Abstract
Messenger RNA (mRNA) markers have been extensively investigated for the identification of forensically relevant body fluids and tissues based on their expression profiles among cell types. As products of the backsplicing of pre-mRNAs, circular RNAs (circRNAs) share exonic sequences with their linear counterparts. The inclusion of circRNAs in mRNA profiling is shown to facilitate the detection of biomarkers in the identification of body fluids. In this study, we identified the expression of circRNAs of 14 out of 45 biomarkers from five body fluid types using outward-facing primer sets and revealed the ratio of circular to total transcripts of biomarkers by RNase R treatment. Furthermore, our results of qPCR analysis show that the inclusion of circRNAs in the detection of biomarkers, including HBA and ALAS2 for blood; MMP7 and MMP10 for menstrual blood; HTN3 for saliva; SPINK5, SERPINB3, ESR1, and CYP2B7P1 for vaginal secretions; TGM4, KLK3, and PRM2 for semen; and SLC22A6 and MIOX for urine, does not impair the specificity of these biomarkers. Additionally, a high copy number of targets from linear transcripts could be employed to increase the detection sensitivity of TGM4 and KLK3 with a low expression level of circRNAs in urine samples. Altogether, these results will help with the development of robust multiplex assays for body fluid identification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The identification of human body fluid stains at a crime scene is of practical significance since it has the potential to provide crucial information in criminal investigations. In recent years, several molecular approaches for cell type inference have been intensively studied, including the detection of the expression of messenger RNAs (mRNAs) [1,2,3,4,5,6,7] and microRNAs (miRNAs) [8,9,10,11], the determination of DNA methylation levels [12,13,14,15,16], and the identification of microbial species [12, 17]. Cells from a single tissue type should have a unique transcriptome or gene expression profile, which facilitates the identification of forensically relevant body fluids. Currently, a number of mRNA markers have been developed for the identification of forensically relevant body fluids and the European DNA profiling group (EDNAP) has widely investigated mRNA profiling of a series of forensic-related body fluids/tissues in the collaborative exercises [18,19,20,21]. However, it should be noted that several biomarkers, such as the vaginal secretion markers HBD1 and MUC4, exhibit detectable cross-reactivity in non-target body fluids [4, 22, 23].
One ongoing challenge for mRNA profiling experiment with body fluid samples is the low quality and quantity of total RNAs. In addition to the optimization of primer sets and PCR amplification conditions, efforts have also been made to promote the identification of body fluids by increasing the recovery of transcripts, purifying PCR products, and employing expression-abundant biomarkers [3, 24,25,26]. In our previous reports, our results show that the inclusion of circular RNAs (circRNAs) in mRNA profiling can facilitate the detection of mRNA markers in forensic body fluid identification [27, 28]. CircRNAs, derived from the backsplicing of pre-mRNAs, have been widely identified as being co-expressed with their linear counterparts [29]. As a matter of fact, the inclusion of circRNAs in mRNA profiling increases the level of transcripts. The determination of circular transcripts of body fluid-specific biomarkers will help with the development of robust multiplex assays for body fluid identification. However, the expression profiles of circRNAs in forensically relevant body fluids are not available. Additionally, it remains unknown whether the inclusion of circRNAs in the detection of these markers impairs the specificity of these biomarkers.
In this study, a total of 45 genes with tissue-specific expression in five body fluids were first obtained from previous reports and/or by retrieving genotype-tissue expression (GTEx) database in UCSC genome browser. The circular transcripts of these biomarkers were identified and the expression level of circRNAs was evaluated. The expression specificity of 14 biomarkers with the inclusion of circular transcripts in six body fluids was further characterized.
Materials and methods
Sample preparation
All samples of each fresh body fluid were collected from healthy volunteers (five males, four females, ranging from 20 to 30 years of age). Peripheral blood (n = 4) was obtained via venipuncture with vacutainer tubes (BD, USA). Saliva (n = 4, 5–7 ml) was collected in sterile 50-ml Corning polyethylene tubes (Corning, USA). Freshly ejaculated semen (n = 4) was gained from fertile men. Vaginal secretions (n = 4) were collected from the vagina by sterile cotton swabs, and four menstrual blood (n = 4, day 2 or 3 of the menstrual cycle) was collected from donors’ sanitary towel. Urine samples (n = 9) were collected from five male and four female individuals, respectively. After collection, all samples were used for the RNA extraction or stored immediately at − 80 °C before use. All procedures were approved by the ethics committee of Shanghai Medical College, Fudan University, and all donors volunteered for this study based on informed consent.
