Abstract
Short tandem repeat (STR) typing from skeletal remains can be a difficult task. Dependent on the environmental conditions of the provenance of the bones, DNA can be degraded and STR typing inhibited. Generally, dense and compact bones are known to preserve DNA better. Several studies already proved that femora and teeth have high DNA typing success rates. Unfortunately, these elements are not present in all cases involving skeletal remains. Processing partial or singular skeletal elements, it is favorable to select bone areas where DNA preservation is comparably higher. Especially, cranial bones are often accidentally discovered during criminal investigations. The cranial bone is composed of multiple parts. In this examination, we evaluated the potential of the petrous bone for human identification of skeletal remains in forensic case work. Material from different sections of eight unknown cranial bones and—where available—additionally other skeletal elements, collected at the DNA department of the Institute of Legal Medicine in Ulm, Germany, from 2010 to 2017, were processed with an optimized DNA extraction and STR typing strategy. The results highlight that STR typing from the petrous bones leads to reportable profiles in all individuals, even in cases where the analysis of the parietal bone failed. Moreover, the comparison of capillary electrophorese (CE) typing to massively parallel sequencing (MPS) analysis shows that MPS has the potential to analyze degraded human remains and is even capable to provide additional information about phenotype and ancestry of unknown individuals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Considering its structure, double-stranded DNA is a stable inert molecule. This advantage enables the reliable use of molecular genetics for the attribution of crime scene traces to a perpetrator in forensic science. Mostly, the forensic molecular analysis comprises biological samples that originate from biofluids and carry enough cellular material for STR analysis [1].
Identifying human remains from autopsies, from mass graves, or for paternity testing, biological material like blood or saliva is either not present or strongly degraded due to advanced putrefaction and cannot be used for DNA analysis anymore. In those cases, remaining/available bones and teeth have to be used for molecular identification. Preservation of DNA in bones is favored because of the binding of the DNA to hydroxyapatite [2]. Ambient temperatures also increase the chance of DNA survival by slowing down hydrolysis and oxidation processes [3]. Unfortunately, the environmental conditions of samples used for forensic identification are often not ideal for DNA recovery. In skeletal remains, the presence of inhibitors found at the excavation side or crime scene, e.g., humic acids, inhibits the amplification process resulting in a reduced amount of amplified PCR product [4]. Additionally, contamination with modern DNA occurring from working staff during exhumation, improper storage, and anthropological investigation makes it even more challenging to gain genuine STR profiles and sequences from skeletal remains [3].
An efficient extraction method for the analysis of challenging bone and teeth samples should remove all inhibitors as well as gain the maximum of the preserved DNA [5].
In the last years, researchers found out that total demineralization is a crucial step in bone and teeth analysis and many extraction protocols are based on this approach [e.g., 6,7,8,9,10].
Also, the choice of the sampling material can influence the STR typing. In general, dense and compact bones are known to preserve DNA better than cancellous and brittle bones do [3, 10, 11]. The results of multiple studies highlight that long bones and teeth are the most appropriate samples for genetic analysis [12,13,14,15,16,17]. Especially, sampling material from the diaphysis of femora and humeri can contain high amounts of endogenous DNA [18]. According to Edson et al. [12] and Milos et al. [13], skull bones are one of the least suitable elements for genetic analysis. Recently, however, ancient DNA (aDNA) studies have shown that the inner part of the petrosal bone can be used for sequencing analysis of ancient remains from Holocene archeological context [19] and even from warm climates from Early Neolithic [20].
To produce investigative leads in human identification cases, additional information about the individuals can be gained from a variety of markers including ancestry-informative markers and phenotype-informative single nucleotide polymorphism (SNP). Massively parallel sequencing (MPS) is a high throughput technology that offers the capacity to analyze a combined marker set of repeat and sequence variants simultaneously without any further sample consumption [21]. The Illumina marker set consist altogether of a set of 229 markers.
The aim of this examination was to evaluate the potential of the petrous bone, one of the most compact bones in the human body, for human identification of skeletal remains in forensic casework. Bone powder was taken from endogenous petrous bones from a selection of eight craniums that have been collected at the DNA department of the Institute of Legal Medicine in Ulm, Germany, from 2010 to 2017 and was processed with an optimized extraction and STR typing strategy. Furthermore, the applicability of MPS for the characterization of individual’s ancestry and external physical traits of the unidentified human remains was evaluated and outcome of MPS STR analysis was compared to the results of conventional PCR/CE analysis.
