Abstract
Endogenous chromosomes contain centromeres to direct equal chromosomal segregation in mitosis and meiosis. The location and function of existing centromeres is usually maintained through cell cycles and generations. Recent studies have investigated how the centromere-specific histone H3 variant CENP-A is assembled and replenished after DNA replication to epigenetically propagate the centromere identity. However, existing centromeres occasionally become inactivated, with or without change in underlying DNA sequences, or lost after chromosomal rearrangements, resulting in acentric chromosomes. New centromeres, known as neocentromeres, may form on ectopic, non-centromeric chromosomal regions to rescue acentric chromosomes from being lost, or form dicentric chromosomes if the original centromere is still active. In addition, de novo centromeres can form after chromatinization of purified DNA that is exogenously introduced into cells. Here, we review the phenomena of naturally occurring and experimentally induced new centromeres and summarize the genetic (DNA sequence) and epigenetic features of these new centromeres. We compare the characteristics of new and native centromeres to understand whether there are different requirements for centromere establishment and propagation. Based on our understanding of the mechanisms of new centromere formation, we discuss the perspectives of developing more stably segregating human artificial chromosomes to facilitate gene delivery in therapeutics and research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During mitosis and meiosis, proper kinetochore assembly on the centromere platform is a critical step to attach chromosomes to microtubules and ensure equal separation of sister chromatids and homologous chromosomes, respectively. Failure to do so will cause aneuploidy and even cell lethality. Recently, our understanding on the molecular mechanism of centromere propagation in different model organisms has advanced. The centromeric DNA sequences are not conserved among eukaryotes. Yet, in most eukaryotes, with a few exceptions (Drinnenberg et al. 2014), centromere identity is epigenetically maintained across the cell cycles by inheriting a centromere-specific histone H3 variant, CENP-A, in the centromeric nucleosomes. Thus, CENP-A is present in most functional centromeres. As DNA replicates in the cell, only half of the CENP-A nucleosomes are segregated to each of the replicated DNA (Jansen et al. 2007; Mellone et al. 2011; Shelby et al. 2000). The replenishment of additional CENP-A nucleosomes and the assembly of kinetochore components occur in a cell cycle–dependent manner. Each step of centromere propagation, including centromere licensing, CENP-A deposition, stabilization, and maintenance of CENP-A, is tightly linked with each other and coupled to the cell cycle. Recently, many excellent reviews have discussed the epigenetic regulation on centromere propagation (Black and Cleveland 2011; Black et al. 2010; Gambogi and Black 2019; McKinley and Cheeseman 2016).
However, how the centromere is first established during evolution, after chromosomal rearrangements, after inactivation of original centromeres, or on newly introduced DNA remains less well understood, partly because centromere establishment does not occur as frequently as centromere propagation, and thus is harder to observe and study. There are two types of new centromere that can be established: neocentromere and de novo centromere. Neocentromeres are new centromeres form on ectopic, non-centromeric DNA sequences on endogenous chromosomes or chromosomal fragments (Amor and Choo 2002; Kalitsis and Choo 2012). The occurrence of neocentromeres indicates that CENP-A can be deposited ectopically into non-centromeric chromatin, indicating that the DNA sequence itself is not an absolute prerequisite for centromere formation. On the other hand, de novo centromeres often refer to new centromeres that are formed on newly introduced, originally naked DNA, after it has become chromatinized in the cells (Harrington et al. 1997). Recently, an outstanding review by Barrey and Heun (2017) has discussed the requirement of de novo centromere formation on naked centromeric DNA and conditions that could artificially induce neocentromere formation on endogenous chromosome. We will continue the discussion by describing the naturally occurring neocentromeres, comparing the de novo centromeres and neocentromeres, and discussing the implications and importance of centromere establishment.
In this review, we will first discuss (1) when and where new centromeres establish and (2) the importance of establishing new centromeres in evolution and diseases. We will further introduce (3) the genetic and (4) epigenetic factors identified at new centromeres in normal or pathological settings, and in experimental conditions, and then discuss how they contribute to the formation of new functional centromeres. Lastly, we will speculate (5) the future directions and potential applications of the research on centromere establishment.
When and where does centromere establishment occur?
A comparison between the findings of neocentromeres and de novo centromeres and their implications are summarized in Table 1.
Neocentromere formation accompanied by deletion or inactivation of the original centromere
Centromere repositioning in natural speciation
Neocentromere formation can be spontaneous, and most of the time, the outcome is deleterious. Dicentric chromosome can undergo chromosomal breakage or rearrangement (Fig. 1a) (McClintock 1939, 1941). If there is a shift of centromere position (simultaneous neocentromere formation on ectopic location and inactivation of the original centromere), the chromosome will be stable. This event, called centromere repositioning (Montefalcone et al. 1999; Schubert 2018), is proposed to be an important mechanism for promoting karyotype evolution and speciation (Henikoff et al. 2001).
When centromere establishment occurs? Centromere establishment can occur either naturally (a–e) or experimentally (f–j). In the natural situation, centromere can be established spontaneously (for unknown reason, a–c) or under pathologically conditions (d, e). a When a neocentromere forms spontaneously without the inactivation of the original centromere, a dicentric chromosome will be formed which will cause chromosomal breakage and rearrangement (McClintock 1941; McClintock 1939). b On the other hand, if the original endogenous centromere is inactivated, the centromere is said to be repositioned, which is one of the reasons for karyotype evolution and speciation (Amor et al. 2004; Baldini et al. 1993; Carbone et al. 2006; Chmatal et al. 2014; Ferreri et al. 2005; Han et al. 2009; Kobayashi et al. 2008; Montefalcone et al. 1999; Nergadze et al. 2018; Piras et al. 2009; Rocchi and Archidiacono 2006; Rocchi et al. 2012; Ventura et al. 2007; Villasante et al. 2007; Wade et al. 2009; Wong and Choo 2001; Yang et al. 2014). c In maize, a special type of neocentromeres (knobs) can be formed at the terminal of the chromosomes (Kattermann 1939). Knobs lack conserved centromeric proteins CENP-A and CENP-C (Dawe et al. 1999; Dawe and Hiatt 2004) and are structurally different from the classical neocentromeres. In a Rhoades model of meiotic drive, a crossover event results in homologous chromosomes, which are heterozygous at the knob locus. In meiosis I, chromosomes with a knob has an advantage to move more rapidly to the pole. Such orientation is maintained in meiosis II, resulting in chromosomes containing the knob often segregating to the outermost megaspores. The basal outermost megaspore always becomes the functional megaspore (egg). Thus, the chromosome with the knob will be transmitted to the progeny at a higher frequency than the expected Mendelian ratio, which is the underlying mechanism for meiotic drive (Manzanero and Puertas 2003; Rhoades 1942; Sandler and Novitski 1957; Yu et al. 1997) (The schematic is adapted from Kanizay et al. (2013)). d Misregulation and overexpression of CENP-A or other kinetochore proteins is observed in cancer cells, leading to ectopic accumulation of CENP-A (Athwal et al. 2015; Tomonaga et al. 2003). If there is a simultaneous ectopic deposition of CENP-C or CENP-T to the mistargeted CENP-A sites, ectopic kinetochore would be formed (Gascoigne et al. 2011), resulting in a dicentric chromosome. On the other hand, mistargeted CENP-A could also weaken the original centromere by recruiting a subset of centromeric and kinetochore proteins, causing chromosomal instability (Shrestha et al. 2017; Van Hooser et al. 2001). e Chromatid breaks and chromosomal rearrangement in ALP-WDLPS tumor cells could force a neocentromere to form on chromatid fragments lacking endogenous centromeric DNA, i.e., rings and giant rod chromosome (Macchia et al. 2015; Sirvent et al. 2000). f In Drosophila, γ-irradiation generates acentric chromosomal fragments and neocentromeres are established on acentric fragments only if the acentric chromosomal fragments were formerly juxtaposed to the native centromeric DNA (Maggert and Karpen 2001; Williams et al. 1998). g Deletion of the original centromere by homologous recombination results in neocentromere formation at different genomic loci in different models; fission yeast: subtelomeric regions (Ishii et al. 2008); C. albicans: (i) large intergenic regions and repeated DNA at the centromere-distal sites; (ii) centromeric-proximal regions with an absence of specific sequence motifs or repeats; (iii) adjacent to telomeres (Ketel et al. 2009; Thakur and Sanyal 2013); chicken DT40 cells: adjacent to the deleted centromere region (Shang et al. 2013) (see Fig. 2). h Introducing exogenous naked DNA (usually including centromeric DNA) results in the formation of artificial chromosomes with de novo centromere formation on naked DNA, i.e., ScYAC in budding yeast (Murray and Szostak 1983), SpYAC in fission yeast (Hahnenberger et al. 1989), HAC in human cells (Harrington et al. 1997; Ohzeki et al. 2012), and WAC in C. elegans (Mello et al. 1991; Stinchcomb et al. 1985). i Experimental overexpression of CENP-A or CENP-A chaperone induces the ectopic loading CENP-A on chromosome arm in HeLa cells (Lacoste et al. 2014; Yu et al. 2015) and near heterochromatin-euchromatin boundaries in fission yeast (Gonzalez et al. 2014) and in Drosophila S2 cells (Olszak et al. 2011). In fission yeast and Drosophila, accumulation of ectopic CENP-A promotes the development of ectopic kinetochores, resulting in chromosomal breakage and missegregation. j CENP-A is enriched in the DNA damage sites when a DNA double-stranded break is induced on the chromatin by laser or I-SceI cleavage in HeLa cells (Zeitlin et al. 2009). Irradiation-induced DNA damage in human pluripotent stem cells also causes uncoupling and relocalization of CENP-A and CENP-C to separated small foci in the nucleus (Ambartsumyan et al. 2010). It is not known if the ectopic CENP-A can seed the formation of neocentromere. We postulated that if an acentric chromatin fragment is generated by a DNA double-stranded break, the formation of a neocentromere may help to rescue the fragment by restoring the centromeric function. Whether CENP-A has a role in DNA damage repair is still in debate, as DNA breaks induced by multiphoton laser micro-irradiation in human osteosarcoma U2OS cells did not result in CENP-A accumulation on the DNA damage sites (Helfricht et al. 2013)
During primate evolution, centromere repositioning can occur in the absence of chromosomal rearrangement and with synteny maintained along the chromosome (Fig. 1b). During centromere repositioning in the primate evolution, it is hypothesized that the ectopic neocentromeric locus rapidly re-acquires repetitive DNA sequences and maintains a large and complex organization, similar to that at the original centromere within a short evolutionary time (Montefalcone et al. 1999; Rocchi et al. 2012; Ventura et al. 2007), whereas the original centromere is inactivated and partially deleted, with remnants of ancestral centromeric DNA (Baldini et al. 1993; Rocchi and Archidiacono 2006; Villasante et al. 2007; Wong and Choo 2001). It is still not known whether centromere repositioning in primates is first initiated by a partial deletion of the original centromeric DNA, rendering the original centromere inactive, or induced merely by epigenetics (Amor et al. 2004). Phylogenetic, karyotypic, and fluorescence in situ hybridization (FISH) analysis also uncovered centromere repositioning during the evolution of many different mammalian species, such as Macropus (Ferreri et al. 2005), Tokudaia (Kobayashi et al. 2008), Equus (Carbone et al. 2006; Nergadze et al. 2018; Piras et al. 2009; Wade et al. 2009), and Bos (De Lorenzi et al. 2017).
Besides animals, centromere reposition is also observed during the evolution of Cucumis. Cucumber (Cucumis sativus L.) and melon (Cucumis melo L.) are diverged from the same ancestor (Schaefer et al. 2009). By comparing their chromosomes with FISH mapping and next-generation sequencing, a centromere repositioning event was identified in the chromosome C7 of cucumber (Han et al. 2009; Yang et al. 2014). Interestingly, the inactivation of the original centromere was associated with a loss of pericentromeric heterochromatin near the original centromeric DNA (Han et al. 2009).
In disease conditions
Marshall et al. (2008) have summarized more than 90 cases of neocentromere reported in human patients with congenital abnormalities or developmental disorders. In some cases, inactivation of the original centromere occurs by unknown reason without any change of centromere sequences. These functionally “acentric” chromosomes would have been lost, unless neocentromere formation rescues them (Amor et al. 2004; Capozzi et al. 2008). In atypical lipoma and well-differentiated liposarcoma (ALP-WDLPS) cells, chromosomal breakages and rearrangements result in the formation of acentric chromosomal fragments that lack the endogenous centromere, known as supernumerary ring (circular) or giant rod-shaped marker (RGM) chromosomes, which are patchworks from 2 to 6 different chromosomes. A neocentromere can form on a non-centromeric region lacking alpha-satellite DNA in these supernumerary ring or RGM chromosomes, enabling such chromosomes to propagate in cancer cells to potentially confer some selective advantage (Fig. 1e) (Sirvent et al. 2000). The neocentromere may be composed of multiple breakpoints from different chromosomes (Macchia et al. 2015). However, as the neocentromeres are often detected long after they are formed, it is not straightforward to conclude whether the formation of the neocentromere is a cause or a result of chromosomal rearrangements. It is important to clarify the causal relationship between chromosomal rearrangement and neocentromere formation and elucidate how they contribute to tumorigenesis. On the other hand, neocentromere formation without original centromere inactivation will result in dicentric chromosomes, causing chromosomal instability, which are commonly found in cancer cells (Fig. 1d) (Beh et al. 2016; Blom et al. 2010).
