Abstract
In order to better characterize and understand the mtDNA population genetics of Central Asia, the mtDNA control regions of over 1,500 individuals from Uzbekistan have been sequenced. Although all samples were obtained from individuals residing in Uzbekistan, individuals with direct ancestry from neighboring Central Asian countries are included. Individuals of Uzbek ancestry represent five distinct geographic regions of Uzbekistan: Fergana, Karakalpakstan, Khorezm, Qashkadarya, and Tashkent. Individuals with direct ancestry in nearby countries originate from Kazakhstan, Kyrgyzstan, Russia, Afghanistan, Turkmenistan, and Tajikistan. Our data reinforce the evidence of distinct clinal patterns that have been described among Central Asian populations with classical, mtDNA, and Y-chromosomal markers. Our data also reveal hallmarks of recent demographic events. Despite their current close geographic proximity, the populations with ancestry in neighboring countries show little sign of admixture and retain the primary mtDNA patterns of their source populations. The genetic distances and haplogroup distributions among the ethnic populations are more indicative of a broad east–west cline among their source populations than of their relatively small geographic distances from one another in Uzbekistan. Given the significant mtDNA heterogeneity detected, our results emphasize the need for heightened caution in the forensic interpretation of mtDNA data in regions as historically rich and genetically diverse as Central Asia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Central Asia, which can generally be defined as the republics of Kazakhstan, Kyrgyzstan, Uzbekistan, Tajikistan, and Turkmenistan has garnered significant attention in studies of human evolutionary history, due to the region’s central geographic location in Eurasia and its resulting importance as a crossroad of human population movement. The Central Asian steppes are believed to have been populated as early as 45–50 KYA by anatomically modern humans [1], and since that time, the area has served as a major corridor for population migrations between Asia, the Middle East, India, and Europe.
The genetics of Central Asian populations have been intensively studied in efforts to elucidate the route(s) by which anatomically modern humans populated Asia and Europe. Because Central Asia is situated in a key geographical position, the genetics of the region have offered some insight into the possibility of a proposed “northern route” out of Africa through Central Asia and into the East [2–5]. Early genetic studies based on classical markers placed Central Asia at an intermediate and central position in a general west–east genetic cline that included western Eurasian, eastern Eurasian, and even southern Asian components [2]. These results were interpreted as a signal of population expansions extending from western Asia through Central Asia and into the East, in general support of a northern route. Subsequent studies of mtDNA markers also clearly detected clinal patterns of eastern and western lineages in the Central Asian gene pool [6–9]. However, in these cases, the high genetic diversity and fine-scale phylogeographic pattern of the mtDNA lineages indicated that the major cline was more likely a result of admixture between already genetically divergent source populations than a reflection of the region’s importance as a “heartland” of diversity for early migrations into Asia, Europe, and India [1]. While a few deeply rooting mitochondrial lineages have been detected in Central Asia [6, 9], the comparatively recent age of these haplogroups relative to lineages found along the southern coast of Asia, combined with an overall absence of basal mtDNA lineages in the region, supports the hypothesis that the original settlement of Central Asia occurred subsequent to and much later than dispersals to Eastern Asia via the southern coastal route [10]. Instead, extensive gene flow resulting from Asian expansions into the west and migrations along the “Silk Road” are believed to have shaped the primary, admixed mtDNA patterns observed, although additional genetic heterogeneity associated with various sociocultural factors has also been detected [6–9, 11–18].
Considering the population dynamics that have shaped the region and the genetic data that reflect this complexity, we have sampled individuals from Uzbekistan in an effort to better understand mtDNA diversity in Uzbekistan, specifically. Uzbekistan is a relatively small country positioned directly in the middle of Central Asia and bordered by Kazakhstan, Turkmenistan, Tajikistan, Afghanistan, and Kyrgyzstan. Prior to the 20th century, Uzbekistan was inhabited by tribes that conquered the region in the 16th century (Library of Congress Federal Research Division Country Profile: Uzbekistan Feb 2007). However, in the late 1800s, Russia’s interest in the commercial potential of current day Uzbekistan led to military conquest of the area and ultimately, industrialization and population growth. In 1924, when the Uzbek Soviet Socialist Republic was established, the population of what is today Uzbekistan was further defined by the boundaries drawn under Soviet rule, which often traversed ethnic and linguistic lines.