Material and methods
Samples
We analyzed eight cases involving cranial bones that have been preserved over the last 7 years. All skulls are accidental discoveries that were introduced to our laboratory by criminal investigation departments. One case was a recent identification and the whole skeleton was available. The state of preservation and the integrity of the skull bones varied. In one case, teeth were additionally available for analysis, while in the other cases, the mandibles were completely missing. In all cases except B3 and B7, alternative skeletal elements, e.g., the parietal bone or a femur, have been analyzed too to show differences in sample selection.
Age, geographical locations, and climate context as well as environmental conditions of the provenance of the specimen are unknown, except for case B1 where 14C radiocarbon dating and a morphological age determination were accomplished. The histomorphological data suggest that the individual from case B1 was between 15 and 30 years old. Radiocarbon dating shows that the 14C concentration lies within the range of an age plateau and can only be vaguely dated to originate in the years between 1640 and 1960. Also, during the investigation of case B8, the cranium could be assigned to a missing person who disappeared in 1998.
Sample preparation
The temporal bone was isolated with an oscillating saw and photo-documented. The selection of the sampling area (see Fig. 1) within the temporal bone was done according to Hines et al. [22] and Pinhasi et al. [19] who performed an intra-sample comparison of three distinct areas within the petrous bone for DNA extraction. Bone powder was obtained intracranially with a rasp after the outer layer was rasped and discarded, because of potential contamination with modern DNA. Alternative sampling was performed simultaneously from a femur, a tooth, or parietal bones with the single difference that the tooth was decontaminated prior to sampling and only 15 mg of tooth powder was used for extraction procedure. For decontamination, the tooth was washed twice with commercial bleach (10%) and shaken for 30 s. The wash was followed by a second one with deionized water that was submerged 5 min. A final wash was accomplished with 96% ethanol. Afterwards, the tooth was placed in a sterilized fume hood for at least 2 h to air-dry. Other precautions regarding contaminations during DNA analysis were arranged, too: Pre-PCR and post-PCR procedures were performed in separated rooms under a particulate air filter. Gloves, a face mask, a disposable laboratory coat, as well as a snood were worn at all experimental steps. Negative extraction and PCR controls were performed in parallel with each extraction.
DNA extraction
Total demineralization was carried out with ~300 mg bone powder in a 2ml tube with 1.5 ml of a demineralization buffer according to [23] containing EDTA (pH 8.0, 0.5 M) with N-laurylsarcosinate, ATL buffer (Qiagen, Hilden), Proteinase K, and DTT. The mixture was incubated at 56 °C in a rotating shaker. After 20 h of incubation, the lysate was shortly centrifuged to separate the remaining bone powder from the solvent phase. To concentrate the DNA, repetitively, 500 μl of supernatant was pipetted on a Microcon DNA Fast Flow Centrifugal Filter Unit (Merck Millipore, Darmstadt) and centrifuged at 500 g until the flow through has passed. The lysate was collected and an automated extraction procedure was carried out using the Maxwell® RSC Blood DNA Kit (Promega, Mannheim) on the Maxwell® RSC instrument (Promega, Mannheim) according to the manufacturer’s recommendations. The elution volume was 50 μl.
Quantification
DNA extracts were quantified with the PowerQuant™ System (Promega, Mannheim) according to the manufacturer’s recommendations using 5 μl of extract. Amplification was carried out on the 7500 Real-Time PCR System (Applied Biosystems, Darmstadt). Data were analyzed with the PowerQuantAnalysis 1.0.0 tool. The PowerQuant™ Kit provides quantitative analysis of human and male DNA and additionally determines a degradation index (DI) using the ratio of one long and one short amplicon and an internal PCR control (IPC), which is highly beneficial for degraded forensic samples like bones.
STR typing and capillary electrophoresis
Amplification was accomplished with two distinct kits, with alleles considered as genuine only if present in both amplifications or if the allele could be clearly assigned. A result was classified as successful if reproducible results were obtained from a minimum of 8 autosomal loci and the sex indicating locus amelogenin. STR typing was performed with the Investigator® ESSplex SE QS Kit (Qiagen, Hilden) and with the PowerPlex® ESX 17 Fast System (Promega, Mannheim) according to the manufacturer’s protocol on a Biometra thermocycler with 30 cycles using 10 μl of DNA extract. For case B1 and case B2, the DNA analysis of the alternative skeletal elements was accomplished in 2010–2011. During this period, the in-house developed Multiplex Kit P11 including 11 markers (based on the Q8 system) was used for STR typing [24].