Chromosomal rearrangement or experimental deletion of the original centromere
Scientists have induced chromosomal rearrangements or manually deleted the original centromere to mimic the loss of the original centromere and investigated the location preference of neocentromere formation on acentric chromosomes. As demonstrated in Drosophila melanogaster, γ-irradiation induced chromosomal breakage and generated acentric chromosomal fragments, in which some may acquire centromere activity (Williams et al. 1998). Intriguingly, neocentromeres are established on DNA fragments which were formerly juxtaposed to the native centromeric DNA, but not on fragments that were far from the centromere.
Maggert and Karpen (2001) suggested that these acentric fragments may acquire new centromeric activity due to the spreading of centromeric epigenetic marks from the neighboring native centromere (Fig. 1f). Similarly, in a human fibroblast cell line, the pericentromere was removed from chromosome 17 by chromosomal rearrangement, resulting in neocentromere formation with spreading of CENP-A from α-satellites into the chromosome arm (Sullivan et al. 2016).
Besides γ-irradiation–induced chromosomal breakage, neocentromeres can also be induced by deletion of the original centromere through genetic manipulation (Fig. 1g). In a number of cell models, such as Schizosaccharomyces pombe, Candida albicans, and chicken DT40 cells, neocentromeres can be established on non-centromeric regions by deleting the original centromeric sequence through homologous recombination and selecting for cells that retain that specific chromosome using a marker gene. The neocentromere rescues the loss of the chromosome and the lethality of cells. Interestingly, these neocentromeres are preferred to establish on certain chromatin regions, indicating the presence of neocentromere hotspots (Fig. 2). In Schizosaccharomyces pombe, centromeric deletion resulted in neocentromere formation at subtelomeric regions (Ishii et al. 2008). Unlike in fission yeast, Candida albicans lacks precentromeric heterochromatin and has smaller centromeres that occupy about 3–4 kb of DNA sequence. In C. albicans, deletion of the native centromere resulted in the formation of neocentromere at intergenic, non-transcribed regions with flanking repeated DNA either at centromere-distal sites (200–450 kb from the original centromere), including telomeres, or in centromeric-proximal regions with an absence of specific sequence motifs or repeats (Ketel et al. 2009; Thakur and Sanyal 2013). Yet, neocentromere formation in centromeric-proximal regions suppresses expression of the newly integrated maker gene, which replaces the original centromere (Thakur and Sanyal 2013). Similarly, in chicken DT40 cells, 76% of neocentromeres form adjacent to the deleted centromere region and suppress the expression of the genes within the neocentromere region (Shang et al. 2013).
Neocentromeres in plants induce multicentromeres in meiosis
A special type of neocentromere, namely terminal neocentromere, or “knob,” is observed in plant species such as rye and maize at the terminal region of all of the chromosomes (Kattermann 1939), which mainly contains heterochromatin formed on tandem repeated sequence with some homology with the centromeric repeats (Fig. 1c) (Hiatt et al. 2002; Peacock et al. 1981). Knobs are structurally different from the classical neocentromere, as they lack conserved centromeric component CENP-A and inner kinetochore protein CENP-C (Dawe et al. 1999; Dawe and Hiatt 2004), and they interact with spindle microtubules in a lateral way, but not the canonical end-on fashion (Yu et al. 1997). These terminal neocentromeres form in strains carrying the maize abnormal chromosome 10 (Ab10). Ab10 contains an extremely large knob and two neocentromere-activating cassettes, controlling the activities of the terminal neocentromeres. How the cassettes evolved on the Ab10 is still unknown (Hiatt et al. 2002). Unlike the neocentromeres of humans and Drosophila, plant terminal neocentromeres is active only in meiosis, together with the native centromere (Manzanero and Puertas 2003). Yet, these dicentric chromosomes do not cause chromosomal breakage in meiosis. Instead, the microtubules attach laterally to the neocentromeres and attach end-on to the native kinetochores. Together, they move the chromosomes to the opposite poles, and thus these chromosomes with “knobs” are preferentially segregate to gametes (see the section “Meiotic drive and speciation” for details), resulting in a meiotic drive that is not fixed yet, potentially because they may also display deleterious consequences (Manzanero and Puertas 2003; Yu et al. 1997).
Neocentromere formation caused by misregulation of centromeric proteins
In pathological conditions or cancer cells
The precise timing of CENP-A assembly on centromere is regulated through phosphorylation by cell cycle–dependent kinase (CDK) and Polo-like kinase (PLK) and dephosphorylation by protein phosphatase 1 alpha (PP1a) (Yu et al. 2015). HJURP/Smc3 (Camahort et al. 2007; Dunleavy et al. 2009; Foltz et al. 2009), CAL1 (Chen et al. 2014), or LIN-53 (Lee et al. 2016) is the CENP-A chaperone for centromere propagation, which directly binds to CENP-A in humans, yeast, flies, or worms, respectively, at specific cell cycle stages. Under the normal condition, the native centromere in human cells maintains existing centromeric proteins across cell cycles to form a functional kinetochore. CENP-A’s protein level is regulated by E3 ubiquitin and SUMO ligases, which can promote the degradation of ectopically incorporated CENP-A (Canzonetta et al. 2015; Collins et al. 2004; Deyter and Biggins 2014; Gonzalez et al. 2014; Hewawasam et al. 2010; Moreno-Moreno et al. 2006; Ohkuni et al. 2016; Ranjitkar et al. 2010). Misregulation of CENP-A level and/or the degradation system in pathological conditions could cause neocentromere formation. For instance, in colorectal cancer cells lines, CENP-A and other kinetochore proteins are overexpressed and deposited on ectopic regions (Athwal et al. 2015; Tomonaga et al. 2003), potentially promoting neocentromere formation and ectopic kinetochores and driving aneuploidy (Fig. 1d). Yet, ectopic CENP-A does not always induce the formation of kinetochore. In another study, when CENP-A is mistargeted to non-centromeric chromatin by overexpression artificially, the mistargeted CENP-A is not sufficient to induce the formation of a complete kinetochore complex in human cells (Van Hooser et al. 2001). It is possible that ectopic CENP-A needs to reach a high threshold level to drive the neocentromere formation, possibly in conditions in which the CENP-A degradation system is disrupted. In addition, a simultaneous ectopic accumulation of CENP-T or CENP-C to the CENP-A–mistargeted sites may also require for ectopic kinetochore formation (Gascoigne et al. 2011). Besides forming ectopic kinetochore, ectopic CENP-A could sequester away a subset of centromeric and kinetochore proteins from the native centromere and weaken the native kinetochore, leading to chromosomal instability (Shrestha et al. 2017; Van Hooser et al. 2001). Nonetheless, the expression level of CENP-A and other kinetochore proteins has been proposed to be used in prognosis to predict the chromosomal instability (CIN) level of cancers and the response to treatments (Zhang et al. 2016). Intriguingly, overexpression of CENP-A can sometimes be beneficial. When challenged with the DNA-damaging agent camptothecin and ionizing radiation, the survival rate of HeLa cells increased when CENP-A is artificially overexpressed (Lacoste et al. 2014). This may be due to a potential role of CENP-A in DNA damage repair, which will be discussed in the section ““Ectopic” CENP-A in response to DNA damage.”
In cancer cells with CENP-A overexpression, the proportion of CENP-A/H4 to H3/H4 dimers/tetramers increased. Other more promiscuous histone chaperones, like DAXX, could bind to heterotetrameric H3.3-CENP-A-H4 and deposit this complex into ectopic chromatin regions (Lacoste et al. 2014). Interestingly, a study in fission yeast showed that chromosomal segregation defect caused by induced CENP-A overexpression can be suppressed by overexpressing histone H3 or histone H4, as this may restore the stoichiometry of CENP-A/H4 to H3/H4 (Gonzalez et al. 2014). In addition, when CENP-A chaperone HJURP is overexpressed in CENP-A-overexpressed cancer cells, ectopic CENP-A localization is reduced, indicating that a balance level between CENP-A and CENP-A chaperone HJURP could help to minimize CENP-A mislocalization (Nye et al. 2018).
Experimentally
The formation of neocentromere can also be induced experimentally by overexpressing CENP-A (as mentioned in some studies in the section “In pathological conditions or cancer cells”) or CENP-A chaperone in many organisms (Fig. 1i). Studies in HeLa cells found that ectopic CENP-A localization on chromosome arms can be stimulated by CENP-A overexpression (Lacoste et al. 2014; Van Hooser et al. 2001; Yu et al. 2015). Similar to human cells, overexpression of CENP-ACnp1 in fission yeast and CENP-ACID in Drosophila S2 cells also result in ectopic CENP-A localization (Gonzalez et al. 2014; Heun et al. 2006). In fission yeast, overexpressed CENP-A is deposited at non-centromeric regions, preferentially at/near heterochromatin regions localized at nuclear periphery (Fig. 2) (Gonzalez et al. 2014). Overexpression of CENP-A in Drosophila S2 cells leads to ectopic CENP-A deposition with neocentromere formation at heterochromatin-euchromatin boundaries near telomeres and pericentromeric heterochromatin (Fig. 2, Table 4) (Olszak et al. 2011). In budding yeast, when CENP-ACse4 is overexpressed, CENP-ACse4 nucleosomes are found in ectopic sequences known as centromere-like regions (CLRs) (Lefrancois et al. 2013).
Neocentromeres can also be formed by artificial tethering of a kinetochore protein. Tethering LacI-CENP-A, CENP-A chaperone, or kinetochore protein to euchromatin-located LacO arrays (or tethering tetR in a TetO system) is commonly used in many species, e.g., human cells (Logsdon et al. 2015), Drosophila (Mendiburo et al. 2011; Roure et al. 2019), chicken DT40 cells (Gascoigne et al. 2011; Hori et al. 2013), and budding yeast (Ho et al. 2014), for inducing ectopic centromere formation. For instance, in budding yeast, a synthetic kinetochore can be formed by tethering an outer kinetochore protein Ask1 to a LacO array on the chromosome. Interestingly, CENP-ACse4 is also localized to this synthetic kinetochore without a centromere sequence (Ho et al. 2014). This technique allows researchers to investigate the determinants of individual CENP-A domains or kinetochore components in centromere establishment and kinetochore formation.
De novo centromere formation by introducing exogenous naked DNA
Yeast artificial chromosomes
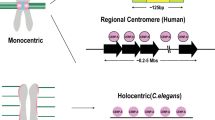
Budding Saccharomyces cerevisiae yeast artificial minichromosome (ScYAC) was the first artificial chromosome constructed (Murray and Szostak 1983). It consists of centromeric DNA sequence, replication origin, and telomere sequences. S. cerevisiae consists of a point centromere. The minimal functional centromeric DNA sequence (~ 125 bp) contains no DNA repeats and is comprised of three conserved DNA domains, namely CDEI, CDEII, and CDEIII. CDEI (~ 8 bp) and CDEIII (~ 25 bp) consist of palindromic motifs. CDEII (~ 90 bp) represents the central domain with approximately 90% of AT content, where CENP-A localizes (Meluh and Koshland 1997). In fungi closely related to S. cerevisiae, their point centromeric DNAs are between 125 and 225 bp, arranged in the conserved CDEI-CDEII-CDEIII structure (Meraldi et al. 2006). Transformation of naked or purified ScYAC DNA results in de novo centromere establishment on the centromeric DNA, and the ~ 125-bp centromeric DNA sequence is sufficient for centromere formation and function (Fig. 1h, Table 2) (Cottarel et al. 1989).
Fission yeast S. pombe centromeric DNA ranges from 35 to 110 kb, consisting of a 4–7-kb central core (cc) domain flanked by two repetitive domains (imr and otr) (Chikashige et al. 1989; Nakaseko et al. 1986). CENP-A localizes to the central domain (cc and imr repeats), while otr repeats constitute the pericentric heterochromatin (Partridge et al. 2000). Transforming purified DNA with centromere DNA sequence (core and surrounding repeated sequences) results in the formation of fission yeast artificial minichromosome (SpYAC) with de novo centromere establishment (Fig. 1h, Table 2) (Hahnenberger et al. 1989). Importantly, both the central and flanking repetitive domains are required for de novo centromere formation (Folco et al. 2008), in which the otr domain has a role in establishing heterochromatin for de novo CENP-ACnp1 recruitment and kinetochore assembly, but not in maintenance of CENP-ACnp1 (Kagansky et al. 2009).