Today, despite its relatively small size, Uzbekistan is the most populous of the five former Central Asian Soviet republics, with a population of approximately 27.3 million. The Uzbek population is comprised primarily of ethnic Uzbeks (76%), Russians of European origin (6%), Tajiks (5%), Kazakhs (4%), and Kyrgyz (1%; Library of Congress Federal Research Division Country Profile: Uzbekistan Feb 2007). Along with the historical events that have shaped the genetics of Central Asia as a whole, political changes in and around Uzbekistan specifically, starting with Russian rule in the 19th century and continuing through Uzbekistan’s independence in the early 1990s, have affected the population composition and likely also the genetic composition of this small Central Asian country. Thus, our primary goal is to gain a better understanding of the population genetics of Uzbekistan at the local level, in terms of mtDNA distributions among various subpopulations, both regional subpopulations with Uzbek ancestry and ethnic populations with ancestry in neighboring countries. Since the genetic composition of Uzbekistan likely reflects not only the general heterogeneity of the Central Asian gene pool, but also the signature of much more recent historical events, fine-scale characterization may reveal unusual patterns in the mtDNA population genetics of the country. From the standpoint of forensic mtDNA testing, capturing this information at the finest level should help reveal the magnitude and significance of any inter-population differentiation and provide a better understanding of the degree to which separate forensic reference databases may need to be maintained.
Materials and methods
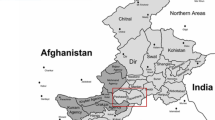
Buccal swabs were collected from 1,575 residents of Uzbekistan with direct ancestry in Afghanistan (n = 98), Kazakhstan (n = 256), Kyrgyzstan (n = 249), Russia (n = 151), Tajikistan (n = 244), Turkmenistan (n = 249), and Uzbekistan (n = 328). The 328 individuals of Uzbek ancestry represent five distinct regions of Uzbekistan: Fergana (53), Karakalpakstan (46), Khorezm (99), Qashkadarya (75), and Tashkent (55). Geographic locations of sample collection are shown in Figs. 1 and 2. The samples were collected specifically for DNA testing purposes and to the best of our knowledge, represent unrelated individuals within at least three generations.
Mitochondrial DNA control region sequencing
For mitochondrial DNA control region typing, samples were extracted and PCR amplified and sequenced as described in [19]. Extracts were amplified for the entire control region, except in rare cases for which three overlapping amplicons were used. For each sample, 16 sequencing primers provided at least double-strand coverage of every base. All electropherograms were analyzed independently by at least three scientists: two at the Armed Forces DNA Identification Laboratory in Rockville, MD and one at the Institute of Legal Medicine, Innsbruck Medical University, Austria. The data generation and analysis approach involved highly redundant data processing and review that was specifically designed to minimize sequence data artifacts and other data analysis and transcription errors [20, 21]. The finalized sequence data for all samples spanned positions 16024-576 with respect to the revised Cambridge Reference Sequence and were aligned to the rCRS according the phylogenetic nomenclature system of [22]. Sequences will be searchable via the EMPOP database under accession numbers EMP00056–EMP00066 and sequence data in electronic form are available from the authors upon request or may be downloaded directly from GenBank (GU069007–GU070581).