An ABI PRISM 3130 Genetic Analyzer was used for product separation and detection of amplicons. For the capillary electrophoresis, 1 μl of amplified DNA product was added to 12 μl Hi-Di™ formamide and 0.5μl-size standard BTO550 for ESSplex SE QS Kit detection or mixed with 10 μl Hi-Di™ formamide and 1 μl of WEN ILS 500 pro for the detection of ESX 17 Fast products. Reaction plates were sealed and incubated for 5 min at 92 °C for denaturation. Samples were injected for 16 s at 1.2 kV using a 36cm array with performance optimized polymer (POP) 7 as a separation matrix. The resulting data were analyzed using GeneMapper ID v3.2 with a defined peak threshold of 50 relative fluorescent units (rfu).
Massive parallel sequencing with the Illumina MiSeq®
Sequencing was performed on the Illumina MiSeq® FGx Forensic Genomics System at the Bavarian State Criminal Police Office in Munich. The ForenSeq® Signature Prep Kit was used to prepare libraries according to the manufacturer’s recommendations [25]. All cases were included into the analysis and consensus profiles were obtained from two replicate analyses. The analysis comprised both primer sets A and B resulting in the overall evaluation of 229 markers: amelogenin, 27 autosomal STRs, 24 Y-STRs, 7 X-STRs, 54 biogeographical ancestry SNPs, 94 identity SNPs, and 22 phenotype-informative SNPs. Thresholds were set up as recommended: analytical threshold was at 4.5% and interpretation threshold at 1.5% of the total reads of the locus. Intralocus balance of STRs and SNP was set to 60%. Stutter bands for STR loci were identified according to predefined % intensity values (ranging from 7.5–33% for autosomal STRs and from 15–50% for X- and Y-STRs) reported in the ForenSeq Universal Analysis Software Guide [25]. Amplifications (PCR1 and PCR2) were carried out in a Mastercycler thermocycler (Eppendorf, Hamburg) according to the manufacturer’s protocol using 5 μl of the DNA extracts. Following normalization, 10 μl of the normalized libraries was pooled and sequenced.
Data analysis
Analysis of the MPS data was accomplished by the ForenSeq® Universal Analysis Software (UAS). Concordance was assessed by the comparison of the overlapping 16 STR loci of the MiSeq system and of the ABI 3130 instrument generated data using the Investigator ESSplex SE QS (Qiagen, Hilden) and the PowerPlex® ESX 17 Fast System (Promega, Mannheim).
Statistics
Principal component analysis (PCA) of ancestry-informative SNP data as well as phenotyping SNP data evaluation was accomplished by the UAS. Phenotyping SNP data for bones 6 and 8 were not evaluated by the UAS system. So SNP data were analyzed with the HIrisPlex eye and hair color DNA phenotyping webtool (http://hirisplex.erasmusmc.nl/#) [26, 27].
Results and discussion
This examination aims to assess the potential of the petrous bone for the individualization of unidentified human skeletal remains in forensic casework. Moreover, the performance of MPS STR analysis was compared to the results of conventional PCR/CE analysis and the applicability of MPS for the characterization of physical traits, and biogeographical ancestry was evaluated.
Optimization of DNA extraction
In the last years, plenty optimized DNA extraction protocols for bone samples have been published. Most of them include a total demineralization step intended to improve the DNA recovery, which is followed by a silica-based column extraction [6, 8, 9]. Some of the presented protocols, especially the procedure published by Huel et al. [23] for the International Commission on Missing Persons (ICMP) in 2012, require huge volumes of reagents and are very time consuming. Based on the workflow of the ICMP, we developed an optimized extraction procedure for degraded skeletal remains in our laboratory. Modifications were done concerning the amounts of the detergents that were used for full demineralization of the bone powder. Furthermore, we combined the down-scaled protocol with an automatization of the extraction procedure using the Maxwell extraction platform, which we also use for the routine case work. The modifications have mainly two advantages. First of all, the implication of an extraction automat minimizes the risk of contamination in comparison to manual protocols. Moreover, using a down-scale approach improves the comfortable handling of bone samples and reduces the cost per sample.
