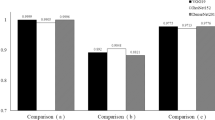
During the 500 performed training steps, the training accuracy and the validation accuracy of the GA classifiers showed a fast increase to 99%/98 and 96%/91%. The cross entropy showed a rapid decrease to a final value of 0.062/0.100 (Fig. 2a–d; Table 1(a, b)).
Performance of the classifier in the final testing
The mean GA probability score of the final testing was 0.981 ± 0.048/ 0.972 ± 0.043 for the GA FAF images and 0.012 ± 0.016/ 0.061 ± 0.072 for the healthy or ORD FAF images (Figs. 3 and 4a, b). According to this, the mean normal FAF probability score/ORD probability score was 0.012 ± 0.017/0.062 ± 0.072 for the GA FAF images and 0.981 ± 0.047/0.972 ± 0.044 for the healthy/ORD FAF images. The GA probability scores were highly significantly different between the two image groups (p < 0.001).
In the two cases, all of the 60 tested FAF images were correctly diagnosed by the GA classifier (Table 2(a, b)). The sensitivity, the specificity, and the accuracy of the GA classifier were 100%.
Repeatability and precision
The mean absolute GA probability score difference of the final testing between the two independently performed deep learning procedures was 0.0004 ± 0.0005/0.001 ± 0.009%, the mean coefficient of variation 0.17 ± 1.44%/0.44 ± 0.69%. The Bland-Altman scattering profiles confirmed this good repeatability by showing a distribution of the values next to the mean of the difference with only a few outliers (Fig. 5a, b).
Diffuse-trickling pattern classifier
Performance of the training process
The increase of the training accuracy and the validation accuracy curves of the dt-GA classifier were lower compared to those of the GA classifier. The training accuracy was 94% and the validation accuracy 77% after the 500 training steps. The cross entropy curve also showed a flatter decrease with a value of 0.16 after the completed training process (Fig. 2e, f; Table 1(c)).
Performance of the classifier in the final testing
The mean dt-GA probability score of the final testing was 0.834 ± 0.123 for the dt-GA FAF images and 0.132 ± 0.121 for the FAF images with ndt-GA (Fig. 4c). According to this, the mean ndt-GA probability score was 0.166 ± 0.123 for dt-GA FAF images and 0.868 ± 0.121 for the ndt-GA FAF images. Between the dt-GA probability scores of the two groups, there was no significant difference (p = 0.353). Comparing the dt-GA probability scores of the dt-GA group and the ndt-GA probability scores of the ndt-GA group, the difference was highly significant (p < 0.001). The dt-GA classifier performed a correct diagnosis in all of the 20 tested FAF images (Table 2(c)). This results in a sensitivity, a specificity, and an accuracy of 100%.
Repeatability and precision
For the dt-GA classifier, the mean absolute difference of the dt-GA probability scores between the two independently performed deep learning procedures was 0.003 ± 0.033, the mean coefficient of variation was 2.53 ± 2.23%. As with the GA classifier, the Bland-Altman scattering profile with an even distribution and only one outlier confirms the good intra-classifier repeatability results (Fig. 5c).
Comparison of the GA classifier and the dt-GA classifier
A comparison of the absolute GA probability scores values of the GA classifier and the dt-GA classifier revealed a highly significant difference (p < 0.001).
Discussion
FAF enables the visualization of the RPE and is an established imaging modality in the diagnosis of GA [3, 8,9,10, 29, 30]. Using the GA classification system for FAF introduced by Bindewald et al. in 2005, a prognosis of the progression rate is possible [3, 10, 29]. Thereby, the dt-GA is known to have a significantly higher progression rate than other patterns [10, 29].
In this context, we created two DCNN classifier approaches in our study, one to automatically detect GA in FAF images and another to automatically detect dt-GA in FAF images. The two classifier approaches showed an excellent performance with a sensitivity, a specificity, and an accuracy of 100%. Focusing on the absolute probability scores of the classifier’s decision process, the GA classifiers achieved significantly better than the dt-GA classifier. The main reasons for this are probably the lower number of FAF images used for the training procedure and the more subtle differences in FAF pattern between dt-GA and ndt-GA compared to GA and healthy retina or GA and ORD. This is also in accordance with the different curves of the training accuracy, the validation accuracy, and the cross entropy that show a more effective training process for the GA classifiers.