Human artificial chromosomes
Human regional centromeres, spanning 3–5 Mb, consist of 171-bp α-satellite DNA (alphoid DNA) arranged in tandem arrays (Waye and Willard 1987; Willard and Waye 1987; Wu and Manuelidis 1980). Based on the bottom-up approach, introducing long arrays of α-satellite DNA (minimum 30 kb and usually 60–70 kb) (Okamoto et al. 2007), together with telomeric DNA, a selectable marker and genomic DNA into human HT1080 cells, followed by concatemerization, can result in an assembly of mitotically stable human artificial minichromosomes with de novo centromere formation efficiency at about 30% (Fig. 1h, Table 2) (Harrington et al. 1997; Ohzeki et al. 2012). However, the formation efficiency of human artificial chromosome (HAC) is rather low in other cell lines and requires several weeks of selection (Macnab and Whitehouse 2009). Alternatively, partial deletion of an existing chromosome based on the top-down approach can also lead to the formation of HAC (Table 2) (Katoh et al. 2004; Yang et al. 2000).
De novo centromere formation on worm artificial chromosomes in Caenorhabditis elegans
C. elegans is becoming a useful model organism to study centromere establishment. Instead of forming a single discrete (monocentric) centromeric domain on the chromosome, C. elegans centromere is diffused along the length of the chromosomes (holocentric) (Gassmann et al. 2012; Steiner and Henikoff 2014). CENP-A is de-enriched in active genes expressed in embryos, and germline genes (Gassmann et al. 2012). DNA injected into C. elegans germline resulted in the formation of worm artificial chromosomes (WACs), also known as extrachromosomal arrays (Ex) (Fig. 1h, Table 2) (Mello et al. 1991; Stinchcomb et al. 1985). Importantly, no worm DNA is required for the WAC formation (Mello et al. 1991; Stinchcomb et al. 1985), possibly due to its holocentric nature, which may be less DNA sequence–dependent and more promiscuous for different DNA sequences. These artificial chromosomes form de novo centromeres and segregate accurately at a high frequency within a few cell divisions after AC formation (Yuen et al. 2011). The formation of de novo centromere on the artificial chromosomes is fast, efficient, and robust (Yuen et al. 2011), facilitating the mechanistic investigation of de novo centromere formation and the interplay between centromere function and transcription.
Plant artificial chromosomes
In plants, de novo centromere formation on naked DNA has not been reported (Gaeta et al. 2012; Houben et al. 2008; Mette and Houben 2015). It is suggested that epigenetic marks that direct centromere formation cannot be reestablished on naked DNA introduced into the plant (Birchler 2015; Gaeta et al. 2012). An alternative approach to create plant artificial chromosomes (PACs) with a functional centromere is to truncate the endogenous chromosome by inserting telomere to the chromosome arms (Farr et al. 1991; Yu et al. 2006, 2007). The telomere-mediated chromosomal truncation generated a minichromosome that can be further engineered to carry desired sequences. PACs generated by this method were reported in maize (Yu et al. 2006, 2007), Arabidopsis (Nelson et al. 2011; Teo et al. 2011), barley (Kapusi et al. 2012), rice (Xu et al. 2012), wheat (Yuan et al. 2017), and Brassica (Yan et al. 2017).
“Ectopic” CENP-A in response to DNA damage
CENP-A enrichment has been reported at sites of laser-induced DNA damage or I-SceI cleavage in HeLa cells, together with other centromeric proteins, such as CENP-N, CENP-U, and DNA repair factors (Fig. 1j) (Zeitlin et al. 2009). Irradiation-induced DNA damage also causes relocalization of CENP-A and CENP-C to separated small foci in the nucleus, resulting in uncoupling of kinetochore proteins and kinetochore weakening (Ambartsumyan et al. 2010) (Fig. 1j). Indeed, the increasing CENP-A expression level in cells increases the survival rate in DNA damage (Lacoste et al. 2014; Lawrence et al. 2015; Zeitlin et al. 2009). Reciprocally, when CENP-A is knocked down in human pluripotent stem cells (hPSCs), DNA damage induces significant apoptosis, possibly because the knockdown reduced the availability of new CENP-A protein to rebuild the weakened kinetochores (Ambartsumyan et al. 2010). The relocalization of CENP-A after DNA damage suggested that CENP-A may participate in the DNA double-stranded break (DSB) repair process (Zeitlin et al. 2009). Alternatively, the enrichment of CENP-A at the DNA damage site could also serve as a mechanism to restore the centromeric function on potentially acentric chromatin fragments. Nonetheless, DNA breaks induced by multiphoton laser micro-irradiation in human osteosarcoma U2OS cells did not show CENP-A accumulation but showed CENP-S and CENP-X assembly on the DNA damage sites (Helfricht et al. 2013). The discrepancy may be due to the use of different cell lines or conditions. Interestingly, another study showed that upon DNA damage by etoposide in the murine NIH/3T3 cells, CENP-A is delocalized from centromere to the periphery and inside of the nucleolus, dependent on ATM kinase and p53 (Hedouin et al. 2017). However, this is a rather late event (~ 24 h, versus within minutes in the study of Zeitlin et al. (2009)) and may not participate in DNA repair per se but may be a consequence of prolonged stress and DNA damage response, promoting maintenance of cell cycle arrest as in senescent cells (Hedouin et al. 2017). Whether CENP-A participates in DNA damage and repair is still in debate.
Why is centromere establishment important in evolution and diseases?
Meiotic drive and speciation
Maize terminal neocentromere plays an important role in meiotic drive (Dawe et al. 2018; Lyttle 1991). Maize chromosome with a terminal neocentromere (knob) has an advantage to move more rapidly to the spindle pole during meiosis I. Such orientation is maintained in meiosis II; thus, chromosomes containing the knob are often segregated to the outermost megaspores. As the basal outermost megaspore always becomes the functional megaspore (equivalent to the egg in mammals) instead of non-functional megaspores (equivalent to the polar body in mammals) (Rhoades 1942), the chromosome with a terminal neocentromere will be transmitted to progenies at a higher frequency than the expected Mendelian ratio, and it is proposed to be a driving force of evolution which favors certain karyotypes (Sandler and Novitski 1957). However, the chromosome with a terminal neocentromere is still not fixed in the population, suggesting that it may also somehow reduce fitness, causing a negative selection and balancing out the meiotic drive effect (Buckler et al. 1999). Centromere repositioning may result in stronger centromeres (with increased kinetochore protein levels and/or enhanced kinetochore-microtubule interaction), which could also promote meiotic drive, fixation, and speciation (Chmatal et al. 2014). Meiotic drive is also demonstrated in mice. Chromosomes with more centromeric DNA repeats form a stronger kinetochore, which increases the chance of that chromosome to segregate to the egg during female meiosis (Iwata-Otsubo et al. 2017; Lampson and Black 2017).
Rescuing acentric chromosomes from loss and affecting growth advantage
Neocentromeres are also associated with certain cancer types. As mentioned, supernumerary ring (circular) or giant rod–shaped marker chromosomes are a feature of ALP-WDLPS. These chromosomes lack α-satellite sequences, but the formation of neocentromeres on non-centromeric sequences (Sirvent et al. 2000) is proposed to confer a selective advantage for such chromosomes to stably segregate in cancer cells. On the other hand, human patients ascertained with neocentromeres often contain chromosomal rearrangements, which result in developmental delays or infertility (Klein et al. 2012). Indeed, the neocentromeres may have rescued the rearranged acentric chromosome from being lost and enabled the survival of the individuals (Marshall et al. 2008).
Inducing chromosome instability in dicentric chromosomes
In colorectal and other solid cancer cells, CENP-A and other kinetochore proteins are commonly overexpressed and deposited on ectopic regions (Athwal et al. 2015; Tomonaga et al. 2003), potentially promoting neocentromere formation and ectopic kinetochores and driving chromosomal instability, resulting in aneuploidy. Therefore, it is important to understand the causes and consequences of centromere protein overexpression and the mechanism of neocentromere formation in cancer cells, as it may unravel how tumors progress and reveal novel molecular markers for cancer diagnosis or prognosis (de Wolf and Kops 2017; Zhang et al. 2016).
DNA sequence preference for centromere establishment
AT content
Studies on human neocentromeres formed on ectopic, non-centromeric DNA regions revealed that they prefer to be localized to AT-rich regions, similar to the feature of native centromeric sequences (Lo et al. 2001, b). Despite that, many AT-rich regions in the human genome have not been correlated to neocentromere formation, possibly due to the low number of cases ascertained (Alonso et al. 2007). Human neocentromeres are usually identified on marker chromosomes in patients with developmental delays or in human cancers (Amor and Choo 2002). Whether these examples represent the general features of human neocentromeres is still in question. Therefore, the exact role of AT content in centromere establishment is still not clear. One possibility is that there is a threshold requirement for AT content (Marshall et al. 2008). Such hypothesis is theoretically testable as de novo centromeres can be established on exogenous, purified centromeric DNA introduced into cells as demonstrated in yeast artificial chromosomes (YACs), HACs, and C. elegans WACs. However, native centromeric sequences are usually required for efficient formation of the de novo centromeres in yeast and humans (Tables 2 and 3). Surprisingly, in holocentric C. elegans, any DNA seems to be able to form artificial chromosomes that establish centromeres at high frequency. Using this model, the preference of AT content in centromere establishment on C. elegans artificial chromosomes is further investigated by injecting DNA with different AT contents into the germline. High AT content is preferred for new centromere formation as co-injection with the AT-rich sequence increases WAC segregation frequency (Lin and Yuen, unpublished).
Sequence repetitiveness
Centromere repeats may be derived from retroviral elements
Native centromeres are commonly found embedded in tandem repeat sequences, such as satellite DNA in mammals and retroelements in some plants that last for kilobases to megabases on a chromosome. In humans, retroelements spread across the centromeres of 15 chromosomes (Zahn et al. 2015). Concerted evolution of centromere tandem repeats promotes the homogenization of centromere DNA across chromosomes within a species (Malik and Henikoff 2009; Melters et al. 2013). The widely observed repetitive sequences at centromeric DNA, in paradox with its high sequence diversity among species, raise a question of what the function of repetitive DNA in centromere establishment is.
While the origin of centromeric tandem repeat sequence is unknown, one hypothesis proposes that they may be derived from subtelomeric elements, which may be, in turn, retroelements (Villasante et al. 2007). Centromeric retrotransposons (CRs) were found in almost all grass species (Jiang et al. 2003), where they were predicted to be integrated into the centromeres million years ago. By sequence alignment, the centromeric retrotransposons in rice and maize reveal their common characteristics of the long terminal repeat (LTR), indicating that they may originate from the same ancestor (Nagaki et al. 2005; Wolfgruber et al. 2009). Retrotransposon repeat integration into the established centromere could be a potential driving force to homogenize the centromeric sequences among chromosomes (Sharma et al. 2013). Despite the sequence differences of the centromeric repeats between Arabidopsis lyrata and Arabidopsis thaliana, transformation of CR element (Tal1), that is enriched in Arabidopsis lyrata centromere, could also target and integrate into the centromeric repeats of A. thaliana (Birchler and Presting 2012; Tsukahara et al. 2012), suggesting that the accumulation of newly integrated CR can eventually change the centromere composition. In maize inbreeds, severe selection for favorable centromere-linked genes increases the transcription level at the centromere, which, in turn, forces the centromere to relocate from the maize centromere–specific satellite repeat, CentC, to its flanking regions, followed by integration and enrichment of centromere-specific retrotransposon, maize CR (CRM). Accumulated CR invasion events would restore the repetitiveness of the neocentromere, stabilize it, and drive the replacement of the centromere repeat context in domestic maize (Schneider et al. 2016). However, expanded centromeric retrotransposons can also form multiple active centromeres on a chromosome, which may cause chromosomal instability and drive karyotype change. For example, in a wheat-rye hybrid, rye centromeric transposons were expanded to the euchromatin of rye chromosome and caused chromosomal breakage and re-joining (Guo et al. 2016).
However, CR expansion or integration into the centromere is not the only driving force that triggers centromere evolution. In an oat-maize hybrid, the maize centromere in oat background could expand to their flanking regions from 1.4–1.8 Mb to 3.3–3.8 Mb, which is not caused by CR invasion. The expansion of maize centromere in oat background is restricted by the actively transcribed regions, suggesting that transcription background on chromosome also shapes the centromere composition. One explanation for this kind of expansion is that centromere sizes tend to be relatively uniform within an organism (plant hybrid), regardless of the chromosome size or the origin of the centromeres (Wang et al. 2014). The expansion of maize centromeres in an oat-maize hybrid, to the size comparable to that of oat centromeres, may improve the stability of maize chromosomes in the oat background.
Indeed, retroviral sequences have been found not only in plants but also in human and tammar centromeres (Zahn et al. 2015). In humans, HIV infection and the presence of HIV Tat protein can trigger endogenous, centromere-localized retroviral K111 expression, and relocation to other genomic locations (Contreras-Galindo et al. 2013). Karyotyping of Macropus species shows that chromosomal rearrangements at the centromeres could have been driven by breakage at a centromere-specific retroviral element (Bulazel et al. 2007; Ferreri et al. 2004).