Statistical analyses
MtDNA control region haplotypes from the ethnic populations (as they will be referred to from here on) of Uzbekistan (Afghanistan, Kazakhstan, Kyrgyzstan, Russia, Tajikistan, Turkmenistan, Uzbekistan; see Fig. 1) were evaluated with basic measures of haplotypic diversity. The five Uzbek subpopulations from different regions of Uzbekistan (Fergana, Karakalpakstan, Qashqadarya, Tashkent, Khorezm; Fig. 2) were characterized for the same indices, in order to assess the extent of geographic structure among subpopulations with Uzbek ancestry. Arlequin, v. 3.1 [23] was used to generate Ф st values. Haplotype diversity indices and random-match probabilities were calculated by hand (1−sum of squares of haplotype frequencies) × (n/n-1); [24]) and random-match probabilities were generated both empirically, via pairwise comparison of the database and as the sum of squares of the haplotype frequencies [25]. Multi-dimensional scaling plots were performed using the STATISTICA 7 package (StatSoft Inc., Tulsa, OK, USA), with the third dimension included to distinguish populations that were superimposed when the distances were summarized in just two dimensions.
For all comparisons, cytosine insertions at positions 16193, 309, and 573 were ignored. For analyses that addressed only our data from Uzbekistan, entire control region sequences (16024-576) were used. However, in order to evaluate these data in the context of previously published Central Asian mtDNA data, the 1,575 sequences were trimmed to a range of 16024–16383 in order to include HVI sequences reported by Comas et al. [6, 9] and Quintana-Murci et al. [8]. The 309 sequences from these three publications represent seven Central Asian populations from Kazakhstan (CKaz; n = 56), Kyrgyzstan Sary-Tash (CKyrS; n = 46), the Talas valley in Kyrgyzstan, (CKyrT; n = 48), Tajikistan Shugnan (QTaj; n = 44), Turkmenistan (QTur; n = 41), Turkmenistan Kurdish (QTurK; n = 32), and Uzbekistan (QUzb; n = 42). Analyses of molecular variance (AMOVA) were conducted using 1,000 permutation replicates, a transition/transversion ratio of 10, an alpha of 0.3, and the Kimura 2-parameter method for calculating distance [26].
mtDNA haplogroup assignment
The 1,575 entire control region sequences were assigned to mitochondrial DNA haplogroups using Phylotree Build 5 as a reference [27]. In 131 samples, for which the haplogroup affiliation could not be determined based on control region data alone, SNPs selected by Alvarez-Iglesias et al. [28], as well as SNPs at positions 8277, 8392, 9123, 11914, 4833, 8473, 9090, 10397, 12372, and 13563 were assayed to clarify haplogroup membership. In order to visualize the contributions of various mtDNA clusters to the Uzbeki mtDNA gene pool and compare this distribution to previous analyses, haplogroups were clustered according to their geographic origin following Quintana-Murci et al. [8] (west Eurasian: R0, N1, JT, UK, W and X; South Asian: M*, U2a-c, U9, R*, R1-R2, R5-R6, N1d; East Eurasian: M-CDGZ, A, B, F, N9a; African: L).
Results and discussion
Population statistics for regional subpopulations of Uzbekistan with Uzbek ancestry (Karakalpakstan, Khorezm, Qashkadarya, Tashkent, Fergana)
A total of 1,575 individuals from Uzbekistan were sequenced for the entire mtDNA control region (Supplementary Table 1). For those subpopulations with Uzbek ancestry (five regional subpopulations from Fergana, Karakalpakstan, Qashqadarya, Tashkent, and Khorezm represented by 328 samples), genetic diversity parameters are shown in Table 1 and haplogroup compositions are shown in Fig. 3. Among the five regional subpopulations, genetic diversities ranged from 0.987 to 0.998, and mean pairwise differences varied only between 11.9 and 12.9. The two most common haplotypes in the entire data set were shared by four of the five subpopulations and thus, the basic diversity measures were very consistent among the populations. AMOVA results mirrored the summary statistics, indicating that 99.55% of the mtDNA variation is accounted for by differences within the populations, while only 0.45% of the variation is due to differences between them. In addition, all pairwise Ф st values, which varied between −0.003 (Fergana-Karakalpakstan) and 0.0094 (Khorezm-Fergana) were low and insignificant after Bonferroni correction (Supplementary Table 2). Given the genetic homogeneity observed in the mtDNA data from regional subpopulations with Uzbek ancestry, the data were pooled together for subsequent analyses.