DNA quantification and STR typing
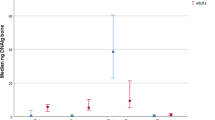
Applying the PowerQuant™ System for quantification has the benefit of an integrated degradation sensor, which shows if the samples are degraded by detecting the presence of a 294 basepair (bp) long amplicon. As expected, the degradation index for all samples was above the threshold of 2 and indicated a high degradation of the samples. The cases B1, B4, and B6 exhibited the highest degradation ratios, reaching 150.25, 24.93, and 20.02, respectively. DNA concentrations recovered from the petrous bones varied widely between individuals and ranged between 5.9 and 102.7 pg/μl. The mean DNA yield was 44.4 pg/μl (±35.4 pg/μl). DNA concentrations from other skeletal elements showed comparably lower amounts. Samples from the parietal bone produced DNA yields under 5 pg/μl, while DNA yields from tooth and femur were high enough to result in full profiles (Table 1).
PCR amplification was carried out with two PCR kits: the recently released Investigator® ESSplex SE QS Kit (Qiagen) and the PowerPlex® ESX 17 Fast System (Promega). Consensus profiles were recorded for the incorporation into the German database using at least 8 loci. Harder et al. [28] have shown the usability of certain STR kits for DNA profiling of highly challenging bones samples. They concluded that especially the ESI and ESX kits (Promega, Mannheim) as well as the NGM Kit (Thermo Fisher, Darmstadt) should be preferred for the analysis of highly degraded ancient material. Zupanič Pajnič et al. [29] also recommend the ESX Kit and the Investigator ESSplex Kit (Qiagen) for bone analysis.
Here, we used the follow-ups of both kits; PowerPlex® ESX 17 Fast System and Investigator® ESSplex SE QS. Our results demonstrate that both kits have proven to be suitable to analyze degraded bone samples. The Investigator® ESSplex SE QS Kit showed high sensitivity and short amplicons, especially for SE33. Additionally, the simultaneous application of the ESX Fast Kit demonstrated concordant results.
All in all, informative profiles were obtained for all eight bone samples and all profiles could be reported and saved for the search in the database (see Table 1). Moreover, alternative samples have been analyzed in all cases, except B3 and B5. Bone powder of the parietal bone was obtained for cases B1 and B2 and B4. A tooth was used for DNA extraction in case B6. Cases B7 and B8 were additionally analyzed using bone powder from long bones. The amplification of the alternative samples (see Table 1) show—as expected—that using a tooth and long bones for STR typing results in complete profiles as well. However, the analysis of bone powder gained from the parietal bones showed only sporadic results that were not sufficient for inclusion into the database or failed totally.
Our results are concordant with data from Edson et al. [12] who could successfully amplify 51% of cranium bones in 2004. Also, Mundorff et al. [14] yielded in 40–50% of their cases profiles using the cranial bones. In 2009, Edson et al. [30] unraveled the results according to the distinct elements of the cranium and reported that temporal bones (90%) yielded comparably higher results than parietal (52%), frontal (68%), or occipital bones (65%). In comparison with the results of Edson et al. [30], our amplification success of temporal bone samples was even higher. We were able to fully analyze the eight cranial bones, including five specimens that have been stored in our department since 2010 and report meaningful profiles to the database.
Furthermore, Rothe et al. analyzed a group of four medieval individuals excavated from St. Peters graveyard in Berlin using temporal bones, teeth and femora. Their results were comparable to our conclusions. In three cases, they were able to produce full profiles using petrous bones and in one case they generated a partial profile. Using femur or teeth for STR profiling produced in three cases partial profiles and in one case even no STR profile at all [31].
Concordance between CE and MPS
Concordance was evaluated between the length-based CE genotype and the length-based NGS genotype for 16 overlapping loci. Consensus profiles of the samples that were analyzed with MPS and CE are shown in Table 2. Autosomal allele calls of the samples between MPS and CE were mostly concordant (see an example of B5 in Supplementary data 1). In cases B1 and B2, MPS provided additional allele calls where CE showed allelic dropout and vice versa (see Table 3). The analysis of sample B6 showed some discrepancies. Some of them may be explained by stochastic phenomena since the DNA amount of sample B6 was comparably low (5.9 pg/μl). This may especially have an impact on the MPS analysis since the input volume of the DNA extract is limited to 5 μl when using the ForenSeq® Signature Prep Kit. Therefore, just the half amount of DNA (less than 30 pg) compared to CE was analyzed with MPS. Other inconsistencies may resulted after stutter evaluation. Stutter interpretation and threshold setting are important issues in STR typing and should be defined carefully. Further validation work will show if the stutter rates proposed by Illumina will fit the needs for the interpretation of forensic challenging samples.