The sequence features of neocentromeres
Unlike endogenous centromeres, neocentromeres identified in chicken DT40 cells (Shang et al. 2013), Drosophila (Olszak et al. 2011; Williams et al. 1998), yeast (Copenhaver et al. 2009), and wheat (Guo et al. 2016) show common localizations at non-repetitive sequences (Figs. 1g and 2, Table 3). Human neocentromeres were also mostly found at non-repetitive regions (Garsed et al. 2014; Marshall et al. 2008; Sullivan et al. 2016), with an exception that localizes at non-centromeric tandem repeats (Hasson et al. 2011). Interestingly, the underlying sequences of neocentromeres also have enriched dyad symmetries and non-B-form DNA, which may facilitate de novo centromere formation (Kasinathan and Henikoff 2018). Most surprisingly, a recent study has shown that a neocentromere sequence, from pseudodicentric-neocentric (PD-NC4) on human chromosome 4 (Amor et al. 2004), which lacks α-satellite repeats, can bypass the need of CENP-B in de novo centromerization and can form HACs (Logsdon et al. 2019).
Features of α-satellite in de novo centromere formation in mammalian cells
A sequence-specific centromeric protein, CENP-B, was found to be required for de novo formation of a HAC or mouse artificial chromosome (MAC) with a functional centromere (Okada et al. 2007). The presence of CENP-B box, a DNA element of the α-satellite DNA, facilitates de novo centromere formation in HACs, and α-satellite DNA with enriched CENP-B box density further enhances the de novo centromere formation efficiency (Tables 2 and 3) (Basu et al. 2005, b; Grimes et al. 2002; Harrington et al. 1997; Ohzeki et al. 2002). On the contrary, CENP-B does not affect endogenous chromosomal stability in mice (Kapoor et al. 1998). Yet, a recent study found that mouse fibroblasts without CENP-B have a chronic elevated rate of chromosomal missegregation (Fachinetti et al. 2015). New characterization approaches allow human α-satellite DNA to be further subdivided, and third-generation long-read DNA sequencing enables more precise mapping of centromeric regions (Jain et al. 2018). By a combination of chromatin immunoprecipitation (ChIP)-seq and de novo HAC formation assay, sequence requirements for centromere formation were further tested. Interestingly, α-satellites that are not associated with CENP-A at endogenous human centromeres, but with high-order repeats (HORs), have centromere formation competency, while those monomeric repeat units do not. It should be noted that both these HORs and monomeric sequences contain CENP-B boxes, which indicates that the repeat structure, rather than the CENP-B box alone, affects centromere formation competency (Hayden et al. 2013). Indeed, sequence abundance and composition within the repeats could affect de novo centromere formation. It is found that sequence variants within HOR reduces CENP-A level and centromere stability (Table 3) (Aldrup-MacDonald et al. 2016). Surprisingly, human alphoid DNA is sufficient to form MACs in mouse cells, despite that their centromeric satellite DNA shares no conserved sequence except for the CENP-B box (Okada et al. 2007; Zeng et al. 2004).
On tracts of repeat sequences, non-B-form DNA secondary structures tend to spontaneously form hairpins, R-loops, and cruciform (Sharma 2011). Non-B-form DNAs have been observed commonly in centromeric DNA from a variety of organisms, including humans, and are recognized by CENP-A chaperones (Kasinathan and Henikoff 2018). A recent study proposed that centromeres are formed at non-B-form DNA, by its dyad symmetries, where inverted repeats of base pair with each other sequences to form cruciform structure. However, some species, such as humans, mice, and budding yeast, that lack dyad symmetries in centromeric DNA recruit sequence-specific DNA-binding proteins, such as CENP-B, to promote the formation of non-B-form DNA secondary structures (Kasinathan and Henikoff 2018).
Sequence specificity
For neocentromeres formed on non-centromeric endogenous DNA, there is no sequence similarity to native centromere sequence, suggesting that DNA sequence is not necessary or sufficient for centromere establishment. It is clear that the localization of neocentromeres can be influenced by the epigenetic environment at the loci. In the section “Epigenetic effect on centromere,” we will discuss the epigenetic environment of centromeric establishment by dissecting the neocentromeric chromatin into two regions as in native centromeres: (1) the core centromeric chromatin where ectopic CENP-A is deposited in the section “Chromatin environment for CENP-A centromeric chromatin establishment” and (2) the flanking chromatin in the section “Flanking chromatin environment for centromeric chromatin establishment.”
Epigenetic effect on centromere
Chromatin environment for CENP-A centromeric chromatin establishment
Histone modifications
A number of studies showed that euchromatic histone marks in the chromatin are favored for centromere establishment (Bergmann et al. 2011; Nakano et al. 2008; Ohzeki et al. 2012, 2015). By tethering different histone modifiers onto a LacO array, which replaces the alternating CENP-B binding site on alphoid DNA in HACs, H3K9ac (a euchromatin mark) is found to be promoting de novo CENP-A incorporation and stable HAC formation, whereas H3K9me3 (the heterochromatin mark) is inhibiting the process (Table 4) (Ohzeki et al. 2012). Yet, heterochromatin marks that are enriched in the flanking regions of centromere could help to determine the centromere boundaries (Sharma et al. 2019). Similarly, histone H3 and H4 acetylations on C. elegans ACs facilitate de novo centromere formation (Zhu et al. 2018) and heterochromatin inhibits the formation of de novo centromeres on AC in C. elegans (Table 4) (Yuen et al. 2011). These findings suggest that a general open chromatin environment is favorable for centromere establishment.
In a human chromosome in which an ectopic alphoid DNA from YAC is integrated at the terminal region, the centromere activity is suppressed in the integrated alphoid DNA. However, inducing histone hyperacetylation by treating cells with histone deacetylase inhibitor trichostatin A (TSA) promotes the assembly of CENP-A on the integrated alphoid DNA and the release of minichromosomes from the YAC integration site (Nakano et al. 2003). Besides the role of centromere establishment, euchromatin is involved in maintaining the stability of de novo centromeres. On HACs which have been propagating through mitoses, depletion of a euchromatin mark H3K4me2, by tethering lysine-specific demethylase 1 or 2 (LSD1/2) to HAC centromeres, causes defective incorporation of CENP-A on the alphoid DNA, resulting in a gradual loss of kinetochore function (Table 4) (Bergmann et al. 2011; Molina et al. 2016). It is proposed that H3K4me2 may help to maintain an open chromatin environment for CENP-A centromeric nucleosome incorporation, facilitating centromere propagation (Stimpson and Sullivan 2011).
Surprisingly, in a study using chicken DT40 cells, in which neocentromere formation is induced by deleting the native centromere on chromosome Z by loxP recombination, chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) indicated that the neocentromeres are not associated with either the euchromatin mark H3K4me2 or the heterochromatin mark H3K9me3 (Shang et al. 2013). The reason for these different epigenetic features is unclear but may be related to differences in organisms (Table 4) or differences between de novo centromeres and neocentromeres (Table 1). Moreover, the epigenetic states may dynamically change during the new centromere establishment process or after they have been stabilized; thus, the experimental results may vary depending on whether the chromatin status is analyzed.
DNA modifications
In maize endogenous B chromosome, hypomethylated centromeric DNA is functioning normally, whereas hypermethylation of centromeric DNA causes centromere inactivation (Koo et al. 2011). On a neocentromere on human chromosome mardel(10) (a complex rearrangement of chromosome 10 with a complete loss of alphoid DNA found in a child patient with developmental delay) (Voullaire et al. 1993), there is an increase of DNA methylation as compared to the locus 10q25 on the normal chromosome 10 (Wong et al. 2006). The presence of DNA hypermethylation at the neocentromere, which may recruit chromatin modifiers for the formation of heterochromatin (Lorincz et al. 2004; Stirzaker et al. 2004), seems to contradict the above hypothesis that heterochromatin antagonizes centromere establishment in HACs (Ohzeki et al. 2012). However, DNA hypermethylation at centromeric satellite repeats can prevent centromeres to illicit mitotic recombination in mice, suggesting that DNA hypermethylation may only be essential for centromere maintenance (Jaco et al. 2008). However, the mardel(10) study also identified hypomethylated pockets within the hypermethylated neocentromeric DNA, suggesting that there is still euchromatic property within the neocentromere, which may support the formation or function of the neocentromere (Table 4) (Wong et al. 2006).
Euchromatin (open chromatin) and non-coding RNA
In line with the notion that de novo centromeric chromatin on HACs is euchromatic (Bergmann et al. 2011; Ohzeki et al. 2012), RNA polymerase II–driven transcription on C. elegans ACs also facilitates de novo centromere formation (Zhu et al. 2018). Neocentromeres are also often found in transcriptionally active DNA or genic regions. In the mardel(10) neocentromere, the CENP-A domain was spanned by the actively expressed ATRNL1 gene, and the neocentromere formation did not affect the expression of the gene significantly (Fig. 2, Table 4) (Saffery et al. 2003). A similar observation has also been found on a natural monocentric chromosome of rice, where at least four active genes are interspersed in Cen8 (Nagaki et al. 2004). In HACs, the CENP-A domain is not only restricted to form on the alphoid DNA, but it also spreads over the neighboring, active selection marker genes (Lam et al. 2006). Colorectal cancer cell lines with overexpression of CENP-A contain ectopic incorporation of CENP-A in DNase I–hypersensitive regions and transcription factor binding sites, suggesting that CENP-A prefers to deposit in euchromatic, transcriptionally active regions (Athwal et al. 2015). Indeed, on endogenous human chromosomes, a low level of ectopic CENP-A was assembled at transcriptionally active sites on euchromatin, but this was removed after DNA replication (Nechemia-Arbely et al. 2019).
On the contrary, C. albicans neocentromeres can be found in large intergenic regions, and neocentromere activity can silence active genes nearby (Ketel et al. 2009). Similarly, in fission yeast, neocentromeres are formed on subtelomeric, heterochromatin-euchromatin boundary regions after deletion of endogenous centromeric DNA (Fig. 2, Table 4) (Ishii et al. 2008). A number of nitrogen starvation-induced genes are mapped in these regions, and the gene expression levels remained low even after the removal of nitrogen from the media (Ishii et al. 2008), indicating that neocentromere activity may not be compatible with high gene expression. However, one should note that neocentromeres generated experimentally are often identified under selective pressure. Therefore, whether the gene within or near the neocentromere is silenced or not may depend on the combined growth advantage conferred by maintaining the neocentromere and, concurrently, silencing or expressing the nearby genes.
If neocentromeres are transcriptionally incompatible in C. albicans and fission yeast, why they prefer to establish on the euchromatin at the first place? In particular for species with relatively compact genomes, there are fewer intergenic regions, which might limit the location options for neocentromere to establish. It is possible that euchromatin possesses a more open environment for initial CENP-A deposition, promoting the formation of neocentromere. However, after the new CENP-A domain is established, the underlying transcriptional activity at the neocentromere may be reduced or even silenced, potentially to maintain its stability. In fact, studies in budding yeast have suggested that a fine balanced level of centromeric transcription is essential for normal centromere function (Hill and Bloom 1987; Ling and Yuen 2019a, b; Ohkuni and Kitagawa 2012). Similarly, tethering either a transcriptional activator or silencer to the propagating HAC centromere can disrupt kinetochore structure, resulting in missegregation of HACs (Cardinale et al. 2009; Nakano et al. 2008).
When euchromatin provides a more open chromatin environment for de novo CENP-A deposition, such environment also concomitantly allows the expression of RNA from the neocentromeric region. In fact, transcripts originated from the centromeric core region may have a direct role in centromere function. In S. pombe, the accumulation of RNAPII at the centromeric DNA at G2 phase destabilizes the H3 nucleosomes, which, in turn, facilitates new CENP-A deposition (Shukla et al. 2018). Despite that the central-core centromere sequences from other Schizosaccharomyces yeasts, Schizosaccharomyces octosporus and Schizosaccharomyces cryophilus, hold non-homologous sequences, they are compatible with de novo CENP-A deposition in S. pombe (Tong et al. 2019). Such result indicates that these centromeric DNAs possess some conserved properties, such as the transcriptional landscape that promotes centromere establishment. In native centromeres, centromeric transcripts promote new CENP-A loading in human cells (Bobkov et al. 2018; McNulty et al. 2017), enhance the interaction between inner kinetochore protein CENP-C with the centromeric DNA in human cells (McNulty et al. 2017) and also in maize (Du et al. 2010) (Table 4), and regulate the chromosome passenger complex (CPC) component Aurora B activity and localization (Blower 2016; Ferri et al. 2009; Ideue et al. 2014). Importantly, too much or too little centromeric transcripts are detrimental. In human cells, SUV39 histone methyltransferases interact with α-satellite RNA transcripts to establish heterochromatin and ensure genomic stability (Johnson et al. 2017). Knockdown of centromeric transcripts resulted in mitotic defects in human cells (Ideue et al. 2014) and loss of minichromosome in budding yeast (Ling and Yuen 2019b). On the other hand, overexpression of centromeric transcript in mouse and human cells also caused chromosomal missegregation (Bouzinba-Segard et al. 2006; Chan et al. 2017) and a decrease of centromeric localization of CENP-A, CENP-C, and Aurora B in budding yeast (Ling and Yuen 2019b). Nonetheless, it is not known if centromeric transcripts are participating directly in centromere establishment as these studies are focusing on existing centromere function.