RNA preparation and reverse transcription
For vaginal secretions and menstrual blood samples, total RNAs were extracted with RNeasy® Mini Kit (Qiagen, Germany) according to the manufacturer’s protocol with the following adaptations: entire cotton swabs or approximately 1 cm2 sanitary napkins were cut into pieces and incubated at 56 °C for 1 h in 350 μl RLT buffer containing 3.5 μl β-mercaptoethanol, and then the complete mixture including the swabs or sanitary napkins pieces was passed over a QiaShredder column (Qiagen, Germany), after which the flow-through was incubated at 56 °C for another 30 min. For urine samples, 40 ml fresh urine in a 50-ml Corning polyethylene tube was centrifuged at 4 °C for 10 min to collect sediments. Urine sediments were then washed twice with sterile cold phosphate buffer (PBS) by centrifugation and resuspension and subjected to total RNA extraction with RNeasy® Mini Kit according to the manufacturer’s protocol. For blood and saliva samples, total RNAs were obtained using RNeasy® Mini Kit. For semen samples, after being liquefied at 37 °C for 30 min, 100-μl semen samples were mixed with an equal volume of PBS and then subjected to total RNA extraction with TRIzol LS (Invitrogen, USA) followed by cleanup using RNeasy® Mini Kit (Qiagen, Germany) according to the manufacturers’ protocols. RNase-free DNase Set (Qiagen, Germany) was used for on-column DNA digestion, and total RNA was eluted in 35 μl RNase-free ddH2O. RNA concentration was determined using a NanoDrop ND-2000 spectrophotometer (Thermo Scientific, USA). RNA integrity number (RIN) assessed using an Agilent RNA 6000 Nano Kit on an Agilent 2100 bioanalyzer (Agilent, Germany) was 4.5–9.1 for peripheral blood, 2.4–4.3 for menstrual blood, 1.0–2.4 for saliva, 2.2–4.8 for vaginal secretion, 2.6–2.7 for semen, and 1.0–1.2 for urine. Total RNA was used for subsequent evaluation or immediately stored at − 80 °C before use. For each reverse transcription reaction, random hexamers were used to synthesize cDNA in a final volume of 20 μL using a QuantiTect Reverse Transcription Kit (Qiagen, Germany) according to the manufacturer’s instructions. Reverse transcription (RT) minus controls without reverse transcriptase were used to rule out potential contamination of gDNA. The cDNA was immediately used for PCR or stored at − 20 °C before use.
Identification of circular transcripts
Divergent primer sets for candidate biomarkers (Table S1) were designed and synthesized as described previously [27]. The primer sequences used in this study as well as product sizes are listed in Table S2. For cDNA synthesis, 30 ng of total RNA from each sample of body fluids was used for reverse transcription. In singleplex PCR, 25 μl of reaction mix contained 1 μl of cDNA or equal volume of sterile water as the non-template control, 2 μl of dNTPs mixture containing 2.5 mM of each dNTP (Takara, Japan), 2.5 μl of 10 × PCR buffer, and 1 U AmpliTaq Gold DNA polymerase (Thermofisher, USA). The cycling conditions were as follows: the initial denaturation was at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, annealing at the indicated temperatures in Table S2 for 35 s, 72 °C for 60 s with a final elongation at 72 °C for 10 min. PCR products of circRNAs were separated by agarose gel electrophoresis (AGE) followed by purification and the junctions of head-to-tail products were determined by Sanger sequencing (Sunny Biotech., China).
Quantitative real-time PCR (qPCR)
In this study, L-primers refer to primer sets that mainly amplify linear transcripts of genes and LC-primers refer to primer sets that can simultaneously amplify linear and circular transcripts of genes. Specificity of all primer sets was checked with melting profiling together with agarose gel electrophoresis, and amplification efficiency of each primer set was evaluated by qPCR (Table S4). To eliminate linear RNA for the detection of circRNAs, total RNA was treated with RNase R (Epicenter, USA) at 37 °C for 1 h with a dose of 3 U per 1 μg of RNA before reverse transcription reaction.
QPCR was performed using QuantiNova™ SYBR® Green PCR Kit (Qiagen, Germany) in a total reaction volume of 10 μl using indicated primer sets (Table S4). Each reaction mixture contained 1 μl of cDNA (equivalent to 1.5 ng of total RNA), 5 μl of SYBR plus ROX reference dye premix (the volume ratio of SYBR to ROX is 100:1), and 0.7 μM for each primer. An equal volume of 1 μl ddH2O or RT minus control was used to rule out potential contamination. qPCR reactions were conducted in triplicate for each RT reaction product. The cycling conditions consisted of an initial denaturation at 95 °C for 2 min, followed by 45 cycles of 95 °C for 15 s and 60 °C for 34 s and were performed on a 7500 Real-Time PCR system (Thermofisher, USA). The amplification curves were analyzed using SDS v1.4 software (Thermofisher, USA), to determine the quantification cycle (Cq) values and perform melting curve analysis. The detection results of markers were considered valid when the Cq value is less than 40 and Cq values deviating less than 0.5 cycle from the median value of the respective technical repeat. To analyze the ratio of circular to total transcripts of each gene, delta Cq (ΔCq) for each gene in a sample was termed as the average Cq from triplicate reactions (RNase R treatment) − the average Cq from triplicate reactions (without treatment). The ratio of circular to total transcripts of each gene was calculated as 100% × \( {\left(1+E\right)}^{-\overline{\Delta \mathrm{Cq}}} \), where\( \overline{\Delta \mathrm{Cq}} \) is the average ΔCq value obtained from four samples and E denotes the amplification efficiency of a primer set.
The expression stability of candidate reference genes (β-actin (ACTB), β2M, 18S rRNA, and GAPDH) was evaluated based on their expression in different body fluids using RefFinder [30], and β-actin was selected as the reference control in this study (Fig. S3). For the specificity analysis of biomarkers, the expression of biomarkers detected using LC-primers in each type of body fluid was evaluated and ΔCq for each marker in one sample was defined as the average Cq from triplicate reactions (marker) − the average Cq from triplicate reactions (β-actin).