Furthermore, the UAS flags data deviating from the thresholds by a series of quality control indicators (QC indicators). QC indicators occur as interpretation threshold, imbalanced, stutter, or allele count. Undetected loci are indicated as inconclusive (INC). The number of flagged loci during MPS analysis of the bones seems to be elevated in comparison to the MPS analysis of buccal swabs that contain high amounts of DNA (data not shown). Inconclusive results due to dropout were also observed. Noticeable dropout during MPS occurred in D12S391, Penta D, and Penta E. D12S391 detection was completely missing in B4 and B6, while partial dropout was observed in two cases (B1 and B2). Likewise, Penta E also shows increased dropout. The locus was absent in five of seven samples (B1, B2, B3, B4 and B6). Penta D dropout was observed in samples B2, B3, and B6. Increased dropout of the three loci can be explained by the lengths of the amplicons. Amplicon length of D12S391 lies between 237 and 281 bp, while Penta D and Penta E produce amplicons between 209 and 293 bp and 362 and 467 bp, respectively.
Generally, STR assays that are routinely used for STR typing consist of 17 loci. Kits containing more than 17 loci are already available, e.g., the PowerPlex® Fusion System from Promega or the GlobalFiler® PCR Amplification Kit from Thermo Fisher. Both kits analyze 24 loci simultaneously. Contrary to the Promega system, the latter requires a new generation of CE sequencers that allow the parallel analysis of six dyes. In addition to the routinely used STR markers, MPS provides even more autosomal STRs: altogether, up to 27 loci and amelogenin can be detected. Furthermore, 24 Y-STRs and 7 X-STRs are implemented into the analysis. However, one deficit of the MPS STR marker set is the absence of marker SE33. Because of its polymorphic nature, it is highly informative and used for the database set in Germany. In contrast to conventional STR typing, MPS provides sequence information of all analyzed alleles. It is expected that a larger population study will provide a diversity of alleles with same length but different sequences (so called isoalleles). First examinations of 120 individuals at the BLKA already show that isoalleles appeared in 53% of all buccal swab samples in at least one locus. In our limited sample data set, we found two isoalleles, one in vWA, and another one in D3S1358 (Table 4). Generally, the presence of an intra-allelic variant can be used to discriminate between stutter and true alleles and can help to deconvolute mixtures in forensics casework.
Forensic DNA phenotyping and ancestry-informative SNPs
Since the analysis of skeletal remains can benefit from the prediction of other informative traits like biogeographical origin and phenotype, we included both primer mixes A and B of the ForenSeq® Signature Prep Kit into the analysis with the llumina MiSeq® system. Altogether, the Illumina marker sets consists of 229 markers including amelogenin, 27 autosomal STRs, 24 Y-STRs, 7 X-STRs, 54 biogeographical ancestry SNPs, 94 iSNPs, and 22 phenotype-informative SNPs. Table 5 shows an overview of the markers that were detected for each samples. In general, it can be concluded that decreased DNA amounts lead to dropout of STRs and SNPs. Especially, longer amplicons tend to be not detected, e.g., iSNP rs1736442, which is 153 bp long is missing in almost all analyzed bone samples.
Eye and hair color as well as biogeographical ancestry of all samples were predicted by the UAS after MPS analysis. The AUS outputs biogeographical results in the form of a PCA predicting the individuals’ general population background. The differentiation is based on data from four distinct populations (European, Ad Mixed American, East Asian, and African). The PCA shows that all analyzed individuals can be assigned either to European or Ad Mixed American populations (see Fig. 2). The results of hair and eye color prediction are summarized in Table 6. Generally, the closer the AUC value is to 1, the accurate the prediction. Except for samples B6 and B8, hair and eye color could be predicted for every sample. Sample B6 had the lowest DNA concentration measured with only 5.9 pg/μl. This minute DNA amounts lead to incomplete MPS analysis: Only 12 SNPs (55%) for hair and eye color prediction were analyzed and only 40 (74%) biogeographical SNPs were detected. While biogeographical predication showed that the individual descended from Europe, hair and eye color prediction failed. Nevertheless, sequenced SNPs could be used for eye and hair color probability estimation applying the HIrisPlex eye and hair color DNA phenotyping webtool, which is available via the publically accessible website (http://hirisplex.erasmusmc.nl/#, access 13.05.17). This tool uses a prediction model, which is based on phenotype and genotype data of 9188 European individuals [26, 27] for eye color estimation. The input of the SNP data produced a valid result estimating that the individuals’ most likely predicted eye color is blue with almost 92%. The data set for hair color estimation consists of 1601 individuals. In addition to the hair color, the tool also gives probabilities for the hair shades. Probability for light hair in this case was 0.97. The combination of hair color and hair shade can be used to make a final prediction. According to Walsh et al. 2013, highest p value is utilized. In this case, the highest p value was obtained with 0.74 for blond hair. The combination of the p values for blond hair and light shade suggests that the individual had blond hair. The same evaluation was also applied for bone sample 8. The results indicate that the individual had blue eyes and blond hair, too.