Flanking chromatin environment for centromeric chromatin establishment
Pericentric heterochromatin
The requirement of flanking heterochromatin for centromere establishment is best illustrated in S. pombe. It was observed that both the central core and at least part of the flanking centromeric domains (otr) are required for de novo centromere formation on the SpYAC (Tables 2 and 3) (Folco et al. 2008). Non-coding RNA transcription at the pericentric heterochromatin and processing of the transcripts by the RNA interference pathway are important for the establishment of the pericentric heterochromatin (Allshire and Ekwall 2015a; Folco et al. 2008; Kagansky et al. 2009). In turn, heterochromatin facilitates the initial recruitment of CENP-ACnp1 to naïve DNA introduced, but once the de novo centromere is established, CENP-ACnp1 can be propagated in the absence of heterochromatin (Folco et al. 2008). Other studies suggested that the cohesin complex, associated with the flanking pericentric heterochromatin, helps to maintain sister chromatid cohesion and may also play a role in centromere function (Bernard et al. 2001; Nonaka et al. 2002). In Drosophila S2 cells in which neocentromeres can be generated by overexpression of CENP-A, the ectopic incorporation of CENP-A is always found to be localized at heterochromatin-euchromatin boundaries (either near telomeres or pericentromeric heterochromatin), suggesting that such heterochromatin junction is favorable for ectopic CENP-A loading (Fig. 2) (Olszak et al. 2011). Besides, in a human cell line, BBB, which contains a neocentromere in band 13q33.1 (Warburton et al. 2000), ChIP analysis of the heterochromatin histone mark H3K9me3 has revealed a small block (~ 15 kb) of heterochromatin domain close to the CENP-A domain (Alonso et al. 2010).
With the plausible function of the flanking heterochromatin in centromere establishment, it is predicted that certain heterochromatin histone marks may favor centromere establishment. However, heterochromatin is not necessary for centromeric protein deposition on the alphoid sequence in HACs (Nakashima et al. 2005). In contrast, inhibition of heterochromatin protein 1 (HP1) accelerated the de novo CENP-A incorporation in HACs (Ohzeki et al. 2012) and WACs (Yuen et al. 2011). Yet, heterochromatin formed outside the centrochromatin is essential for establishing a stable HAC (Nakashima et al. 2005). Indeed, other than the neocentromere in BBB cell line, it is not common to see a heterochromatin domain at human neocentromeres, indicating that heterochromatin or its associated cohesion activity is not a prerequisite for centromere establishment per se (Alonso et al. 2010). However, as a result, the neocentric sister chromatids always separate prematurely (Alonso et al. 2010).
Gene context, transcription, and non-coding RNA
Although the importance of flanking heterochromatin on centromere establishment is not well understood, a study in S. pombe has identified the role of pericentromeric transcripts in regulating heterochromatin formation (Verdel et al. 2004), which, in turn, governs de novo centromere formation (Folco et al. 2008). Double-stranded non-coding RNAs (ncRNAs) are found to be expressed from the pericentromeric region (outer repeats, otr), and these ncRNAs are processed into small interfering RNA (siRNA) by Dicer (Dcr1) and the RNA interference (RNAi) effector complex. RNA-induced initiation of transcriptional gene silencing (RITS) complex is recruited onto the pericentromeric region by the siRNAs. The RITS complex then recruits the methyltransferase Clr4 for promoting H3K9 methylation and heterochromatin formation through HP1Swi6 (Verdel et al. 2004; Zhang et al. 2008). If the formation of pericentromeric heterochromatin is disrupted, either by deletion of HP1Swi6 or the RNAi components (Chp1 or Dcr1), CENP-A deposition on the introduced naked centromeric DNA will be disrupted (Folco et al. 2008), indicating that the integrity of pericentromeric heterochromatin is important for centromere establishment and the non-coding pericentromeric transcript is the key initial player behind this process. By tethering H3K9 methyltransferase Clr4 to the region flanking the centromeric DNA, pericentromeric heterochromatin can be formed without RNAi. This synthetic heterochromatin bypasses the need for RNAi in promoting de novo centromere formation (Kagansky et al. 2009).
Although pericentromeric transcripts are also found in many different organisms such as humans and mice, they seem to be in the form of long ncRNA (> 1 kb, up to 5 kb) instead of siRNA as in S. pombe (Jolly et al. 2004; Lu and Gilbert 2007; Rudert et al. 1995; Saksouk et al. 2015; Valgardsdottir et al. 2008). Dicer-deficient cells show defects in cohesin localization in chicken DT40 cells (Fukagawa et al. 2004) and cell proliferation, viability, and differentiation in mice (Kanellopoulou et al. 2005; Maison et al. 2002; Murchison et al. 2005). However, it is unclear whether these pericentromeric lncRNA transcripts are processed by the RNAi machinery as in S. pombe and if RNAi-mediated pericentromeric heterochromatin formation is conserved in all regional monocentromeres. Nonetheless, long pericentromeric transcripts are also able to interact with HP1 in mouse cells (Maison et al. 2011), suggesting that pericentromeric transcripts may still regulate heterochromatin formation in an RNAi-independent manner. It will be exciting to see more studies investigating the function of these long pericentromeric transcripts on centromere establishment in the future.
Future perspectives
Interplay between centromeric and the flanking chromatin in centromere establishment
Taken together, one common characteristic in neocentromeres in multiple models is that they are established on DNA which shows no homology to native centromeric DNA sequences, preferably close to the original centromere location or repeats, indicating that the preference of ectopic centromere is not merely controlled by sequence but also affected by position and chromatin environment. The results from Drosophila, fission yeast, and humans suggest that neocentromeres may prefer to form at the junction of euchromatin and heterochromatin (Fig. 2) (Ishii et al. 2008; Olszak et al. 2011; Wong et al. 2006). A euchromatic chromatin environment allowing transcription is observed in new CENP-A domains in human cells (Bergmann et al. 2011; Ohzeki et al. 2012; Saffery et al. 2003). On the other hand, experimental data in Candida and chicken cells suggest an inverse relationship between robust gene expression and neocentromere formation (Shang et al. 2013; Thakur and Sanyal 2013). Moreover, while the flanking heterochromatin is important for de novo centromere formation in fission yeast (Allshire and Ekwall 2015a; Folco et al. 2008; Kagansky et al. 2009), it is not always a prerequisite for de novo centromere formation (Ohzeki et al. 2012; Yuen et al. 2011). For a cis chromosomal locus that is critical for organism survival, such diversity in structure is surprising. How centromeric and the flanking chromatin environments interplay with each other for centromere establishment still needs further investigation.
Cancer patient–derived cell models for neocentromere identification
Chromosomal instability has been suggested to promote tumorigenesis, but the exact mechanism of how a normal cell progresses to develop into a cancer cell is still unclear. Cancer patient–derived cell lines show a high frequency of genome rearrangements, neochromosomes, and neocentromere (Goodspeed et al. 2016). A recent study uses fluorescent dyes to mark neochromosomes, followed by flow sorting of chromosomes to isolate and enrich neochromosomes and high-throughput sequencing to identify chromosomal rearrangement positions at single base pair resolution (Garsed et al. 2014). These new methods would help to elucidate the composition of neochromosomes, tracing the chromosomal breakage and rearrangement events (Papenfuss and Thomas 2015). Moreover, the long reads from third-generation DNA sequencing technique not only enable researchers to resolve the context of highly repetitive DNA in centromere (Jain et al. 2018; Mahajan et al. 2018) but also more reliably identify indels and chromosomal rearrangements of the tumor genome and describe their architecture in detail.
A combination of centromere protein immunofluorescence, centromere fluorescence in situ hybridization, and multicolor FISH would also enhance the identification of neocentromeres (Beh et al. 2016). In addition, the neocentromere on neochromosomes or RGM chromosomes can also be precisely mapped by high-resolution ChIP-seq using antibodies against centromeric protein, e.g., CENP-A. ChIP-seq data can resolve neocentromere in higher resolution, which provides a complement of the traditional FISH method. Such a combination of cytological and high-throughput sequencing techniques would accelerate the unraveling of the relationship between chromosomal rearrangement and neocentromere formation.
New model organisms for studying centromere establishment on artificial chromosomes
One should note that it is sometimes technically difficult to dissect between the process of centromere establishment and maintenance. Many neocentromeres analyzed were under selective pressure or have already been propagated in the cells for many rounds of mitotic divisions or generations. It will be interesting to analyze the chromatin requirements for neocentromere formation in a live cell condition and in real time, preferably without arbitrary selection, to follow the process of centromere establishment and the fate of the neocentromere, distinguishing this establishment process from the maintenance of existing centromere through mitosis or meiosis. In C. elegans, we can trace the artificial chromosomes specifically by injecting LacO DNA and expressing LacI::GFP fusion protein (Yuen et al. 2011; Zhu et al. 2018), similar to the TetO/TetR system used in HACs (Bergmann et al. 2011, 2012; Ohzeki et al. 2012). The centromere establishment process is more efficient in artificial chromosomes in C. elegans than in HACs. Live-cell imaging without selection can be used to observe the artificial chromosomes before and after de novo centromere formation. The epigenetic environment of WACs can be manipulated as in HACs by tethering histone modifiers (Bergmann et al. 2011, 2012; Ohzeki et al. 2012). With such complementary setups, it is expected that systematic cracking of the genetic and epigenetic requirement for centromere establishment could be accomplished in the future.
Improving HACs for gene therapy
HACs can be used as a vector to deliver the functional copy of a gene for gene therapy. Compared with some virus-based vectors, HACs can host a large gene insert, and HACs do not integrate into the host genome, avoiding insertional mutagenesis. For successful and sustainable gene therapy, it is important for the HAC to maintain a functional centromere and segregate faithfully. To avoid potential side effects from introduced DNA, it is advantageous to minimize the sequence requirement of HAC. In the past, most HACs require the use of alphoid DNA sequence for efficient chromosomal assembly. In the bottom-up approach, this alphoid DNA, usually 60–70 kb in length (Okamoto et al. 2007), is transfected into cells and multimerized and forms HACs with various sizes. The location or the copy number of the gene being assembled in the HAC cannot be predetermined, which may limit the use of such HACs in gene therapy (Kim et al. 2011). In the top-down approach, the alphoid array, telomeric sequences, and gene insertion site are cloned (Kazuki and Oshimura 2011). By elucidating the de novo centromere formation mechanism, smaller de novo centromere-based HACs with a defined centromere sequence may be generated. The most recent HACs contain non-repetitive centromere sequence, bypassing the need of centromeric DNA and CENP-B (Logsdon et al. 2019). This has revolutionized and streamlined the construction and characterization of HACs. By understanding the epigenetic environment for centromere formation, it may even be possible to manipulate the histone environment to facilitate the establishment of centromere in the HAC or improve the stability of the centromere in the HAC. With the improvement of microcell-mediated chromosome transfer (MMCT) for introduction of HACs into the target cells (Brown et al. 2017; Hiratsuka et al. 2015; Liskovykh et al. 2016; Suzuki et al. 2016), we anticipate that these efforts will contribute to the development of an effective vector for gene delivery in therapeutic and research use.