Population statistics for the ethnic subpopulations of Uzbekistan (Russia, Kazakhstan, Afghanistan, Tajikistan, Kyrgyzstan, Uzbekistan, Turkmenistan)
Table 2 summarizes a number of relevant parameters for the various populations originating from Afghanistan, Kazakhstan, Kyrgyzstan, Russia, Tajikistan, Turkmenistan, and Uzbekistan (pooled regional populations with Uzbek ancestry). Of the seven population samples, Afghanistan and Kazakhstan appear to be the least and most genetically diverse, respectively. Kazakhstan had a large number of unique haplotypes and thus a low random-match probability (0.13%). Afghanistan, on the other hand, had the fewest total number of haplotypes relative to population size and the highest random-match probability, at 5.5%. mtDNA haplotype diversities for all populations ranged from 0.946 to 0.999.
For all population samples, the average number of pairwise differences varied between 9.3 for the Russian population and 13.0 for the Tajik population. Notably, although the Tajik population exhibited the highest average pairwise difference, other summary statistics for this population suggested relatively low genetic variation. The Tajik pairwise random-match probability and haplotype diversity values were second only to Afghanistan in terms of reduced genetic variability, at 2.3% and 0.9826, respectively. However, the generally ragged (as opposed to smooth and unimodal) shape of the Tajik mismatch distribution (data not shown) suggests that the high mean pairwise difference is primarily a reflection of the presence of highly divergent lineages within the population. The Tajik mtDNA gene pool harbors nearly equal proportions of eastern Eurasian and western Eurasian haplotypes.
The haplogroup compositions of the seven ethnic populations showed distinct patterns (Fig. 4), with an obvious difference being the contributions of eastern Eurasian and western Eurasian mtDNA lineages in any given case. A gradient from west to east is clearly distinguishable. In addition, Ф st values confirmed that significant genetic heterogeneity exists between all of the populations (significant at the 0.05 level, after Bonferroni correction; Supplementary Table 3). Among the various pairwise comparisons, genetic differentiation comprised between 0.6% (Kazakhstan and Kyrgyzstan) and 8.4% (Kyrgyzstan and Russia) of the total genetic variation among the respective population pairs. The large genetic distance estimated for the Russian and Kyrgyz populations can largely be explained by the huge disparity in western Eurasian and eastern Eurasian/South Asian lineages between the two populations (see Fig. 4). Indeed, the differences between most of the populations are due to the dramatic differences in eastern Eurasian and western Eurasian haplogroup composition. As can be seen in the MDS plot (Fig. 5), the first dimension largely reflects the variable contributions of Western Eurasian and Eastern Eurasian mtDNA lineages, with Russia and Kyrgyzstan sharing opposite positions in that spectrum. The first two dimensions place both the Russian and Afghani populations (large western Eurasian composition) apart from the other populations with much greater Eastern Eurasian mtDNA contributions. In fact, the Russian population, with nearly 99% of the samples representing western Eurasian haplogroups, was by far the most genetically distinct population. This was reflected in all of its pairwise population comparisons. Its haplogroup composition and low mean pairwise differences (quite similar to European populations) clearly reflect its demographic history. This particular population from Tashkent descends from individuals who were evacuated from western Russia during the Second World War.