However, phenotyping as well as biogeographic data should be treated with caution since in Germany, e.g., it is prohibited by the German Code of Criminal Procedure to use that information for case work except in cases of identification (§88 StPO). Additionally, the comparison of known physical traits in a study in our institutions (data not shown) showed that the results were not always concordant. Hence, further input and validation are needed to increase the robustness of the MPS technology, especially of the phenotyping and biogeographical SNPs.
Concluding remarks
By optimizing the DNA extraction procedure as well as by using the sensitive Investigator® ESSplex SE QS, we created a highly effective strategy for the DNA analysis of skeletal remains (see Fig. 3) in our department. In comparison to the workflow that was previously used in our laboratory (non-optimized DNA extraction from parietal bone and analysis with in-house kit), the optimized procedure yielded higher DNA amounts and resulted in a greater amplification success. Additionally, the risk of contamination was diminished by the automatization of the extraction process. Further, the successful analysis of bone material could be improved by the specific selection of sample material. Using bone powder of the endogenous petrous bone, one of the hardest bones in the human body, showed in all cases reproducible profiles, which we were able to report to the database. Hereafter, considering this remarkable STR typing results, our work supports the hypothesis that DNA analysis from dense petrous bone is likely to provide reportable DNA profiles. Therefore, we recommend to use the petrous bone for DNA analysis in casework where only cranial bones are available. Additionally, the implementation of MPS showed concordant STR results to the CE-based analysis. This finding is in accordance with the results of comparable studies that where applied with reference samples [32, 33], aged buccal swab samples [34], and DNA samples artificially enriched in apurinic-apyrimidinic sites [35] that also demonstrated that MPS and conventional STR and Y-STR typing are concordant. The concordance of the results shows that MPS has the potential to reach the same sensitivity as current DNA typing technologies. Higher sensitivity of the MPS analysis may be achieved by slightly adapting the ForenSeq® Signature Prep Kit protocol and workflow for low template DNA analysis conditions, e.g., by raising the input volume for the Illumina kit to at least 10 μl of the DNA extract, which is standard for most CE-based STR kits.
In most cases, the conventional STR typing provides sufficient discrimination power. However, the analysis of skeletal remains could frequently benefit from the simultaneous detection of additional STR loci on autosomes and sex chromosomes and the analysis of SNPs related to ancestry and physical characteristics like eye and hair color. Additional information can be used to narrow down the assignment of potential missing persons to skeletal remains making it possible for criminal investigations to evolve from the “passive comparison” into the “active search” stage [36]. Therefore, it can be concluded that with its capacity for simultaneous analysis of multiple types of DNA markers, MPS technology is a promising platform for the genetic analysis of human remains.
References
Wiegand P, Rolf B (2003) Analyse biologischer Spuren Teil II: DNA-Typisierung. Rechtsmedizin 13:375–383. doi:10.1007/s00194-003-0230-6
Kemp BM, Smith DG (2005) Use of bleach to eliminate contaminating DNA from the surface of bones and teeth. Forensic Sci Int 154:53–61. doi:10.1016/j.forsciint.2004.11.017
Keyser-Tracqui C, Ludes B (2005) Methods for the study of ancient DNA. Methods Mol Biol 297:253–264
Alaeddini R (2012) Forensic implications of PCR inhibition—a review. Forensic Sci Int Genet 6:297–305. doi:10.1016/j.fsigen.2011.08.006