References
Aldrup-MacDonald ME, Kuo ME, Sullivan LL, Chew K, Sullivan BA (2016) Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Research 26:1301–1311
Allshire RC, Ekwall K (2015a) Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb Perspect Biol 7:a018770
Alonso A, Fritz B, Hasson D, Abrusan G, Cheung F, Yoda K, Radlwimmer B, Ladurner AG, Warburton PE (2007) Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol 8:R148
Alonso A, Hasson D, Cheung F, Warburton PE (2010) A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin 3:6
Alonso A, Mahmood R, Li S, Cheung F, Yoda K, Warburton PE (2003) Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Human Mol Genet 12:2711–2721
Ambartsumyan G, Gill RK, Perez SD, Conway D, Vincent J, Dalal Y, Clark AT (2010) Centromere protein A dynamics in human pluripotent stem cell self-renewal, differentiation and DNA damage. Hum Mol Genet 19:3970–3982
Amor DJ, Bentley K, Ryan J, Perry J, Wong L, Slater H, Choo KH (2004) Human centromere repositioning “in progress”. Proc Natl Acad Sci U S A 101:6542–6547
Amor DJ, Choo KH (2002) Neocentromeres: role in human disease, evolution, and centromere study. Am J Hum Genet 71:695–714
Athwal RK, Walkiewicz MP, Baek S, Fu S, Bui M, Camps J, Ried T, Sung MH, Dalal Y (2015) CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenetics Chromatin 8:2
Baker RE, Rogers K (2005) Genetic and genomic analysis of the AT-rich centromere DNA element II of Saccharomyces cerevisiae. Genetics 171:1463–1475
Baldini A, Ried T, Shridhar V, Ogura K, D’Aiuto L, Rocchi M, Ward DC (1993) An alphoid DNA sequence conserved in all human and great ape chromosomes: evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum Genet 90:577–583
Barrey EJ, Heun P (2017) Artificial chromosomes and strategies to initiate epigenetic centromere establishment. Prog Mol Subcell Biol 56:193–212
Basu J, Compitello G, Stromberg G, Willard HF, Van Bokkelen G (2005) Efficient assembly of de novo human artificial chromosomes from large genomic loci. BMC Biotechnol 5:21
Basu J, Stromberg G, Compitello G, Willard HF, Van Bokkelen G (2005) Rapid creation of BAC-based human artificial chromosome vectors by transposition with synthetic alpha-satellite arrays. Nucleic Acids Res 33:587–596
Beh TT, MacKinnon RN, Kalitsis P (2016) Active centromere and chromosome identification in fixed cell lines. Mol Cytogenet 9:28
Bergmann JH, Jakubsche JN, Martins NM, Kagansky A, Nakano M, Kimura H, Kelly DA, Turner BM, Masumoto H, Larionov V, Earnshaw WC (2012) Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J Cell Sci 125:411–421
Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC (2011) Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 30:328–340
Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294:2539–2542
Birchler JA, Presting GG (2012) Retrotransposon insertion targeting: a mechanism for homogenization of centromere sequences on nonhomologous chromosomes. Genes Dev 26:638–640
Birchler JA (2015) Engineered minichromosomes in plants. Chromosome Res 23:77–85
Black BE, Cleveland DW (2011) Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 144:471–479
Black BE, Jansen LE, Foltz DR, Cleveland DW (2010) Centromere identity, function, and epigenetic propagation across cell divisions. Cold Spring Harb Symp Quant Biol 75:403–418
Blom E, Heyning FH, Kroes WG (2010) A case of angioimmunoblastic T-cell non-Hodgkin lymphoma with a neocentric inv dup(1). Cancer Genet Cytogenet 202:38–42
Blower MD (2016) Centromeric transcription regulates Aurora-B localization and activation. Cell Rep 15:1624–1633
Bobkov GOM, Gilbert N, Heun P (2018) Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J Cell Biol 217:1957–1972
Bouzinba-Segard H, Guais A, Francastel C (2006) Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci U S A 103:8709–8714
Brown KE, Barnett MA, Burgtorf C, Shaw P, Buckle VJ, Brown WR (1994) Dissecting the centromere of the human Y chromosome with cloned telomeric DNA. Human Molecular Genetics 3:1227–1237
Brown DM, Chan YA, Desai PJ, Grzesik P, Oldfield LM, Vashee S, Way JC, Silver PA, Glass JI (2017) Efficient size-independent chromosome delivery from yeast to cultured cell lines. Nucleic Acids Res 45(7):e50
Buckler ES, Phelps-Durr TL, Buckler CS, Dawe RK, Doebley JF, Holtsford TP (1999) Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153:415–426
Bulazel KV, Ferreri GC, Eldridge MD, O’Neill RJ (2007) Species-specific shifts in centromere sequence composition are coincident with breakpoint reuse in karyotypically divergent lineages. Genome Biol 8:R170
Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL (2007) Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Molecular Cell 26:853–865
Canzonetta C, Vernarecci S, Iuliani M, Marracino C, Belloni C, Ballario P, Filetici P (2015) SAGA DUB-Ubp8 deubiquitylates centromeric histone variant Cse4. G3 (Bethesda) 6:287–298
Capozzi O, Purgato S, Verdun di Cantogno L, Grosso E, Ciccone R, Zuffardi O, Della Valle G, Rocchi M (2008) Evolutionary and clinical neocentromeres: two faces of the same coin? Chromosoma 117:339–344
Carbone L, Nergadze SG, Magnani E, Misceo D, Francesca Cardone M, Roberto R, Bertoni L, Attolini C, Francesca Piras M, de Jong P, Raudsepp T, Chowdhary BP, Guerin G, Archidiacono N, Rocchi M, Giulotto E (2006) Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics 87:777–782
Cardinale S, Bergmann JH, Kelly D, Nakano M, Valdivia MM, Kimura H, Masumoto H, Larionov V, Earnshaw WC (2009) Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell 20:4194–4204
Catania S, Pidoux AL, Allshire RC (2015) Sequence features and transcriptional stalling within centromere DNA promote establishment of CENP-A chromatin. PLoS Genet 11:e1004986
Chan DYL, Moralli D, Khoja S, Monaco ZL (2017) Noncoding centromeric RNA expression impairs chromosome stability in human and murine stem cells. Dis Markers 2017:7506976
Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG (2014) CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol 204:313–329
Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M (1989) Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell 57:739–751
Chmatal L, Gabriel SI, Mitsainas GP, Martinez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA (2014) Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol 24:2295–2300
Choi ES, Stralfors A, Catania S, Castillo AG, Svensson JP, Pidoux AL, Ekwall K, Allshire RC (2012) Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A(Cnp1) in fission yeast. PLoS Genetics 8:e1002985
Collins KA, Furuyama S, Biggins S (2004) Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Current Biology: CB 14:1968–1972
Contreras-Galindo R, Kaplan MH, He S, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Kappes F, Dube D, Chan SM, Robinson D, Meng F, Dai M, Gitlin SD, Chinnaiyan AM, Omenn GS, Markovitz DM (2013) HIV infection reveals widespread expansion of novel centromeric human endogenous retroviruses. Genome Research 23:1505–1513
Copenhaver GP, Ketel C, Wang HSW, McClellan M, Bouchonville K, Selmecki A, Lahav T, Gerami-Nejad M, Berman J (2009) Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genetics 5:e1000400
Cottarel G, Shero JH, Hieter P, Hegemann JH (1989) A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol Cell Biol 9:3342–3349
Dawe RK, Lowry EG, Gent JI, Stitzer MC, Swentowsky KW, Higgins DM, Ross-Ibarra J, Wallace JG, Kanizay LB, Alabady M, Qiu W, Tseng KF, Wang N, Gao Z, Birchler JA, Harkess AE, Hodges AL, Hiatt EN (2018) A kinesin-14 motor activates neocentromeres to promote meiotic drive in maize. Cell 173:839–850 e818
De Lorenzi L, Iannuzzi A, Rossi E, Bonacina S, Parma P (2017) Centromere repositioning in cattle (Bos taurus) chromosome 17. Cytogenetic and Genome Res 151(4):191–197
de Wolf B, Kops G (2017) Kinetochore malfunction in human pathologies. Adv Exp Med Biol 1002:69–91
Deyter GM, Biggins S (2014) The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes & Development 28:1815–1826
Drinnenberg IA, de Young D, Henikoff S, Malik HS (2014) Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. Elife 3
Du Y, Topp CN, Dawe RK (2010) DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet 6:e1000835
Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G (2009) HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137:485–497
Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW (2015) DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev Cell 33:314–327
Farr C, Fantes J, Goodfellow P, Cooke H (1991) Functional reintroduction of human telomeres into mammalian cells. Proceedings of the National Academy of Sciences of the United States of America 88:7006–7010
Farr CJ, Stevanovic M, Thomson EJ, Goodfellow PN, Cooke HJ (1992) Telomere-associated chromosome fragmentation: applications in genome manipulation and analysis. Nature Genetics 2:275–282
Ferreri GC, Liscinsky DM, Mack JA, Eldridge MD, O’Neill RJ (2005) Retention of latent centromeres in the mammalian genome. J Hered 96:217–224
Ferreri GC, Marzelli M, Rens W, O’Neill RJ (2004) A centromere-specific retroviral element associated with breaks of synteny in macropodine marsupials. Cytogenetic and Genome Research 107:115–118
Ferri F, Bouzinba-Segard H, Velasco G, Hube F, Francastel C (2009) Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res 37:5071–5080
Folco HD, Pidoux AL, Urano T, Allshire RC (2008) Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319:94–97
Foltz DR, Jansen LE, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, Black BE, Cleveland DW (2009) Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137:472–484
Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol 6:784–791
Gaeta RT, Masonbrink RE, Krishnaswamy L, Zhao C, Birchler JA (2012) Synthetic chromosome platforms in plants. Annu Rev Plant Biol 63:307–330
Gambogi CW, Black BE (2019) The nucleosomes that mark centromere location on chromosomes old and new. Essays Biochem 63:15–27
Garsed DW, Marshall OJ, Corbin VD, Hsu A, Di Stefano L, Schroder J, Li J, Feng ZP, Kim BW, Kowarsky M, Lansdell B, Brookwell R, Myklebost O, Meza-Zepeda L, Holloway AJ, Pedeutour F, Choo KH, Damore MA, Deans AJ, Papenfuss AT, Thomas DM (2014) The architecture and evolution of cancer neochromosomes. Cancer Cell 26:653–667
Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM (2011) Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145:410–422
Gassmann R, Rechtsteiner A, Yuen KW, Muroyama A, Egelhofer T, Gaydos L, Barron F, Maddox P, Essex A, Monen J, Ercan S, Lieb JD, Oegema K, Strome S, Desai A (2012) An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature 484:534–537
Giordano-Santini R, Milstein S, Svrzikapa N, Tu D, Johnsen R, Baillie D, Vidal M, Dupuy D (2010) An antibiotic selection marker for nematode transgenesis. Nature Methods 7:721–723
Gonzalez M, He H, Dong Q, Sun S, Li F (2014) Ectopic centromere nucleation by CENP—a in fission yeast. Genetics 198:1433–1446
Goodspeed A, Heiser LM, Gray JW, Costello JC (2016) Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol Cancer Res 14:3–13
Grimes BR, Rhoades AA, Willard HF (2002) Alpha-satellite DNA and vector composition influence rates of human artificial chromosome formation. Mol Ther 5:798–805
Guo X, Su H, Shi Q, Fu S, Wang J, Zhang X, Hu Z, Han F (2016) De novo centromere formation and centromeric sequence expansion in wheat and its wide hybrids. PLoS Genetics 12:e1005997
Hahnenberger KM, Baum MP, Polizzi CM, Carbon J, Clarke L (1989) Construction of functional artificial minichromosomes in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A 86:577–581
Han Y, Zhang Z, Liu C, Liu J, Huang S, Jiang J, Jin W (2009) Centromere repositioning in cucurbit species: implication of the genomic impact from centromere activation and inactivation. Proc Natl Acad Sci U S A 106:14937–14941
Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF (1997) Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15:345–355
Hasson D, Alonso A, Cheung F, Tepperberg JH, Papenhausen PR, Engelen JJ, Warburton PE (2011) Formation of novel CENP-A domains on tandem repetitive DNA and across chromosome breakpoints on human chromosome 8q21 neocentromeres. Chromosoma 120(6):621–632
Hayden KE, Strome ED, Merrett SL, Lee HR, Rudd MK, Willard HF (2013) Sequences associated with centromere competency in the human genome. Molecular and Cellular Biology 33:763–772
Hedouin S, Grillo G, Ivkovic I, Velasco G, Francastel C (2017) CENP-A chromatin disassembly in stressed and senescent murine cells. Sci Rep 7:42520
Helfricht A, Wiegant W, Thijssen P, Vertegaal A, Luijsterburg M, Van Attikum H (2013) Remodeling and spacing factor 1 (RSF1) deposits centromere proteins at DNA double-strand breaks to promote non-homologous end-joining. Cell Cycle 12:3070–3082
Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098–1102
Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH (2006) Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell 10:303–315
Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL (2010) Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Molecular Cell 40:444–454
Hiatt EN, Kentner EK, Dawe RK (2002) Independently regulated neocentromere activity of two classes of tandem repeat arrays. Plant Cell 14:407–420
Hill A, Bloom K (1987) Genetic manipulation of centromere function. Mol Cell Biol 7:2397–2405