Capillary electrophoresis and profile analysis
PCR amplification was performed using fluorescein-labeled primers as described previously [27]. Briefly, the initial denaturation was at 95 °C for 5 min, followed by 28 cycles of 95 °C for 30 s, 60 °C for 35 s, 72 °C for 60 s with a final elongation at 72 °C for 10 min, and then CE was performed. Raw data were analyzed using GeneMapper ID software v3.2. The threshold for a positive result was set to 100 RFUs (relative fluorescence units). Negative controls did not show amplification signals. For samples with negative results in the sensitivity analysis, PCR amplification using LC-primers and L-primers were replicated followed by CE.
Results
Identification of circular transcripts of biomarkers
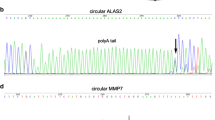
A total of 45 candidate genes with tissue-specific expression in five body fluids including menstrual blood, semen, saliva, vaginal secretions, and urine were first obtained (Table S1). Divergent primer sets based on the combination of exons of each biomarker were used for PCR amplification, and the circular transcripts are determined by Sanger sequencing (Table S2). As a result, 38 major circRNAs from 14 genes were identified (Fig. 1) and the covalent junctions in circular transcripts are shown in Fig. S1. Most of these exon-derived circular transcripts contained two to five exons, with an average length of 530 bp (Table S3), which can facilitate the development of LC-primers in mRNA profiling. Additionally, most of these circular transcripts are composed of incomplete exons, which implies a difficulty in the prediction of circular transcripts. Furthermore, the circular forms of MMP10 showed a significant difference from the results in circBase database. The expression of circRNAs of several biomarkers, such as PRM2, HTN3, MIOX, and SLC22A6, was not observed in previous reports. It is a possibility that circular transcripts can be erroneously assembled or missed out by the analysis of RNA-Seq data.
Identification of circRNAs of tissue-specific biomarkers. a Semen-specific markers PRM2, TGM4, SEGM1, MSMB, KLK3, and LDHC. b Urine-specific markers MIOX and SLC22A6. c Vaginal secretions-specific markers ESR1, SERPINB3, SPINK5, and CYP2B7P1. d Saliva-specific marker HTN3. e Menstrual blood-specific marker MMP10. The curved line indicates the downstream 3′ end of an exon is covalently linked with the upstream 5′ end of an exon, which results in the formation of a circular transcript. All junction sequences of the head-to-tail circRNAs are shown in Fig. S1
Evaluation of the expression content of circRNAs
Previous reports show that the backsplicing of pre-mRNAs competes with the splicing of pre-mRNAs [31]. In fact, a high abundance of circRNAs implicates a low expression level of mRNA from a single gene. Therefore, the evaluation of the ratio of circular to total transcripts of a single gene can help the selection of biomarkers in body fluid identification. To this end, primer sets for biomarkers used in previous reports were first collected and primer sets for the amplification of linear transcripts (L-primers) or for simultaneous amplification of linear and circular transcripts (LC-primers) were developed (Table S4 and Fig. S2). Total RNA with or without RNase R treatment was used for reverse transcription followed by qPCR with L-primers or LC-primers. As shown in Fig. 2, RNase R treatment could significantly remove linear transcripts. RNase R-resistant transcripts of several biomarkers, such as ALAS2, TGM4, PRM2, and KLK3, accounted for a high proportion of transcripts. In contrast, several biomarkers, such as HTN3 and SLC22A6, harbored a low expression level of circular transcripts. Overall, semen-specific markers have higher circular-to-total RNA expression ratios relative to other body fluid markers (Table S5).
Evaluation of the RNase R-resistant transcripts to all transcripts using L-primers or LC-primers of tissue-specific biomarkers. qPCR was performed using L-primers (L) and LC-primers (LC) on cDNA from 4 total RNA samples with or without RNase R treatment. The percentage of RNase R-resistant transcripts to all transcripts of each gene was evaluated as 100% × \( {\left(1+\mathrm{E}\right)}^{-\overline{\Delta \mathrm{Cq}}} \), where \( \overline{\Delta \mathrm{Cq}} \) was termed as the average Cq from triplicate reactions (RNase R treatment) − the average Cq from triplicate reactions (without treatment) and E denotes the mean amplification efficiency of a primer set. a HBA and ALAS2 in blood. b MMP7 and MMP10 in menstrual blood. c SPINK5, SERPINB3, ESR1, and CYP2B7P1 in vaginal secretions. d HTN3, SPINK5, and SERPINB3 in saliva. e TGM4, KLK3, and PRM2 in semen. f SLC22A6 and MIOX in urine. An asterisk indicates that the Cq value of a marker was more than 40 after RNase R treatment. Blue and purple indicate results obtained using L-primers and LC-primers, respectively
Evaluation of the specificity of biomarkers with the inclusion of circRNAs
It is unknown whether the promoter leakage in controlling expression dynamics of genes together with the backsplicing of pre-mRNAs lowers the specificity of biomarkers with the inclusion of circRNAs. To this end, qPCR was performed using LC-primers to test the expression of transcripts in six body fluids including peripheral and menstrual blood, semen, saliva, vaginal secretions, and urine samples. Raw Cq values were normalized relative to the expression of the reference gene β-actin. On the whole, tissue-specific biomarkers were observed to have lower ΔCq values in expected body fluids (Fig. 3). Especially, MIOX, a newly developed biomarker for urine, showed high level of expression with a tissue-specific pattern. ESR1 was only detected in vaginal secretions and menstrual blood, which suggests high degree of expression specificity in body fluids. However, SPINK5 and SERPINB3, two markers for vaginal secretions, showed cross-reactivity with salvia. Additionally, four biomarkers including HBA, MMP7, TGM4, and KLK3 showed cross-reactivity with male urine samples (Fig. 3f). In contrast to HBA and MMP7, TGM4, and KLK3, two prostate-specific biomarkers were not detectable in female urine samples (data not shown), which is consistent with previous reports and our investigation [32, 33]. PRM2 exhibits high semen specificity and is undetectable in urine samples, making it more suitable for differentiating semen and urine samples (Fig. 3e, f). Therefore, the simultaneous detection of linear and circular transcripts shows a similar specificity of biomarkers to previous reports.