Zupanič Pajnič I (2016) Extraction of DNA from human skeletal material. In: William Goodwin (ed) Forensic DNA typing protocols, methods in molecular biology, 1420, Springer Science+Business Media, New York, pp 89–108. doi: 10.1007/978-1-4939-3597-0_6
Loreille OM, Diegoli TM, Irwin JA, Coble MD, Parsons TJ (2007) High efficiency DNA extraction from bone by total demineralization. Forensic Sci Int Genet 1:191–195. doi:10.1016/j.fsigen.2007.02.006
Jakubowska J, Maciejewska A, Pawlowski R (2012) Comparison of three methods of DNA extraction from human bones with different degrees of degradation. Int J Legal Med 126:173–178. doi:10.1007/s00414-011-0590-5
Amory S, Huel R, Bilic A, Loreille O, Parsons TJ (2012) Automatable full demineralization DNA extraction procedure from degraded skeletal remains. Forensic Sci Int Genet 6:398–406. doi:10.1016/j.fsigen.2011.08.004
Lee HY, Park MJ, Kim NY, Sim JE, Yang WI, Shin KJ (2010) Simple and highly effective DNA extraction methods from of skeletal remains using silica columns. Forensic Sci Int Genet 4:275–280. doi:10.1016/j.fsigen.2009.10.014
Rohland N, Hofreiter M (2007) Ancient DNA extraction from bones and teeth. Nat Protoc 2:1756–1762. doi:10.1038/nprot.2007.247
Parsons TJ, Weedn VW (1997) Preservation and recovery of DNA in postmortem specimens and trace samples. In: Haglund WD, Sorg MS (eds) Forensic taphonomy: the postmortem fate of human remains. CRC Press, Boca Raton, FL, pp 109–138
Edson S, Ross JP, Coble MD, Parsons TJ, Barritt SM (2004) Naming the dead—confronting the realities of rapid identification of degraded skeletal remains. Forensic Sci Rev 16:64–89
Milos A, Selmanovic A, Smajlovic L, Huel RLM, Katzmarzyk C, Rizvic A, Parsons TJ (2007) Success rates of nuclear short tandem repeat typing from different skeletal elements. Croat Med J 48:486–493
Mundorff AZ, Bartelink EJ, Mar-Cash E (2009) DNA preservation in skeletal elements from the world trade center disaster: recommendations for mass fatality management. J Forensic Sci 54(4):739–745. doi:10.1111/j.1556-4029.2009.01045.x
Misner LM, Halvorson AC, Dreier JL, Ubelaker DH, Foran DR (2009) The correlation between skeletal weathering and DNA quality and quantity. J Forensic Sci 54(4):822–828. doi:10.1111/j.1556-4029.2009.01043.x
Alt KW, Brandt G, Knipper C, Lehn C (2013) Empfehlungen für die Probenentnahme in der forensischen Anthropologie – Untersuchung von DNA und Stabilisotopen. Rechtsmedizin 24:179–185. doi:10.1007/s00194-014-0950-9
Alonso A, Andelinovic S, Martin P, Sutlovic D, Erceg I, Huffine E, de Simón LF, Albarrán C, Definis-Gojanović M, Fernández-Rodriguez A, Carciá P, Drmić I, Rezić B, Kuret S, Sancho M, Primorac D (2001) DNA typing from skeletal remains: evaluation of multiplex and megaplex STR systems on DNA isolated from bone and teeth samples. Croat Med J 42(3):260–266
Burger J, Bollongino R (2010) Richtlinien zur Bergung, Entnahme und Archivierung von Skelettproben für palaeogenetische Analysen [Guidelines for the recovery, acquisition and storage of skeletal samples for palaeogenetical analyses]. Bulletin der Schweizerischen Gesellschaft für Anthropologie 16(1–2):71–78
Pinhasi R, Fernandes D, Silak K, Novak M, Connell S, Alpaslan-Roodenberg S, Gerritsen F, Moiseyev V, Gromow A, Raczky P, Anders A, Pietrusewsky M, Rollefson G, Jovanovic M, Trinhhoang H, Bar-Oz G, Oxenham M, Matsumara H, Hofreiter M (2015) Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLoS One 10(6):e0129102. doi:10.1371/journal.pone.0129102
Gamba C, Jones ER, Teasdale MD, Mclaughlin RL, Gonzales-Fortes G, Mattiangeli V, Domboróczki L, Kóvári I, Pap I, Anders A, Whittle A, Dani J, Raczky P, Higham TFG, Hofreiter M, Bradley DG, Pinhasi R (2014) Genome flux and stasis in a five millennium transect of European prehistory, Nat Commun 5. doi: 10.1038/ncomms6257
Børsting C, Morling (2015) Next generation sequencing and its applications in forensic genetics. Forensic Sci Int Genet 18:78–89. doi:10.1016/j.fsigen.2015.02.002
Hines DZC, Vennemeyer M, Amory S, Huel RLM, Hanson I, Katzmaryk C, Parsons TJ (2014) Prioritized sampling of bone and teeth for DNA analysis in commingled cases. In: Adams BJ, Byrd JE (eds) Commingled human remains—methods in recovery, analysis, and identification. Elsevier, Amsterdam, pp 275–305