Hiratsuka M, Ueda K, Uno N, Uno K, Fukuhara S, Kurosaki H, Takehara S, Osaki M, Kazuki Y, Kurosawa Y, Nakamura T, Katoh M, Oshimura M (2015) Retargeting of microcell fusion towards recipient cell-oriented transfer of human artificial chromosome. BMC Biotechnol 15:58
Ho KH, Tsuchiya D, Oliger AC, Lacefield S (2014) Localization and function of budding yeast CENP-A depends upon kinetochore protein interactions and is independent of canonical centromere sequence. Cell Rep 9:2027–2033
Hori T, Shang WH, Takeuchi K, Fukagawa T (2013) The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. The Journal of Cell Biology 200:45–60
Houben A, Dawe RK, Jiang J, Schubert I (2008) Engineered plant minichromosomes: a bottom-up success? The Plant Cell 20:8–10
Ideue T, Cho Y, Nishimura K, Tani T (2014) Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes Cells 19:528–538
Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K (2008) Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321:1088–1091
Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmatal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, Black BE (2017) Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr Biol 27:2365–2373 e2368
Jaco I, Canela A, Vera E, Blasco MA (2008) Centromere mitotic recombination in mammalian cells. Journal of Cell Biology 181:885–892
Jain M, Olsen HE, Turner DJ, Stoddart D, Bulazel KV, Paten B, Haussler D, Willard HF, Akeson M, Miga KH (2018) Linear assembly of a human centromere on the Y chromosome. Nat Biotechnol 36:321–323
Jansen LE, Black BE, Foltz DR, Cleveland DW (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176:795–805
Jiang J, Birchler JA, Parrott WA, Dawe RK (2003) A molecular view of plant centromeres. Trends Plant Sci 8:570–575
Johnson WL, Yewdell WT, Bell JC, McNulty SM, Duda Z, O’Neill RJ, Sullivan BA, Straight AF (2017) RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. eLife 6
Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc’h C (2004) Stress-induced transcription of satellite III repeats. J Cell Biol 164:25–33
Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC (2009) Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 324:1716–1719
Kalitsis P, Choo KH (2012) The evolutionary life cycle of the resilient centromere. Chromosoma 121:327–340
Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19:489–501
Kanizay LB, Albert PS, Birchler JA, Dawe RK (2013) Intragenomic conflict between the two major knob repeats of maize. Genetics 194:81–89
Kapoor M, Montes de Oca Luna R, Liu G, Lozano G, Cummings C, Mancini M, Ouspenski I, Brinkley BR, May GS (1998) The cenpB gene is not essential in mice. Chromosoma 107:570–576
Kapusi E, Ma L, Teo CH, Hensel G, Himmelbach A, Schubert I, Mette MF, Kumlehn J, Houben A (2012) Telomere-mediated truncation of barley chromosomes. Chromosoma 121:181–190
Kasinathan S, Henikoff S (2018) Non-B-form DNA is enriched at centromeres. Molecular Biology and Evolution 35:949–962
Katoh M, Ayabe F, Norikane S, Okada T, Masumoto H, Horike S, Shirayoshi Y, Oshimura M (2004) Construction of a novel human artificial chromosome vector for gene delivery. Biochem Biophys Res Commun 321:280–290
Kattermann G (1939) Ein neuer Karyotyp bei Roggen. Zeitschrift für Zellforschung und Mikroskopische Anatomie Abt B Chromosoma 1:284–299
Kazuki Y, Oshimura M (2011) Human artificial chromosomes for gene delivery and the development of animal models. Mol Ther 19:1591–1601
Ketel C, Wang HS, McClellan M, Bouchonville K, Selmecki A, Lahav T, Gerami-Nejad M, Berman J (2009) Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet 5:e1000400
Kim JH, Kononenko A, Erliandri I, Kim TA, Nakano M, Iida Y, Barrett JC, Oshimura M, Masumoto H, Earnshaw WC, Larionov V, Kouprina N (2011) Human artificial chromosome (HAC) vector with a conditional centromere for correction of genetic deficiencies in human cells. Proc Natl Acad Sci U S A 108:20048–20053
Klein E, Rocchi M, Ovens-Raeder A, Kosyakova N, Weise A, Ziegler M, Meins M, Morlot S, Fischer W, Volleth M, Polityko A, Ogilvie CM, Kraus C, Liehr T (2012) Five novel locations of neocentromeres in human: 18q22.1, Xq27.1 approximately 27.2, Acro p13, Acro p12, and heterochromatin of unknown origin. Cytogenet Genome Res 136:163–166
Kobayashi T, Yamada F, Hashimoto T, Abe S, Matsuda Y, Kuroiwa A (2008) Centromere repositioning in the X chromosome of XO/XO mammals, Ryukyu spiny rat. Chromosome Res 16:587–593
Koo DH, Han F, Birchler JA, Jiang J (2011) Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome. Genome Res 21:908–914
Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, Cantaloube S, Kurumizaka H, Imhof A, Almouzni G (2014) Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol Cell 53:631–644
Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA (2006) Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci U S A 103:4186–4191
Lampson MA, Black BE (2017) Cellular and molecular mechanisms of centromere drive. Cold Spring Harb Symp Quant Biol 82:249–257
Lawrence KS, Chau T, Engebrecht J (2015) DNA damage response and spindle assembly checkpoint function throughout the cell cycle to ensure genomic integrity. PLoS Genet 11:e1005150
Lee BC, Lin Z, Yuen KW (2016) RbAp46/48(LIN-53) Is required for holocentromere assembly in Caenorhabditis elegans. Cell Rep 14:1819–1828
Lefrancois P, Auerbach RK, Yellman CM, Roeder GS, Snyder M (2013) Centromere-like regions in the budding yeast genome. PLoS Genet 9:e1003209
Ling YH, Yuen KWY (2019a) Centromeric non-coding RNA as a hidden epigenetic factor of the point centromere. Curr Genet 65(5):1165–1171
Ling YH, Yuen KWY (2019b) Point centromere activity requires an optimal level of centromeric noncoding RNA. Proc Natl Acad Sci U S A 116:6270–6279
Liskovykh M, Lee NC, Larionov V, Kouprina N (2016) Moving toward a higher efficiency of microcell-mediated chromosome transfer. Mol Ther Methods Clin Dev 3:16043
Lo AW, Craig JM, Saffery R, Kalitsis P, Irvine DV, Earle E, Magliano DJ, Choo KH (2001) A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J 20:2087–2096
Lo AW, Magliano DJ, Sibson MC, Kalitsis P, Craig JM, Choo KH (2001) A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res 11:448–457
Logsdon GA, Barrey EJ, Bassett EA, DeNizio JE, Guo LY, Panchenko T, Dawicki-McKenna JM, Heun P, Black BE (2015) Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. The Journal of Cell Biology 208:521–531
Logsdon GA, Gambogi CW, Liskovykh MA, Barrey EJ, Larionov V, Miga KH, Heun P, Black BE (2019) Human artificial chromosomes that bypass centromeric DNA. Cell 178:624–639 e619
Lorincz MC, Dickerson DR, Schmitt M, Groudine M (2004) Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol 11:1068–1075
Lu J, Gilbert DM (2007) Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J Cell Biol 179:411–421
Lyttle TW (1991) Segregation distorters. Annu Rev Genet 25:511–557
Macchia G, Nord KH, Zoli M, Purgato S, D’Addabbo P, Whelan CW, Carbone L, Perini G, Mertens F, Rocchi M, Storlazzi CT (2015) Ring chromosomes, breakpoint clusters, and neocentromeres in sarcomas. Genes Chromosomes Cancer 54:156–167
Macnab S, Whitehouse A (2009) Progress and prospects: human artificial chromosomes. Gene Ther 16:1180–1188
Maggert KA, Karpen GH (2001) The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics 158:1615–1628
Mahajan S, Wei KH, Nalley MJ, Gibilisco L, Bachtrog D (2018) De novo assembly of a young Drosophila Y chromosome using single-molecule sequencing and chromatin conformation capture. PLoS Biology 16:e2006348
Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G (2002) Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nature Genetics 30:329–334
Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, Almouzni G (2011) SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat Genet 43:220–227
Malik HS, Henikoff S (2009) Major evolutionary transitions in centromere complexity. Cell 138:1067–1082
Maloney KA, Sullivan LL, Matheny JE, Strome ED, Merrett SL, Ferris A, Sullivan BA (2012) Functional epialleles at an endogenous human centromere. Proceedings of the National Academy of Sciences of the United States of America 109:13704–13709
Manzanero S, Puertas MJ (2003) Rye terminal neocentromeres: characterisation of the underlying DNA and chromatin structure. Chromosoma 111:408–415
Marshall OJ, Chueh AC, Wong LH, Choo KH (2008) Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82:261–282
McClintock B (1939) The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci U S A 25:405–416
McClintock B (1941) The stability of broken ends of chromosomes in Zea mays. Genetics 26:234–282
McKinley KL, Cheeseman IM (2016) The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol 17:16–29
McNulty SM, Sullivan LL, Sullivan BA (2017) Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Developmental Cell 42:226–+
Mello CC, Kramer JM, Stinchcomb D, Ambros V (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10:3959–3970
Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH (2011) Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genetics 7:e1002068
Melters DP, Bradnam KR, Young HA, Telis N, May MR, Ruby JG, Sebra R, Peluso P, Eid J, Rank D, Garcia JF, Derisi JL, Smith T, Tobias C, Ross-Ibarra J, Korf I, Chan SW (2013) Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol 14:R10
Meluh PB, Koshland D (1997) Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev 11:3401–3412
Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P (2011) Drosophila CENH3 is sufficient for centromere formation. Science 334:686–690
Meraldi P, McAinsh AD, Rheinbay E, Sorger PK (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol 7:R23
Mette MF, Houben A (2015) Engineering of plant chromosomes. Chromosome Res 23:69–76
Molina O, Vargiu G, Abad MA, Zhiteneva A, Jeyaprakash AA, Masumoto H, Kouprina N, Larionov V, Earnshaw WC (2016) Epigenetic engineering reveals a balance between histone modifications and transcription in kinetochore maintenance. Nat Commun 7:13334
Montefalcone G, Tempesta S, Rocchi M, Archidiacono N (1999) Centromere repositioning. Genome Res 9:1184–1188
Moreno-Moreno O, Torras-Llort M, Azorin F (2006) Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Research 34:6247–6255
Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ (2005) Characterization of Dicer-deficient murine embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America 102:12135–12140
Murray AW, Schultes NP, Szostak JW (1986) Chromosome length controls mitotic chromosome segregation in yeast. Cell 45:529–536
Murray AW, Szostak JW (1983) Construction of artificial chromosomes in yeast. Nature 305:189–193
Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J (2004) Sequencing of a rice centromere uncovers active genes. Nature Genetics 36:138–145
Nagaki K, Neumann P, Zhang D, Ouyang S, Buell CR, Cheng Z, Jiang J (2005) Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol Biol Evol 22: 845–855
Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H (2008) Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell 14:507–522
Nakano M, Okamoto Y, Ohzeki J, Masumoto H (2003) Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes. J Cell Sci 116:4021–4034
Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M (1986) Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J 5:1011–1021
Nakashima H, Nakano M, Ohnishi R, Hiraoka Y, Kaneda Y, Sugino A, Masumoto H (2005) Assembly of additional heterochromatin distinct from centromere-kinetochore chromatin is required for de novo formation of human artificial chromosome. Journal of Cell Science 118:5885–5898
Nechemia-Arbely Y, Miga KH, Shoshani O, Aslanian A, McMahon MA, Lee AY, Fachinetti D, Yates JR 3rd, Ren B, Cleveland DW (2019) DNA replication acts as an error correction mechanism to maintain centromere identity by restricting CENP-A to centromeres. Nature Cell Biology 21:743–754
Nelson AD, Lamb JC, Kobrossly PS, Shippen DE (2011) Parameters affecting telomere-mediated chromosomal truncation in Arabidopsis. The Plant Cell 23:2263–2272
Nergadze SG, Piras FM, Gamba R, Corbo M, Cerutti F, McCarter JGW, Cappelletti E, Gozzo F, Harman RM, Antczak DF, Miller D, Scharfe M, Pavesi G, Raimondi E, Sullivan KF, Giulotto E (2018) Birth, evolution, and transmission of satellite-free mammalian centromeric domains. Genome Res 28:789–799
Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y (2002) Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4:89–93
Nye J, Sturgill D, Athwal R, Dalal Y (2018) HJURP antagonizes CENP-A mislocalization driven by the H3.3 chaperones HIRA and DAXX. PLoS One 13:e0205948
Ohkuni K, Kitagawa K (2012) Role of transcription at centromeres in budding yeast. Transcription 3:193–197
Ohkuni K, Takahashi Y, Fulp A, Lawrimore J, Au WC, Pasupala N, Levy-Myers R, Warren J, Strunnikov A, Baker RE, Kerscher O, Bloom K, Basrai MA (2016) SUMO-targeted ubiquitin ligase (STUbL) Slx5 regulates proteolysis of centromeric histone H3 variant Cse4 and prevents its mislocalization to euchromatin. Mol Biol Cell
Ohzeki J, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H (2012) Breaking the HAC barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J 31:2391–2402
Ohzeki J, Larionov V, Earnshaw WC, Masumoto H (2015) Genetic and epigenetic regulation of centromeres: a look at HAC formation. Chromosome Research: an International Journal on the Molecular, Supramolecular and Evolutionary Aspects of Chromosome Biology 23:87–103