Evaluation of the specificity of genes with the inclusion of circRNAs using qPCR. qPCR assay was carried out on four total RNA samples for each body fluid. Expression patterns of 14 genes including HBA and ALAS2 for blood; MMP7 and MMP10 in menstrual blood; HTN3 in saliva; SPINK5, SERPINB3, ESR1, and CYP2B7P1 in vaginal secretions; TGM4, KLK3, and PRM2 in semen; and SLC22A6 and MIOX in urine were measured by qPCR using LC-primers and presented as a scatter plot. ΔCq was termed as the average Cq from triplicate reactions (marker) − the average Cq from triplicate reactions (β-actin). a Blood. b Menstrual blood. c Saliva. d Vaginal secretions. e Semen. f Urine. Data are displayed by scatter plot (the black horizontal line represents the mean value of ΔCq). A dashed line indicates the position of ΔCq value equal to 5 and markers with the median value less than 5 were marked in colors. The markers for peripheral blood, menstrual blood, saliva, vaginal secretions, semen, and urine are indicated using red, spring green, green, purple, blue, and pink, respectively. An asterisk shows that the Cq value of a marker was more than 40 in the indicated tissue
The detection of TGM4 and KLK3 in urine samples
Since the positive detection of TGM4 and KLK3 in urine samples can contribute to gender determination, we subsequently investigated the detection sensitivity of TGM4 and KLK3 using L-primers and LC-primers. The amplification efficiency of L-primers for TGM4 and KLK3 were similar to that of LC-primers (Fig. S4). Sensitivity testing of TGM4 and KLK3 was next assessed in a quantitative approach (input total RNA). A dilution series of total RNA from different individuals (0.2–0.006 ng) was reversely transcribed. PCR amplification was performed using L-primers and LC-primers, respectively, and results are summarized in Table 1. The detection limits of both TGM4 and KLK3 determined using L-primers were up to 0.2 ng of input total RNA (Table 1). In contrast, TGM4 and KLK3 could be detectable with as little as 0.012 ng of input total RNA using LC-primers in all tested samples (Table 1). Furthermore, in contrast to L-primers, the amplification using LC-primers of TGM4 and KLK3 produced considerably higher peaks (Fig. 4a, b).
The detection of TGM4 and KLK3 using L-primers and LC-primers in urine samples. a The average RFU values from the detection of TGM4 and KLK3 using L-primers and LC-primers in three urine samples. PCR amplification of TGM4 and KLK3 was performed on cDNA from 0.2 ng total RNA using L-primers and LC-primers followed by CE. b One representative electrophoretogram out of five samples in a for TGM4 (upper panel) and KLK3 (low panel). The peak height is indicated in each panel. c The ΔCq values obtained using L-primers and LC-primers in urine samples. The reverse transcription was performed using five total RNA samples followed by qPCR using L-primers and LC-primers, respective. ΔCq was termed as the average Cq from triplicate reactions (marker) − the average Cq from triplicate reactions (β-actin)
Since the circRNAs of TGM4 and KLK3 had a high expression level in semen (Fig. 2e and Table S5), we subsequently determined whether the increased detection sensitivity of TGM4 and KLK3 using LC-primers attributed to the inclusion of circRNAs. The expression of circRNAs of TGM4 and KLK3 was evaluated using urine total RNA with or without RNase R treatment. Unexpectedly, the ratio of RNase R-resistant transcripts to total transcripts in urine samples accounted for 0.52% for TGM4 and 1.94% for KLK3, which implies that the increased detection sensitivity of TGM4 and KLK3 using LC-primers did not mainly result from the inclusion of circRNAs. In fact, compared with LC-primers of TGM4 and KLK3, L-primers of TGM4 and KLK3 had similar amplification efficiency (Fig. S4a, b) and however obtained significantly higher Cq values using the same amount of cDNA as template in urine samples (Fig. 4c). Despite a high expression level of circRNAs of TGM4 and KLK3 in semen samples, similar results still could be observed when we performed the evaluation of amplification efficiency of L-primers and LC-primers (Fig. S4a, b). Therefore, linear transcripts of TGM4 and KLK3 with the region LC-primers amplified should have a higher copy number than that with the region L-primers amplified in exfoliated cells, which could attribute to the increased detection sensitivity of TGM4 and KLK3 in urine samples.