Mitochondrial DNA haplogroup distributions among the ethnic populations sampled in this study. The samples with Uzbek ancestry, summarized in Fig. 3 are combined here as a single Uzbek population. Uzb Uzbekistan, Tur Turkmenistan, Rus Russia, Afg Afghanistan, Taj Tajikistan, Kaz Kazakhstan, Kyr Kyrgyzstan
The genetic features of other ethnic populations likely also reflect their documented demographic histories. For instance, the small mtDNA distance between the Tajik and Uzbek populations suggests a recent shared history. Tajiks and Uzbeks were only formally differentiated in 1929 when the Tajik Soviet Socialist Republic was established, and up to 40% of the current Uzbek population is of Tajik ancestry (Library of Congress Federal Research Division Country Profile: Uzbekistan Feb 2007). Although the Ф st value for these two populations suggested significant differentiation (at the 0.05 level after Bonferroni correction), it was among the smallest of the 21 pairwise distances calculated.
An extremely common haplotype was found in the Afghani subpopulation that has never been previously observed in other published mtDNA data. The frequency of this otherwise rare sequence raised questions regarding the randomness of these samples. The specific K1a haplotype was shared among nearly 20% of that population. Furthermore, over a quarter of the K1a individuals share a point heteroplasmy at position 16093. However, careful evaluation of the sample collection records confirmed that these individuals are unrelated within at least three generations. Thus, the more likely explanation is that this population is exhibiting the genetic hallmarks of a recent population bottleneck resulting from founder events and/or endogamy, which is consistent with its documented demographic history. This population was established in Uzbekistan within the last century and was subsequently isolated from the Afghani source populations. Thus, founder effect likely accounts for not only the high frequency of the heteroplasmic K1a haplotype, but also the low haplotype diversity and high random-match probability values observed for this population.
Comparisons to other published data
Given the founder effect suspected in the Afghan population of Uzbekistan, we were interested in determining whether or not other ethnic subpopulations from Uzbekistan exhibited significant differences in mtDNA composition from their source populations. We suspected that founder effect and/or drift may have affected the other sampled ethnic groups as well, since borders established after the Russian revolution likely limited migration and genetic exchange with their source populations. Unfortunately, due to a lack of available mtDNA data from Afghanistan, we were not able to directly assess the degree to which our sample differed from its parent populations. For the other ethnic populations, however, the results were contrary to our expectations. Pairwise Ф st values between the ethnic populations in Uzbekistan and available data from their source populations indicated minimal and statistically insignificant differences in their mtDNA distributions (a Ф st value of 0.014 being the greatest difference between any two populations). The MDS plot of the Ф st values (Fig. 6) positions the population samples from the current study in close proximity to previously reported data from their source populations, particularly across the first dimension. The first dimension largely describes the variable contributions of eastern and western Eurasian haplogroups among the populations, with the Russian and Afghan populations from this study positioned together with a Kurdish population from Turkmenistan in the west Eurasian extreme of this spectrum. Generally speaking, the ethnic populations from the current study are only distinguishable from their source populations in the second dimension. The obvious similarities in mtDNA haplogroup distributions, as well as the non-significant genetic distances, between the ethnic populations from the current study and data from their source populations, suggest that the Afghan population sample from Uzbekistan is the only ethnic population showing evidence of a population bottleneck and founder effect.
MDS plot of Uzbek populations reported here and previously published Central Asian data. Distances were calculated in Arlequin v. 3.1, using the Kimura 2-parameter model. The values are based on HVI data spanning positions 16024–16383, and thus the positioning of populations also represented in Fig. 5 may be slightly different. Stress = 0.08
Overall, despite the close geographic proximity of the sampling areas for the ethnic populations of Uzbekistan, their genetic distances and haplogroup distributions were clear reflections of the broad east–west clines that characterize their source populations. Although the various regional subpopulations with Uzbek ancestry spanned twice the geographic distance of the ethnic populations sampled, the Uzbek subpopulations exhibited genetic distances an order of magnitude smaller than those observed among the ethnic populations.