With a continuous rising in the number of retinal imaging modalities, both a sufficient expertise in the appraisal of these images and time to include the image information in the diagnosis process become the limiting factors in the daily routine [31]. Additionally, a variable interobserver agreement is a relevant task within retinal imaging [30, 32, 33]. Biarnés et al. (2012) described this problem for the classification of different GA patterns in FAF imaging. In their study, a high intraobserver agreement was reached, while interobserver agreement was described as “variable” [30]. A deep learning-based tool to achieve an automated diagnosis and classification might possibly be a future solution for these problems. As an example, the classifiers in our study reached an extremely good repeatability and precision.
Deep Learning algorithms can hierarchically process a huge amount of image data in a way that is comparable to the neuron microstructure of the brain. In analogy to this, the performance of these algorithms increases during the “learning” process [12, 24]. Therefore, a sufficient amount of classified images is necessary. In case of a lack of training images, overfitting is a phenomenon that can occur during the deep learning process. Hereby, the ability of the classifier to correctly classify unknown images is dramatically reduced [12, 14, 34]. Therefore, it is desirable to expand the amount of training images to further improve the performance of the classifiers and to enable finer subdivisions. To reach this aim, multicenter studies should be aspired.
To the best of our knowledge, our study is the first study that uses a deep learning algorithm in the detection and classification of GA in FAF.
Holz et al. (2007) showed that FAF imaging is the only imaging modality that enables a prognostic view in the progression of GA [29, 30]. In their study, they used the classification system by Bindewald et al. (2005) and compared the progression rates of the different patterns after a median follow-up period of nearly 2 years. Thereby, the diffuse patterns, especially dt-GA, was shown to have the highest spread rate [3, 29]. In literature, this is explained with an increased accumulation of lipofuscin in postmiotic RPE cells as an important factor for cell death in the pathogenesis of GA [35, 36]. An automated classification of different FAF patterns in GA is therefore of tremendous interest in ophthalmology. On the one hand, in the context of a more individualized medicine and on the other hand, in the context of better understanding the pathology of GA. For a GA classifier, the ultimate goal is to implement an algorithm that correctly detects GA and classifies its phenotype in a first step, performs an accurate calculations of the lesion size in a second step, and provide information on the disease progression. The first step in this direction was taken with this work.
There are some limitations of our study that have to be considered. One is the relatively low number of included FAF images, although it is in the range of other studies using machine learning in ophthalmology [14, 16, 19, 22]. This may influence the quality of the deep learning process. Nevertheless, the continuous increase of the course of the training and validation accuracy curves suggests a good working learning progress. As a confirmation of this, the classifiers show no sign of overfitting. Overfitting would lead to an increasing gap between the training and validation accuracy curves [12, 14, 34]. Nevertheless, including more FAF images would probably improve the training performance of the dt-GA classifier.
Another limitation is that in order to receive a sufficient number of images to build our DCNN classifier, in some cases, follow-up images or images of the other patient’s eye were used for training. Therefore, a possible effect on the results cannot completely be excluded, as FAF follow-up images or FAF images of the partner eyes are less different with respect to anatomical features compared to FAF images of other patients. Nevertheless, due to the operating mode of the DCNN, we believe that this effect can be ignored for our study since the portion of images coming from one patient is low. Our used DCNN classifier works by detecting patterns in the images that can be recognized in the majority of these images and that differ in the both classified groups. Therefore, when just a few of the FAF images show the same anatomic features, this is ignored by the DCNN. In this context, we believe that the number of images that were obtained from the same patient should be less than 5% of the training dataset of each class dataset and consequently 2.5% of the entire training dataset of a 2-class classifier. Additionally, overfitting is a sensitive marker of the quality of the dataset. If the classifier recognizes image features due to an insufficient training data set (e.g., due to inter-eye or intra-eye correlation) overfitting will occur. As mentioned above, overfitting did not occur in the classifiers used in this study, so that the composition of the image groups can be considered as sufficient.
In the context of its clinical relevance, a limitation is that the differentiation was only performed between healthy FAF images or ORD FAF images and FAF images with GA as well as between dt-GA pattern and ndt-GA pattern. Therefore, in a following study, it is necessary to extend the classifier in order to recognize more different retinal diseases. In this context, a multicenter design is recommended. The feasibility was already shown by Burlina et al. (2017) by extending a 2-class classifier to a 3- and 4-class classifier with still valid results [21].
Conclusions
In conclusion, we created for the first time a deep learning-based classifier for the automated detection and classification of GA in FAF images. Thereby, our approach showed excellent performance results of the classifier and a very good repeatability.
GA is a progressive, sight-threatening disease, with a divergent progression rate depending on the pattern in FAF [10, 29, 30, 36]. Therefore, this approach may be helpful in the prediction of the individual progression risk of the GA, the identification of biomarkers, and the gain of further information for possible future therapeutic approaches. To expand and improve the performance of the classifier multicenter studies is desirable.