Discussion
Several databases for circular RNAs, such as circBase, CIRCpedia, CircNet, and TS_circ database [34,35,36,37], have been established, while there is a lack of datasets for forensically relevant body fluids. Furthermore, although the expression profile of circular transcripts in forensically relevant body fluids can be revealed by next-generation sequencing (NGS), the validation of the head-to-tail products is still required by Sanger sequencing, since the analysis of datasets from NGS can present false-positive results. Especially, on account of a small amount of RNA from these body fluids, NGS and the bioinformatics analysis of datasets also might leave out circular transcripts. Therefore, to directly determine potential circular transcripts of biomarkers, we used outward-facing primer sets based on the combination of exons for PCR amplification followed by Sanger sequencing and identified the expression of circular transcripts from 14 out of 45 genes with tissue-specific expression in the present study. In fact, several circular transcripts of genes, such as MIOX and SLC22A6, are not identified in four databases mentioned above, although the expression of circRNAs in kidney tissues was widely investigated [34, 38]. It should be noted that the low resolution of gel analysis for PCR products might result in the omission of circRNAs with a low expression level. However, the inclusion of circRNAs at a low expression level might not substantially promote the detection of biomarkers in mRNA profiling. Consequently, the determination of each potential circular transcript with a relatively high expression level in this study will help in the selection of target regions for PCR amplification. Nonetheless, some established markers without the expression of or with a very low expression level of circRNAs, such as PRM1 and UMOD, are still considered as important candidates in body fluid identification.
The expression variation of circRNAs occurs in individuals because of genetic context, age, sex, and so on. Because circRNAs and mRNAs of a single gene are from pre-mRNA, the simultaneous detection of circRNAs and mRNAs of biomarkers should not be impaired by the expression variation of circRNAs. Notably, the competition of circRNA biogenesis might result in undetectable expression of linear mRNA. Biomarkers with undetectable expression of linear mRNA in non-target tissues might harbor the detectable expression of circRNAs in non-target tissues. Previous studies reported hundreds of cases where only the expression of circular transcripts were detected but corresponding linear transcripts expression were not detectable [39]. Therefore, it is essential to clarify whether the inclusion of circRNAs impair the specificity of these biomarkers. To address this issue, we investigated the expression specificity of 14 biomarkers from six body fluids using LC-primers. Our results show that the expression specificity of these biomarkers is similar to previous reports. Therefore, circRNAs of mRNA markers in non-target tissues might be absent or have a very low level of expression and the inclusion of circRNAs does not interfere with the specificity of these mRNA markers. For those biomarkers, such as SPINK5 and SERPINB3, cross-reactivity with other body fluids does not attribute to the expression of circRNAs, since the expression of mRNA can be detected in more than one body fluid.
Among the six body fluid types, cross-reactivity of several biomarkers with urine samples was observed. In fact, the identification of urine stains attracts little attention to forensic scientists. HBA had a high and constant expression in all urine samples; however, the reason remains to be clarified. There might be not due to microscopic hematuria since ALAS2 had a low expression level in urine samples. Furthermore, our previous reports show a high expression level of HBB, a blood marker, in urine samples [33]. Matrix metalloproteinases (MMPs) are the key effectors of cell differentiation, cyclic growth, and cell death of the endometrium and play an intriguing role in apoptosis [40,41,42]. The expression of MMP7 in urine samples might attribute to the presence of exfoliated cells harboring the transcripts of MMP7 [43]. Additionally, since male urethra travels through the prostate, it is reasonable to demonstrate the detection of TGM4 and KLK3 in male urine samples. In contrast, we did not observe the expression of TGM4 and KLK3 in female urine samples and the expression of PRM2 in male urine samples. Especially, the use of different primer sets in this study and our previous studies obtained similar results, which can further validate the cross-reactivity of these mRNA biomarkers with urine samples.
The inclusion of circRNAs in mRNA profiling can facilitate the detection of biomarkers. In this study, in contrast to the expression of circRNAs of TGM4 and KLK3 in semen (Fig. 2e), circRNAs of TGM4 and KLK3 in urine samples had a significantly low expression level, which might weakly increase the detection sensitivity of TGM4 and KLK3 in urine samples. The increased detection sensitivity of TGM4 and KLK3 using LC-primers should not result from the alternative splicing, since the alternative splicing does not result in the deletion of the regions L-primers of TGM4 and KLK3 amplify [44, 45]. In fact, exfoliated cells will undergo or might be undergoing apoptosis when they detach from the surrounding extracellular matrix and the constituents of urine might promote the process of cell death, which can trigger global decay of mRNA molecules [46]. Since mRNA can decay from either the 5′ or 3′ end exonucleolytic decay [46], the regions at 5′ and/or 3′ end of linear transcripts might be removed during cell death, which results in the loss of primer binding sites at 5′ and/ or 3′ end. In contrast, a higher copy number of targets from linear transcripts might be employed for PCR amplification by virtue of the primer binding sites in the middle of linear transcripts. Therefore, the development of primer sets to increase the detection sensitivity of mRNA biomarkers in exfoliated cells requires more attention.