Implications for forensic mtDNA testing
Population databases are used to indicate the relative rarity of mtDNA sequences encountered in forensic casework, and in most cases, the evidentiary significance of mtDNA matching improves as database size increases. As a result, large mtDNA databases are generally desirable, and pooling of genetically similar databases can be done in order to establish larger databases—if this is warranted by an absence of population differentiation.
In order to assess the practical implications of the mtDNA heterogeneity found among the ethnic populations in Uzbekistan from the standpoint of forensic mtDNA testing, we looked for each population’s most common mtDNA haplotype in each of the other populations, as well as a pooled “Uzbekistan” database that included all of the samples. Generally, the most common mitochondrial haplotype in each population was rarely seen in other populations (Table 3). For instance, the most common type in the Turkmen population (11 individuals) was only seen in one other individual—someone from the Tajik population. In addition, the most common haplotype in any given population was, more often than not, completely absent in the other populations. Furthermore, when a pooled “Uzbekistan” database comprising data from all seven populations was used to assess mtDNA haplotype frequencies, the frequency of each population’s most common haplotype was significantly underestimated, suggesting that frequency estimates would be most conservative if populations were considered separately.
Conclusions
In order to better understand the fine-scale mtDNA population genetics of Uzbekistan, we examined the degree of mtDNA differentiation between geographically sampled autochthonous Uzbek subpopulations, as well as endogamous ethnic populations with ancestry from neighboring countries. The geographically sampled subpopulations with Uzbek ancestry did not exhibit a significant degree of population substructure with mtDNA control region data. It is therefore appropriate that the subpopulations of Uzbekistan be pooled together and treated as a single database for forensic mtDNA purposes. However, seven ethnic populations of Uzbekistan, representing individuals with direct ancestry in Kazakhstan, Turkmenistan, Tajikistan, Afghanistan, Russia, Uzbekistan, and Kyrgyzstan exhibit clear differences in their mitochondrial DNA compositions. In some cases, the genetic distances detected were relatively small. Yet, in all cases, the differences between populations were statistically significant. Given these results, as well as our observations regarding the relative rarity of each ethnic population’s most common haplotype in each of the remaining populations, mtDNA frequency estimates would likely be most conservative if the ethnic populations were considered separately, instead of pooled together and treated as a single “Uzbekistan” database. Indeed, the ethnic populations of Uzbekistan largely reflected the mtDNA compositions of their source populations as opposed to the mtDNA gene pool of autochthonous Uzbeks.
The data here provide a basic structure for the interpretation of forensic mtDNA testing in Central Asia. A number of the interesting features of these data call for additional study, including newly observed mtDNA haplotypes, significant inter-population heterogeneity, and likely founder effects. Our results highlight significant departures from genetic uniformity over small geographic distances—the result of recent historical events. These data emphasize the need for heightened caution in the forensic interpretation of mtDNA data in regions as historically rich and genetically diverse as Central Asia and highlight the complexities of contemporary populations from the standpoint of forensic DNA testing. Such mosaic populations of recently introduced yet separate groups, each with varying degrees of admixture, are common and widespread in today’s world. They represent a serious general challenge to both the establishment of forensic mtDNA reference databases and the use of those data for interpretation.