References
Smith W, Assink J, Klein R, Mitchell P, Klaver C, Klein B, Hofman A, Jensen S, Wang J, de Jong P (2001) Risk factors for age-related macular degeneration. Ophthalmology 108:697–704
Herrmann P, Holz FG, Charbel Issa P (2013) Etiology and pathogenesis of age-related macular degeneration. Ophthalmologe 110:377–387
Bindewald A, Schmitz-Valckenberg S, Jorzik J, Dolar-Szczasny J, Sieber H, Keilhauer C, Weinberger A, Dithmar S, Pauleikhoff D, Mansmann U, Wolf S, Holz F (2005) Classification of abnormal fundus autofluorescence patterns in the junctional zone of geographic atrophy in patients with age related macular degeneration. Br J Ophthalmol 89:874–878
Ferris FL 3rd, Wilkinson CP, Bird A, Chakravarthy U, Chew E, Csaky K, Sadda SR, Beckman Initiative for Macular Research Classification C (2013) Clinical classification of age-related macular degeneration. Ophthalmology 120:844–851
Cole E, Ferrara D, Novais E, Louzada R, Waheed N (2016) Clinical trial endpoints for optical coherence tomography angiography in neovascular age-related macular degeneration. Retina 36(Suppl 1):S83–S92
Regatieri CV, Branchini L, Duker JS (2011) The role of spectral-domain OCT in the diagnosis and management of neovascular age-related macular degeneration. Ophthalmic Surg Lasers Imaging 42(Suppl):S56–S66
Khurana RN, Dupas B, Bressler NM (2010) Agreement of time-domain and spectral-domain optical coherence tomography with fluorescein leakage from choroidal neovascularization. Ophthalmology 117:1376–1380
Ly A, Nivison-Smith L, Assaad N, Kalloniatis M (2017) Fundus autofluorescence in age-related macular degeneration. Optom Vis Sci 94:246–259
Yung M, Klufas MA, Sarraf D (2016) Clinical applications of fundus autofluorescence in retinal disease. Int J Retina Vitreous. https://doi.org/10.1186/s40942-016-0035-x
Fleckenstein M, Mitchell P, Freund KB, Sadda S, Holz FG, Brittain C, Henry EC, Ferrara D (2017) The progression of geographic atrophy secondary to age-related macular degeneration. Ophthalmology. https://doi.org/10.1016/j.ophtha.2017.08.038
Batıoğlu F, Gedik Oğuz Y, Demirel S, Ozmert E (2014) Geographic atrophy progression in eyes with age-related macular degeneration: role of fundus autofluorescence patterns, fellow eye and baseline atrophy area. Ophthalmic Res 52:53–59
Angermueller C, Parnamaa T, Parts L, Stegle O (2016) Deep learning for computational biology. Mol Syst Biol 12:878
Feeny AK, Tadarati M, Freund DE, Bressler NM, Burlina P (2015) Automated segmentation of geographic atrophy of the retinal epithelium via random forests in AREDS color fundus images. Comput Biol Med 65:124–136
Treder M, Lauermann JL, Eter N (2017) Automated detection of exudative age-related macular degeneration in spectral domain optical coherence tomography using deep learning. Graefes Arch Clin Exp Ophthalmol 256:259–265
Wang Y, Zhang Y, Yao Z, Zhao R, Zhou F (2016) Machine learning based detection of age-related macular degeneration (AMD) and diabetic macular edema (DME) from optical coherence tomography (OCT) images. Biomed Opt Express 7:4928–4940
Sun Y, Li S, Sun Z (2017) Fully automated macular pathology detection in retina optical coherence tomography images using sparse coding and dictionary learning. J Biomed Opt 22:16012
Burlina P, Pacheco KD, Joshi N, Freund DE, Bressler NM (2017) Comparing humans and deep learning performance for grading AMD: a study in using universal deep features and transfer learning for automated AMD analysis. Comput Biol Med 82:80–86
Venhuizen F, van Ginneken B, van Asten F, van Grinsven M, Fauser S, Hoyng C, Theelen T, Sánchez C (2017) Automated staging of age-related macular degeneration using optical coherence tomography. Invest Ophthalmol Vis Sci 58:2318–2328
Bogunovic H, Montuoro A, Baratsits M, Karantonis M, Waldstein S, Schlanitz F, Schmidt-Erfurth U (2017) Machine learning of the progression of intermediate age-related macular degeneration based on OCT imaging. Invest Ophthalmol Vis Sci 58:BIO141–BIO150
Bogunovic H, Waldstein S, Schlegl T, Langs G, Sadeghipour A, Liu X, Gerendas B, Osborne A, Schmidt-Erfurth U (2017) Prediction of anti-VEGF treatment requirements in neovascular AMD using a machine learning approach. Invest Ophthalmol Vis Sci 58:3240–3248