References
Roeder AD, Haas C (2013) MRNA profiling using a minimum of five mRNA markers per body fluid and a novel scoring method for body fluid identification. Int J Legal Med 127:707–721. https://doi.org/10.1007/s00414-012-0794-3
Juusola J, Ballantyne J (2005) Multiplex mRNA profiling for the identification of body fluids. Forensic Sci Int 152:1–12. https://doi.org/10.1016/j.forsciint.2005.02.020
Hanson EK, Ballantyne J (2013) Highly specific mRNA biomarkers for the identification of vaginal secretions in sexual assault investigations. Sci Justice 53:14–22. https://doi.org/10.1016/j.scijus.2012.03.007
Zubakov D, Hanekamp E, Kokshoorn M, van IJcken W, Kayser M (2008) Stable RNA markers for identification of blood and saliva stains revealed from whole genome expression analysis of time-wise degraded samples. Int J Legal Med 122:135–142. https://doi.org/10.1007/s00414-007-0182-6
Lindenbergh A, Maaskant P, Sijen T (2013) Implementation of RNA profiling in forensic casework. Forensic Sci Int Genet 7:159–166. https://doi.org/10.1016/j.fsigen.2012.09.003
Lindenbergh A, De Pagter M, Ramdayal G et al (2012) A multiplex (m)RNA-profiling system for the forensic identification of body fluids and contact traces. Forensic Sci Int Genet 6:565–577. https://doi.org/10.1016/j.fsigen.2012.01.009
Visser M, Zubakov D, Ballantyne KN, Kayser M (2011) MRNA-based skin identification for forensic applications. Int J Legal Med 125:253–263. https://doi.org/10.1007/s00414-010-0545-2
Park JL, Park SM, Kwon OH, Lee HC, Kim JY, Seok HH, Lee WS, Lee SH, Kim YS, Woo KM, Kim SY (2014) Microarray screening and qRT-PCR evaluation of microRNA markers for forensic body fluid identification. Electrophoresis 35:3062–3068. https://doi.org/10.1002/elps.201400075
Sauer E, Reinke AK, Courts C (2016) Differentiation of five body fluids from forensic samples by expression analysis of four microRNAs using quantitative PCR. Forensic Sci Int Genet 22:89–99. https://doi.org/10.1016/j.fsigen.2016.01.018
Wang Z, Zhang J, Luo H, Ye Y, Yan J, Hou Y (2013) Screening and confirmation of microRNA markers for forensic body fluid identification. Forensic Sci Int Genet 7:116–123. https://doi.org/10.1016/j.fsigen.2012.07.006
Sauer E, Babion I, Madea B, Courts C (2014) An evidence based strategy for normalization of quantitative PCR data from miRNA expression analysis in forensically relevant body fluids. Forensic Sci Int Genet 13:217–223. https://doi.org/10.1016/j.fsigen.2014.08.005
Choi A, Shin KJ, Yang WI, Lee HY (2014) Body fluid identification by integrated analysis of DNA methylation and body fluid-specific microbial DNA. Int J Legal Med 128:33–41. https://doi.org/10.1007/s00414-013-0918-4
Kader F, Ghai M (2015) DNA methylation and application in forensic sciences. Forensic Sci Int 249:255–265. https://doi.org/10.1016/j.forsciint.2015.01.037
Vidaki A, Giangasparo F, Syndercombe Court D (2016) Discovery of potential DNA methylation markers for forensic tissue identification using bisulphite pyrosequencing. Electrophoresis 37:2767–2779. https://doi.org/10.1002/elps.201600261
Lee HY, Jung SE, Lee EH, Yang WI, Shin KJ (2016) DNA methylation profiling for a confirmatory test for blood, saliva, semen, vaginal fluid and menstrual blood. Forensic Sci Int Genet 24:75–82. https://doi.org/10.1016/j.fsigen.2016.06.007
An JH, Choi A, Shin KJ, Yang WI, Lee HY (2013) DNA methylation-specific multiplex assays for body fluid identification. Int J Legal Med 127:35–43. https://doi.org/10.1007/s00414-012-0719-1
Hanssen EN, Avershina E, Rudi K, Gill P, Snipen L (2017) Body fluid prediction from microbial patterns for forensic application. Forensic Sci Int Genet 30:10–17. https://doi.org/10.1016/j.fsigen.2017.05.009
Ingold S, Dørum G, Hanson E, Berti A, Branicki W, Brito P, Elsmore P, Gettings KB, Giangasparo F, Gross TE, Hansen S, Hanssen EN, Kampmann ML, Kayser M, Laurent FX, Morling N, Mosquera-Miguel A, Parson W, Phillips C, Porto MJ, Pośpiech E, Roeder AD, Schneider PM, Schulze Johann K, Steffen CR, Syndercombe-Court D, Trautmann M, van den Berge M, van der Gaag KJ, Vannier J, Verdoliva V, Vidaki A, Xavier C, Ballantyne J, Haas C (2018) Body fluid identification using a targeted mRNA massively parallel sequencing approach—results of a EUROFORGEN/EDNAP collaborative exercise. Forensic Sci Int Genet 34:105–115. https://doi.org/10.1016/j.fsigen.2018.01.002
Haas C, Hanson E, Anjos MJ, Ballantyne KN, Banemann R, Bhoelai B, Borges E, Carvalho M, Courts C, de Cock G, Drobnic K, Dötsch M, Fleming R, Franchi C, Gomes I, Hadzic G, Harbison SA, Harteveld J, Hjort B, Hollard C, Hoff-Olsen P, Hüls C, Keyser C, Maroñas O, McCallum N, Moore D, Morling N, Niederstätter H, Noël F, Parson W, Phillips C, Popielarz C, Roeder AD, Salvaderi L, Sauer E, Schneider PM, Shanthan G, Court DS, Turanská M, van Oorschot RAH, Vennemann M, Vidaki A, Zatkalíková L, Ballantyne J (2014) RNA/DNA co-analysis from human menstrual blood and vaginal secretion stains: results of a fourth and fifth collaborative EDNAP exercise. Forensic Sci Int Genet 8:203–212. https://doi.org/10.1016/j.fsigen.2013.09.009