References
Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, Su B, Pitchappan R, Shanmugalakshmi S, Balakrishnan K, Read M, Pearson NM, Zerjal T, Webster MT, Zholoshvili I, Jamarjashvili E, Gambarov S, Nikbin B, Dostiev A, Aknazarov O, Zalloua P, Tsoy I, Kitaev M, Mirrakhimov M, Chariew A, Bodmer WF (2001) The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci 98:10244–49
Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, New Jersey
Lahr MM, Foley R (1994) Multiple dispersals and modern human origins. Evol Anthropol 3:48–60
Maca-Meyer N, Gonzalez AM, Larruga J, Flores C, Cabrer V (2001) Major genomic mitochondrial lineages delineate early human expansions. BMC Genetics 2:13
Tanaka M, Cabrera VM, González AM, Larruga JM, Takeyasu T, Fuku N, Guo LJ, Hirose R, Fujita Y, Kurata M, Shinoda K, Umetsu K, Yamada Y, Oshida Y, Sato Y, Hattori N, Mizuno Y, Arai Y, Hirose N, Ohta S, Ogawa O, Tanaka Y, Kawamori R, Shamoto-Nagai M, Maruyama W, Shimokata H, Suzuki R, Shimodaira H (2004) Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res 14:1832–1850
Comas D, Calafell F, Mateu E, Perez-Lezaun A, Bosch E, Martinez-Arias R, Clarimon J, Facchini F, Giovanni F, Luiselli D, Pettener D, Pertranpetit B (1998) Trading genes along the Silk Road: mtDNA sequences and the origin of Central Asian populations. Am J Hum Genet 63:1824–1838
Calafell F, Comas D, Perez-Lezaun A, Bertranpetit J (2000) Genetics and the population history of Central Asia. In: Renfrew C, Boyle K (eds) Archaeogenetics: DNA and the population history of Europe. McDonald Institute for Archaeological Research, Cambridge
Quintana-Murci L, Chaix R, Wells S, Behar D, Sayar H, Scozzari R, Rengo C, Al-Zahery N, Semino O, Santachiara-Benerecetti S, Coppa A, Ayub Q, Mohyuddin A, Tyler-Smith C, Mehdi Q, Torroni A, McElreavey K (2004) Where west meets east: the complex mtDNA landscape of the Southwest and Central Asian corridor. Am J Hum Genet 74:827–45
Comas D, Plaza S, Wells RS, Yuldaseva N, Lao O, Calafell F, Bertranpetit J (2004) Admixture, migrations, and dispersals in Central Asia: evidence from maternal DNA lineages. Eur J Hum Genet 12:495–504
Macaulay V, Hill C, Achilli A, Rengo C, Clarke D, Meehan W, Blackburn J, Semino O, Scozzari R, Cruciani F, Taha A, Shaari NK, Raja JM, Ismail P, Zainuddin Z, Goodwin W, Bulbeck D, Bandelt HJ, Oppenheimer S, Torroni A, Richards M (2005) Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science 308:1034–36
Laluela-Fox C, Sampietro ML, Gilbert MTP, Castri L, Facchini F, Pettener D, Bertranpetit J (2004) Unravelling migrations in the steppe: mitochondrial DNA sequences from ancient Central Asians. Proc Biol Sci 271:941–7
Chaix R, Quintana-Murci L, Hegay T, Hammer MF, Mobasher Z, Austerlitz F, Heyer E (2007) From social to genetic structures in Central Asia. Curr Biol 17:43–8
Pérez-Lezaun A, Calafell F, Comas D, Mateu E, Bosch E, Martínez-Arias R, Clarimón J, Fiori G, Luiselli D, Facchini F, Pettener D, Bertranpetit J (1999) Sex-specific migration patterns in Central Asian populations, revealed by analysis of Y-chromosome short tandem repeats and mtDNA. Am J Hum Genet 65:208–19
Yao YG, Lu XM, Luo HR, Li WS, Zhang YP (2000) Gene admixture in the Silk Road region of China: evidence from mtDNA and melanocortin 1 receptor polymorphism. Genes Genet Syst 75:173–178
Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C (2002) A genetic landscape reshaped by recent events: Y-chromosomal insights into Central Asia. Am J Hum Genet 71:466–482
Chaix R, Austerlitz F, Hegay T, Quintana-Murci L, Heyer E (2008) Genetic traces of east-to-west human expansion waves in Eurasia. Am J Phys Anthropol 136:309–317
Segurel L, Martinez-Cruz B, Quintana-Murci L, Balaresque P, Georges M, Hegay T, Aldashev A, Nasyrova F, Jobling MA, Heyer E, Vitalis R (2008) PLoS Genet 4:e1000200