Burlina P, Joshi N, Pekala M, Pacheco K, Freund D, Bressler N (2017) Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol 135:1170–1176
Aslam TM, Zaki HR, Mahmood S, Ali ZC, Ahmad NA, Thorell MR, Balaskas K (2017) Use of a neural net to model the impact of optical coherence tomography abnormalities on vision in age-related macular degeneration. Am J Ophthalmol. https://doi.org/10.1016/j.ajo.2017.10.015
Prahs P, Radeck V, Mayer C, Cvetkov Y, Cvetkova N, Helbig H, Marker D (2018) OCT-based deep learning algorithm for the evaluation of treatment indication with anti-vascular endothelial growth factor medications. Graefes Arch Clin Exp Ophthalmol 256:91–98
Rampasek L, Goldenberg A (2016) TensorFlow: biology’s gateway to deep learning? Cell Syst 2:12–14
Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L (2009) ImageNet- a large-scale hierarchical image database. CVPR 2009—IEEE conference on computer vision and. Pattern Recogn 2009:248–255
Szegedy C, Vanhoucke V, Ioffe S, Shlens J (2016) Rethinking the inception architecture for computer vision. IEEE Conf Comput Vis Pattern Recognit 2016:2818–2826
TensorFlow (2017) http://www.tensorflow.org/tutorials/image_recognition. TensorFlow. Accessed 30 Jan 2018
Google Developers (2017) https://codelabs.developers.google.com/codelabs/tensorflow-for-poets/#0. Google Developers. Accessed 4 July 2017
Holz FG, Bindewald-Wittich A, Fleckenstein M, Dreyhaupt J, Scholl HP, Schmitz-Valckenberg S, Group FA-S (2007) Progression of geographic atrophy and impact of fundus autofluorescence patterns in age-related macular degeneration. Am J Ophthalmol 143:463–472
Biarnes M, Mones J, Trindade F, Alonso J, Arias L (2012) Intra and interobserver agreement in the classification of fundus autofluorescence patterns in geographic atrophy secondary to age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol 250:485–490
Schmitz-Valckenberg S, Gobel AP, Saur SC, Steinberg JS, Thiele S, Wojek C, Russmann C, Holz FG, For The Modiamd-Study G (2016) Automated retinal image analysis for evaluation of focal hyperpigmentary changes in intermediate age-related macular degeneration. Transl Vis Sci Technol 5:3
Bearelly S, Chau FY, Koreishi A, Stinnett SS, Izatt JA, Toth CA (2009) Spectral domain optical coherence tomography imaging of geographic atrophy margins. Ophthalmology 116:1762–1769
Holz FG, Jorzik J, Schutt F, Flach U, Unnebrink K (2003) Agreement among ophthalmologists in evaluating fluorescein angiograms in patients with neovascular age-related macular degeneration for photodynamic therapy eligibility (FLAP-study). Ophthalmology 110:400–405
Lakhani P, Sundaram B (2017) Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 284:574–582
Holz F, Bellman C, Staudt S, Schütt F, Völcker H (2001) Fundus autofluorescence and development of geographic atrophy in age-related macular degeneration. Invest Ophthalmol Vis Sci 42:1051–1056
Schmitz-Valckenberg S, Bindewald-Wittich A, Dolar-Szczasny J, Dreyhaupt J, Wolf S, Scholl HP, Holz FG, Fundus Autofluorescence in Age-Related Macular Degeneration Study G (2006) Correlation between the area of increased autofluorescence surrounding geographic atrophy and disease progression in patients with AMD. Invest Ophthalmol Vis Sci 47:2648–2654
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors certify that they have no affiliations with/or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements) or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Financial disclosure
M. Treder: Allergan, Novartis; J.L. Lauermann: Bayer, Novartis; N. Eter: Allergan, Alimera, Bausch and Lomb, Bayer, Heidelberg Engineering, Novartis, Roche.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
For this type of study formal consent is not required.
Rights and permissions
About this article
Cite this article
Treder, M., Lauermann, J.L. & Eter, N. Deep learning-based detection and classification of geographic atrophy using a deep convolutional neural network classifier. Graefes Arch Clin Exp Ophthalmol 256, 2053–2060 (2018). https://doi.org/10.1007/s00417-018-4098-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00417-018-4098-2