Haas C, Hanson E, Banemann R, Bento AM, Berti A, Carracedo Á, Courts C, Cock GD, Drobnic K, Fleming R, Franchi C, Gomes I, Hadzic G, Harbison SA, Hjort B, Hollard C, Hoff-Olsen P, Keyser C, Kondili A, Maroñas O, McCallum N, Miniati P, Morling N, Niederstätter H, Noël F, Parson W, Porto MJ, Roeder AD, Sauer E, Schneider PM, Shanthan G, Sijen T, Syndercombe Court D, Turanská M, van den Berge M, Vennemann M, Vidaki A, Zatkalíková L, Ballantyne J (2015) RNA/DNA co-analysis from human skin and contact traces—results of a sixth collaborative EDNAP exercise. Forensic Sci Int Genet 16:139–147. https://doi.org/10.1016/j.fsigen.2015.01.002
Haas C, Hanson E, Anjos MJ, Banemann R, Berti A, Borges E, Carracedo A, Carvalho M, Courts C, de Cock G, Dötsch M, Flynn S, Gomes I, Hollard C, Hjort B, Hoff-Olsen P, Hríbiková K, Lindenbergh A, Ludes B, Maroñas O, McCallum N, Moore D, Morling N, Niederstätter H, Noel F, Parson W, Popielarz C, Rapone C, Roeder AD, Ruiz Y, Sauer E, Schneider PM, Sijen T, Court DS, Sviežená B, Turanská M, Vidaki A, Zatkalíková L, Ballantyne J (2013) RNA/DNA co-analysis from human saliva and semen stains—results of a third collaborative EDNAP exercise. Forensic Sci Int Genet 7:230–239. https://doi.org/10.1016/j.fsigen.2012.10.011
Cossu C, Germann U, Kratzer A, Bär W, Haas C (2009) How specific are the vaginal secretion mRNA-markers HBD1 and MUC4? Forensic Sci Int Genet Suppl Ser 2:536–537. https://doi.org/10.1016/j.fsigss.2009.08.063
Richard MLL, Harper KA, Craig RL, Onorato AJ, Robertson JM, Donfack J (2012) Evaluation of mRNA marker specificity for the identification of five human body fluids by capillary electrophoresis. Forensic Sci Int Genet 6:452–460. https://doi.org/10.1016/j.fsigen.2011.09.007
Sirker M, Schneider PM, Gomes I (2016) A 17-month time course study of human RNA and DNA degradation in body fluids under dry and humid environmental conditions. Int J Legal Med 130:1431–1438. https://doi.org/10.1007/s00414-016-1373-9
Setzer M, Juusola J, Ballantyne J (2008) Recovery and stability of RNA in vaginal swabs and blood, semen, and saliva stains. J Forensic Sci 53:296–305. https://doi.org/10.1111/j.1556-4029.2007.00652.x
Haas C, Hanson E, Kratzer A, Bär W, Ballantyne J (2011) Selection of highly specific and sensitive mRNA biomarkers for the identification of blood. Forensic Sci Int Genet 5:449–458. https://doi.org/10.1016/j.fsigen.2010.09.006
Zhang Y, Liu B, Shao C, Xu H, Xue A, Zhao Z, Shen Y, Tang Q, Xie J (2018) Evaluation of the inclusion of circular RNAs in mRNA profiling in forensic body fluid identification. Int J Legal Med 132:43–52. https://doi.org/10.1007/s00414-017-1690-7
Song F, Luo H, Xie M, Zhu H, Hou Y (2017) Microarray expression profile of circular RNAs in human body fluids. Forensic Sci Int Genet Suppl Ser 6:e55–e56. https://doi.org/10.1016/j.fsigss.2017.09.005
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. https://doi.org/10.1038/nature11928
Xie F, Xiao P, Chen D, Xu L, Zhang B (2012) miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol 80:75–84. https://doi.org/10.1007/s11103-012-9885-2
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) CircRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66. https://doi.org/10.1016/j.molcel.2014.08.019
Hochmeister MN, Budowle B, Rudin O, Gehrig C, Borer U, Thali M, Dirnhofer R (1999) Evaluation of prostate-specific antigen (PSA) membrane test assays for the forensic identification of seminal fluid. J Forensic Sci 44:1057–1060. https://doi.org/10.1520/JFS12042J
Xu Y, Xie J, Cao Y, Zhou H, Ping Y, Chen L, Gu L, Hu W, Bi G, Ge J, Chen X, Zhao Z (2014) Development of highly sensitive and specific mRNA multiplex system (XCYR1) for forensic human body fluids and tissues identification. PLoS One 9:7–13. https://doi.org/10.1371/journal.pone.0100123
Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, Xiang Y, Liu L, Zhong S, Han L, He C (2017) Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform 18:984–992. https://doi.org/10.1093/bib/bbw081
Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, Liu CC, Huang HD (2016) CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res 44:D209–D215. https://doi.org/10.1093/nar/gkv940
Glazar P, Papavasileiou P, Rajewsky N, et al (2014) circBase : a database for circular RNAs circBase : a database for circular RNAs. 0–5. https://doi.org/10.1261/rna.043687.113.overview