Heyer E, Balaresque P, Jobling MA, Quintana-Murci L, Chaix R, Segurel L, Aldashev A, Hegay T (2009) BMC Genetics 10:49. doi:10.1186/1471-2156-10-49
Irwin J, Saunier J, Strouss K, Sturk K, Diegoli T, Just R, Coble M, Parson W, Parsons T (2007) Development and expansion of high quality control region databases to improve forensic mtDNA evidence interpretation. Forensic Sci Int: Genetics 1:154–157
Bandelt HJ, Lahermo P, Richards M, Macaulay V (2001) Detecting errors in mtDNA data by phylogenetic analysis. Int J Legal Med 115:64–69
Bandelt HJ, Quintana-Murci L, Salas A, Macaulay V (2002) The fingerprint of phantom mutations in mitochondrial DNA data. Am J Hum Genet 71:1150–1160
Bandelt HJ, Parson W (2008) Consistent treatment of length variants in the human mtDNA control region: a reappraisal. Int J Legal Med 122:11–21
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Stoneking M, Hedgecock D, Higuchi RG, Vigilant L, Erlich HA (1991) Population variation of human mtDNA control region sequences detected by enzymatic amplification and sequence-specific oligonucleotide probes. Am J Hum Genet 48:370–382
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.1: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
van Oven M, Kayser M (2009) Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 30(2):E386–E394 http://www.phylotree.org
Alvarez-Iglesias V, Jaime JC, Carracedo A, Salas A (2007) Coding region mitochondrial DNA SNPs: targeting East Asian and Native American haplogroups. Forensic Sci Int: Genetics 1:44–55
Acknowledgements
The authors would like to thank Kimberly Sturk, Toni Diegoli, Katharine Strouss, Carla Paintner, Daniela Niederwieser, Bettina Zimmermann, and Gabriela Huber for excellent technical assistance; collaborators Dilobar Akhmedova, and Nizom Rakhmatullaev; Nina Duftner for data analysis; Rebecca Just, Michael Coble, and Ibrokhim Abdurakhmanov for helpful discussion; and the Armed Forces Institute of Pathology, the American Registry of Pathology, James Canik, Brion Smith, and Louis Finelli for logistical and administrative support. Part of this research received support from the FWF Austrian Science Fund (L397). The opinions and assertions contained herein are solely those of the authors and are not to be construed as official or as views of the U.S. Department of Defense, the U.S. Department of the Army, or the U.S. Department of Justice.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
MtDNA haplotypes from 1,575 individuals residing in Uzbekistan, listed as differences from the rCRS. Haplotypes are preceded by a header line (denoted with a #! symbol) which specifies the sites assayed for a given set of samples. In all cases, the entire control region was sequenced. Samples are labeled according to: AFG (Afghanistan), KYR (Kyrgyzstan), KAZ (Kazakhstan), RUS (Russia), TAJ (Tajikistan), TUR (Turkmenistan), FER (Uzbekistan–Fergana), KAR (Uzbekistan–Karakalpakstan), QAS (Uzbekistan–Qashkadarya), TAS (Uzbekitan–Tashkent), XOR (Uzbekistan–Khorezm) (PDF 339 kb)
Supplementary Table 2
Фst values for regional subpopulations of Uzbekistan with Uzbek ancestry. Values are not significant at the 0.05 level after Bonferroni correction. (DOC 148 kb)
Supplementary Table 3
Фst values for ethnic populations of Uzbekistan. All values are statistically significant at the 0.05 level after Bonferroni correction (DOC 12.0 kb)
Rights and permissions
About this article
Cite this article
Irwin, J.A., Ikramov, A., Saunier, J. et al. The mtDNA composition of Uzbekistan: a microcosm of Central Asian patterns. Int J Legal Med 124, 195–204 (2010). https://doi.org/10.1007/s00414-009-0406-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-009-0406-z