Huel R, Amory S, Bilić A, Vidović S, Jasaragić E, Parsons TJ (2012) DNA extraction from aged skeletal samples for STR typing by capillary electrophoresis. In: Alonso A (ed) DNA electrophoresis protocols for forensic genetics, methods in molecular biology, 830. Springer Science+Business Media, LLC, Berlin. doi:10.1007/978-1-61779-461-2_13
Müller K, Klein R, Miltner E, Wiegand P (2007) Q8—a short amplicon multiplex including the German DNA database systems. Forensic Sci Int Genet 1:205–207. doi:10.1016/j.fsigen.2007.02.005
Illumina ForenSeq™ DNA Signature Prep Guide. 2014. http://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/forenseq/forenseq-dna-signature-prep-guide-15049528-c.pdf
Walsh S, Liu F, Wollstein A, Kovatsi L, Ralf A, Kosiniak-Kamysz A, Branicki W, Kayser M (2013) The HIrisPlex system for simultaneous prediction of hair and eye color from DNA. Forensic Sci Int Genet 7:98–115. doi:10.1016/j.fsigen.2012.07.005
Walsh S, Chaitanya L, Clarisse L, Wirken L, Draus-Barini J, Kovatsi L, Maeda H, Ishikawa T, Sijen T, de Knijff P, Branicki W, Liu F, Kayser M (2014) Developmental validation of the HIrisPlex system: DNA-based eye and hair colour prediction for forensic and anthropological usage. Forensic Sci Int Genet 9:150–161. doi:10.1016/j.fsigen.2013.12.006
Harder M, Renneberg R, Meyer P, Krause-Kyora B, von Wurmb-Schwark N (2012) STR-typing of ancient skeletal remains: which multiplex-PCR is the best? Croat Med J 53:416–422. doi:10.3325/cmj.2012.53.416
Zupanič Pajnič I, Pogorelc BG, Balažic J, Zupanc T, Štefanič B (2012) Highly efficient nuclear DNA typing of the World War II skeletal remains using three new autosomal short tandem repeat amplification kits with the extended European standard set of loci. Croat Med J 53:17–23. doi:10.3325/cmj.2012.53.17
Edson SM, Christensen AF, Barritt SM, Meehan A, Leney MD, Finelli LN (2009) Sampling of the cranium for mitochondrial DNA analysis of human skeletal remains. Forensic Sci. Int. Genet 2:269–270. doi:10.1016/j.fsigss.2009.09.029
Rothe J, Melisch C, Powers N, Geppert M, Zander J, Purps J, Spors B, Nagy M (2015) Genetic research at a fivefold children’s burial from medieval Berlin. Forensic Sci Int Genet 15:90–97. doi:10.1016/j.fsigen.2014.10.022
Just RS, Moreno LI, Smerick JB, Irwin JA (2017) Performance and concordance of the ForenSeq™ system for autosomal and Y chromosome short tandem repeat sequencing of reference-type specimens. Forensic Sci Int Genet 28:1–9. doi:10.1016/j.fsigen.2017.01.001
Van der Gaag KJ, de Leeuw RH, Hoogenboom J, Patel J, Storts DR, Laros JFJ, de Knijff P (2016) Massively parallel sequencing of short tandem repeats—population data and mixture analysis results for the PowerSeq™ system. Forensic Sci Int Genet 24:86–96. doi:10.1016/j.fsigen.2016.05.016
Churchill JD, Schmedes SE, King JL, Budowle B (2016) Evaluation of the Illumina® beta version ForenSeq™ DNA signature prep kit for use in genetic profiling. Forensic Sci Int Genet 20:20–29. doi:10.1016/j.fsigen.2015.09.009
Fattorini P, Previderé C, Carboni I, Marrubini G, Sorçaburu-Cigliero S, Grignani P, Bertoglio B, Vatta P, Ricci U (2017) Performance of the ForenSeq™ DNA signature prep kit on highly degraded samples. Electrophoresis 38:1163–1174. doi:10.1002/elps.201600290
Yang Y, Xie B, Yan J (2014) Application of next-generation sequencing technology in forensic science. Genomics, Proteomics, Bioinformatics 12:190–197. doi:10.1016/j.gpb.2014.09.001
Acknowledgements
We kindly thank our dissector Gabriele Kottmair for the helping hand with the bone preparation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Kulstein, G., Hadrys, T. & Wiegand, P. As solid as a rock—comparison of CE- and MPS-based analyses of the petrosal bone as a source of DNA for forensic identification of challenging cranial bones. Int J Legal Med 132, 13–24 (2018). https://doi.org/10.1007/s00414-017-1653-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-017-1653-z