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L (2016) Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 26:1277–1287. https://doi.org/10.1101/gr.202895.115
Xu T, Wu J, Han P, Zhao Z, Song X (2017) Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics 18(Suppl 6):680. https://doi.org/10.1186/s12864-017-4029-3
Memczak S, Papavasileiou P, Peters O, Rajewsky N (2015) Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 10:1–13. https://doi.org/10.1371/journal.pone.0141214
Goffin F, Munaut C, Frankenne F, Perrier d’Hauterive S, Béliard A, Fridman V, Nervo P, Colige A, Foidart JM (2003) Expression pattern of metalloproteinases and tissue inhibitors of matrix-metalloproteinases in cycling human endometrium. Biol Reprod 69:976–984. https://doi.org/10.1095/biolreprod.103.015933
Mannello F, Luchetti F, Falcieri E, Papa S (2005) Multiple roles of matrix metalloproteinases during. Apoptosis 10:19–24. https://doi.org/10.1007/s10495-005-6058-7
Verma RP, Hansch C (2007) Matrix metalloproteinases (MMPs): chemical-biological functions and (Q)SARs. Bioorganic Med Chem 15:2223–2268. https://doi.org/10.1016/j.bmc.2007.01.011
Miranda KC, Bond DT, McKee M, Skog J, Păunescu TG, da Silva N, Brown D, Russo LM (2010) Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int 78:191–199. https://doi.org/10.1038/ki.2010.106
Pampalakis G, Scorilas A, Sotiropoulou G (2008) Novel splice variants of prostate-specific antigen and applications in diagnosis of prostate cancer. Clin Biochem 41:591–597. https://doi.org/10.1016/j.clinbiochem.2007.12.022
Cho S-Y, Choi K, Jeon J-H, Kim CW, Shin DM, Lee JB, Lee SE, Kim CS, Park JS, Jeong EM, Jang GY, Song KY, Kim IG (2010) Differential alternative splicing of human transglutaminase 4 in benign prostate hyperplasia and prostate cancer. Exp Mol Med 42:310–318. https://doi.org/10.3858/emm.2010.42.4.031
Del Prete MJ, Robles MS, Guáo A et al (2002) Degradation of cellular mRNA is a general early apoptosis-induced event. FASEB J 16:2003–2005. https://doi.org/10.1096/fj.02-0392fje
Funding
This work was supported by National Natural Science Foundation of China (81571853 and 81701866).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Figure S1
The junction sequences of head-to-tail circRNAs. Results from Sanger sequencing and the arrow indicates the junction sites of head-to-tail products. The locations of circular RNAs in the genome are indicated above each sequence. (PNG 1758 kb)
Figure S2
Target regions of genes for PCR amplification. (A) HBA and ALAS2 for blood. (B) MMP7 and MMP10 for menstrual blood. (C) HTN3 for saliva. (D) ESR1, SPINK5, SERPINB3 and CYP2B7P1 for vaginal secretions. (E) PRM2, TGM4, and KLK3 for semen. (F) MIOX and SLC22A6 for urine. The curved line indicates the downstream 3′ end of an exon is covalently linked with the upstream 5′ end of an exon, which results in the formation of a circular transcript. The regions covered by the horizontal lines represent the position of the amplicons. The blue horizontal lines indicate regions amplified by previously reported primer sets and the red horizontal lines indicate regions amplified by primer sets in this study. Asterisk and triangle indicate regions amplified by L-primers and LC-primers, respectively, in this study. (PNG 219 kb)
Figure S3
The evaluation of expression stability of candidate reference genes. The Cq values of β-actin, β2M, GAPDH and 18S rRNA for samples (n = 4) from each body fluid were used as input data in RefFinder to determine the most stable reference gene by evaluating their stability to generate scores for all candidate genes. Lower score represented more stable genes. (PNG 359 kb)
Figure S4
Dissociation curves from the real-time PCR assay using L-primers (left) and LC-primers (right) of TGM4 (A) and KLK3 (B). The amplification efficiencies were determined using L-primers of TGM4 and KLK3 as well as using LC-primers of TGM4 and KLK3 in qPCR. The cDNA from total RNA of semen was subjected to 10-fold serial dilutions. The standard curves were linear over four to five orders of magnitude with an R2 value of more than 0.98. The slope of the linear equation was used to calculate the efficiency with the following equation: E = 10–1/slope- 1. Each point represents the mean of three replicates. (PNG 362 kb)
Table S1
(DOCX 15 kb)
Table S2
(DOCX 17 kb)
Table S3
(DOCX 17 kb)
Table S4
(DOCX 24 kb)
Table S5
(DOCX 17 kb)
Rights and permissions
About this article
Cite this article
Liu, B., Song, F., Yang, Q. et al. Characterization of tissue-specific biomarkers with the expression of circRNAs in forensically relevant body fluids. Int J Legal Med 133, 1321–1331 (2019). https://doi.org/10.1007/s00414-019-02027-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-019-02027-y