Ohzeki J, Nakano M, Okada T, Masumoto H (2002) CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159:765–775
Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H (2007) CENP-B controls centromere formation depending on the chromatin context. Cell 131:1287–1300
Okamoto Y, Nakano M, Ohzeki J, Larionov V, Masumoto H (2007) A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. EMBO J 26:1279–1291
Olszak AM, van Essen D, Pereira AJ, Diehl S, Manke T, Maiato H, Saccani S, Heun P (2011) Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol 13:799–808
Papenfuss AT, Thomas DM (2015) The life history of neochromosomes revealed. Mol Cell Oncol 2:e1000698
Partridge JF, Borgstrom B, Allshire RC (2000) Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev 14:783–791
Peacock WJ, Dennis ES, Rhoades MM, Pryor AJ (1981) Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc Natl Acad Sci U S A 78:4490–4494
Piras FM, Nergadze SG, Poletto V, Cerutti F, Ryder OA, Leeb T, Raimondi E, Giulotto E (2009) Phylogeny of horse chromosome 5q in the genus Equus and centromere repositioning. Cytogenet Genome Res 126:165–172
Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S (2010) An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell 40:455–464
Ren X, Katoh M, Hoshiya H, Kurimasa A, Inoue T, Ayabe F, Shibata K, Toguchida J, Oshimura M (2005) A novel human artificial chromosome vector provides effective cell lineage-specific transgene expression in human mesenchymal stem cells. Stem Cells 23:1608–1616
Rhoades MM (1942) Preferential segregation in maize. Genetics 27:395–407
Rocchi M, Archidiacono N (2006) Genome plasticity in evolution. In: Lupski JR, Stankiewicz P (eds) Genomic disorders: the genomic basis of disease. Humana, Totowa, pp 153–165
Rocchi M, Archidiacono N, Schempp W, Capozzi O, Stanyon R (2012) Centromere repositioning in mammals. Heredity (Edinb) 108:59–67
Roure V, Medina-Pritchard B, Anselm E, Jeyaprakash AA, Heun P (2019) Epigenetic inheritance of centromere identity in a heterologous system. biorxiv
Rudert F, Bronner S, Garnier JM, Dolle P (1995) Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm Genome 6:76–83
Saffery R, Sumer H, Hassan S, Wong LH, Craig JM, Todokoro K, Anderson M, Stafford A, Choo KH (2003) Transcription within a functional human centromere. Mol Cell 12:509–516
Saksouk N, Simboeck E, Dejardin J (2015) Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 8:3
Sandler L, Novitski E (1957) Meiotic drive as an evolutionary force. The American Naturalist 91:105–110
Schaefer H, Heibl C, Renner SS (2009) Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc Biol Sci 276:843–851
Schneider KL, Xie Z, Wolfgruber TK, Presting GG (2016) Inbreeding drives maize centromere evolution. Proceedings of the National Academy of Sciences of the United States of America 113:E987–E996
Schubert I (2018) What is behind “centromere repositioning”? Chromosoma 127:229–234
Semple JI, Garcia-Verdugo R, Lehner B (2010) Rapid selection of transgenic C. elegans using antibiotic resistance. Nature Methods 7:725–727
Shang WH, H, Ori T, Martins NM, Toyoda A, Misu S, Monma N, Hiratani I, Maeshima K, Ikeo K, Fujiyama A, Kimura H, Earnshaw WC, Fukagawa T (2013) Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev Cell 24:635–648
Shang WH, Hori T, Martins NM, Toyoda A, Misu S, Monma N, Hiratani I, Maeshima K, Ikeo K, Fujiyama A, Kimura H, Earnshaw WC, Fukagawa T (2013) Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev Cell 24:635–648
Sharma S (2011) Non-B DNA secondary structures and their resolution by RecQ helicases. Journal of Nucleic Acids 2011:Article ID 724215 15 pages
Sharma AB, Dimitrov S, Hamiche A, Van Dyck E (2019) Centromeric and ectopic assembly of CENP-A chromatin in health and cancer: old marks and new tracks. Nucleic Acids Research 47:1051–1069
Sharma A, Wolfgruber TK, Presting GG (2013) Tandem repeats derived from centromeric retrotransposons. BMC Genomics 14:142
Shelby RD, Monier K, Sullivan KF (2000) Chromatin assembly at kinetochores is uncoupled from DNA replication. The Journal of Cell Biology 151:1113–1118
Shrestha RL, Ahn GS, Staples MI, Sathyan KM, Karpova TS, Foltz DR, Basrai MA (2017) Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget 8:46781–46800
Shukla M, Tong P, White SA, Singh PP, Reid AM, Catania S, Pidoux AL, Allshire RC (2018) Centromere DNA destabilizes H3 nucleosomes to promote CENP-A deposition during the cell cycle. Current Biology: CB 28:3924–3936 e3924
Sirvent N, Forus A, Lescaut W, Burel F, Benzaken S, Chazal M, Bourgeon A, Vermeesch JR, Myklebost O, Turc-Carel C, Ayraud N, Coindre JM, Pedeutour F (2000) Characterization of centromere alterations in liposarcomas. Genes Chromosomes Cancer 29:117–129
Steiner FA, Henikoff S (2014) Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. Elife 3:e02025
Stimpson KM, Sullivan BA (2011) Histone H3K4 methylation keeps centromeres open for business. EMBO J 30:233–234
Stinchcomb DT, Shaw JE, Carr SH, Hirsh D (1985) Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol Cell Biol 5:3484–3496
Stirzaker C, Song JZ, Davidson B, Clark SJ (2004) Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res 64:3871–3877
Sullivan LL, Maloney KA, Towers AJ, Gregory SG, Sullivan BA (2016) Human centromere repositioning within euchromatin after partial chromosome deletion. Chromosome Research: an International Journal on the Molecular, Supramolecular and Evolutionary Aspects of Chromosome Biology 24:451–466
Suzuki T, Kazuki Y, Oshimura M, Hara T (2016) Highly efficient transfer of chromosomes to a broad range of target cells using Chinese hamster ovary cells expressing murine leukemia virus-derived envelope proteins. PLoS One 11:e0157187
Teo CH, Ma L, Kapusi E, Hensel G, Kumlehn J, Schubert I, Houben A, Mette MF (2011) Induction of telomere-mediated chromosomal truncation and stability of truncated chromosomes in Arabidopsis thaliana. Plant J 68:28–39
Thakur J, Sanyal K (2013) Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function at native physical chromosomal loci in Candida albicans. Genome Res 23:638–652
Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F (2003) Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res 63:3511–3516
Tong P, Pidoux AL, Toda NRT, Ard R, Berger H, Shukla M, Torres-Garcia J, Muller CA, Nieduszynski CA, Allshire RC (2019) Interspecies conservation of organisation and function between nonhomologous regional centromeres. Nat Commun 10:2343
Tsukahara S, Kawabe A, Kobayashi A, Ito T, Aizu T, Shin-i T, Toyoda A, Fujiyama A, Tarutani Y, Kakutani T (2012) Centromere-targeted de novo integrations of an LTR retrotransposon of Arabidopsis lyrata. Genes Dev 26:705–713
Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C, Riva S, Biamonti G (2008) Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res 36:423–434
Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR (2001) Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci 114:3529–3542
Ventura M, Antonacci F, Cardone MF, Stanyon R, D’Addabbo P, Cellamare A, Sprague LJ, Eichler EE, Archidiacono N, Rocchi M (2007) Evolutionary formation of new centromeres in macaque. Science 316:243–246
Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672–676
Villasante A, Abad JP, Mendez-Lago M (2007) Centromeres were derived from telomeres during the evolution of the eukaryotic chromosome. Proceedings of the National Academy of Sciences of the United States of America 104:10542–10547
Villasante A, Mendez-Lago M, Abad JP, Montejo de Garcini E (2007) The birth of the centromere. Cell Cycle 6:2872–2876
Voullaire LE, Slater HR, Petrovic V, Choo KH (1993) A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet 52:1153–1163
Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, Blocker H, Distl O, Edgar RC, Garber M, Leeb T, Mauceli E, MacLeod JN, Penedo MC, Raison JM, Sharpe T, Vogel J, Andersson L, Antczak DF, Biagi T, Binns MM, Chowdhary BP, Coleman SJ, Della Valle G, Fryc S, Guerin G, Hasegawa T, Hill EW, Jurka J, Kiialainen A, Lindgren G, Liu J, Magnani E, Mickelson JR, Murray J, Nergadze SG, Onofrio R, Pedroni S, Piras MF, Raudsepp T, Rocchi M, Roed KH, Ryder OA, Searle S, Skow L, Swinburne JE, Syvanen AC, Tozaki T, Valberg SJ, Vaudin M, White JR, Zody MC, Broad Institute Genome Sequencing P, Broad Institute Whole Genome Assembly T, Lander ES, Lindblad-Toh K (2009) Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326:865–867
Wang K, Wu Y, Zhang W, Dawe RK, Jiang J (2014) Maize centromeres expand and adopt a uniform size in the genetic background of oat. Genome Research 24:107–116
Warburton PE, Dolled M, Mahmood R, Alonso A, Li S, Naritomi K, Tohma T, Nagai T, Hasegawa T, Ohashi H, Govaerts LC, Eussen BH, Van Hemel JO, Lozzio C, Schwartz S, Dowhanick-Morrissette JJ, Spinner NB, Rivera H, Crolla JA, Yu C, Warburton D (2000) Molecular cytogenetic analysis of eight inversion duplications of human chromosome 13q that each contain a neocentromere. Am J Hum Genet 66:1794–1806
Waye JS, Willard HF (1987) Nucleotide sequence heterogeneity of alpha satellite repetitive DNA: a survey of alphoid sequences from different human chromosomes. Nucleic Acids Res 15:7549–7569
Willard HF, Waye JS (1987) Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol 25:207–214
Williams BC, Murphy TD, Goldberg ML, Karpen GH (1998) Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet 18:30–37
Wolfgruber TK, Sharma A, Schneider KL, Albert PS, Koo DH, Shi J, Gao Z, Han F, Lee H, Xu R, Allison J, Birchler JA, Jiang J, Dawe RK, Presting GG (2009) Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic Loci shaped primarily by retrotransposons. PLoS Genet 5: e1000743
Wong LH, Choo KH (2001) Centromere on the move. Genome Res 11:513–516
Wong NC, Wong LH, Quach JM, Canham P, Craig JM, Song JZ, Clark SJ, Choo KH (2006) Permissive transcriptional activity at the centromere through pockets of DNA hypomethylation. PLoS Genet 2:e17
Wu JC, Manuelidis L (1980) Sequence definition and organization of a human repeated DNA. J Mol Biol 142:363–386
Xu C, Cheng Z, Yu W (2012) Construction of rice mini-chromosomes by telomere-mediated chromosomal truncation. Plant J 70:1070–1079
Yan X, Li C, Yang J, Wang L, Jiang C, Wei W (2017) Induction of telomere-mediated chromosomal truncation and behavior of truncated chromosomes in Brassica napus. Plant J 91:700–713
Yang L, Koo DH, Li D, Zhang T, Jiang J, Luan F, Renner SS, Henaff E, Sanseverino W, Garcia-Mas J, Casacuberta J, Senalik DA, Simon PW, Chen J, Weng Y (2014) Next-generation sequencing, FISH mapping and synteny-based modeling reveal mechanisms of decreasing dysploidy in Cucumis. Plant J 77:16–30
Yang JW, Pendon C, Yang J, Haywood N, Chand A, Brown WR (2000) Human mini-chromosomes with minimal centromeres. Human Molecular Genetics 9:1891–1902
Yu W, Han F, Gao Z, Vega JM, Birchler JA (2007) Construction and behavior of engineered minichromosomes in maize. Proceedings of the National Academy of Sciences of the United States of America 104:8924–8929
Yu HG, Hiatt EN, Chan A, Sweeney M, Dawe RK (1997) Neocentromere-mediated chromosome movement in maize. J Cell Biol 139:831–840
Yu W, Lamb JC, Han F, Birchler JA (2006) Telomere-mediated chromosomal truncation in maize. Proceedings of the National Academy of Sciences of the United States of America 103:17331–17336
Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu H, Wang Z, Li S, Cui L, Shen J, Zhai L, Peng S, Wong J, Dong S, Yuan Z, Ou G, Zhang X, Xu P, Lou J, Yang N, Chen P, Xu RM, Li G (2015) Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev Cell 32:68–81
Yuan J, Shi Q, Guo X, Liu Y, Su H, Guo X, Lv Z, Han F (2017) Site-specific transfer of chromosomal segments and genes in wheat engineered chromosomes. J Genet Genomics 44:531–539
Yuen KW, Nabeshima K, Oegema K, Desai A (2011) Rapid de novo centromere formation occurs independently of heterochromatin protein 1 in C. elegans embryos. Curr Biol 21:1800–1807
Zahn J, Kaplan MH, Fischer S, Dai M, Meng F, Saha AK, Cervantes P, Chan SM, Dube D, Omenn GS, Markovitz DM, Contreras-Galindo R (2015) Expansion of a novel endogenous retrovirus throughout the pericentromeres of modern humans. Genome Biology 16:74
Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JY, Berns MW, Cleveland DW (2009) Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci U S A 106:15762–15767
Zeng K, de las Heras JI, Ross A, Yang J, Cooke H, Shen MH (2004) Localisation of centromeric proteins to a fraction of mouse minor satellite DNA on a mini-chromosome in human, mouse and chicken cells. Chromosoma 113:84–91
Zhang W, Mao JH, Zhu W, Jain AK, Liu K, Brown JB, Karpen GH (2016) Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat Commun 7:12619
Zhang K, Mosch K, Fischle W, Grewal SI (2008) Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15:381–388
Zhu J, Cheng KCL, Yuen KWY (2018) Histone H3K9 and H4 acetylations and transcription facilitate the initial CENP-A(HCP-3) deposition and de novo centromere establishment in Caenorhabditis elegans artificial chromosomes. Epigenetics Chromatin 11:16
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ling, Y.H., Lin, Z. & Yuen, K.W.Y. Genetic and epigenetic effects on centromere establishment. Chromosoma 129, 1–24 (2020). https://doi.org/10.1007/s00412-019-00727-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-019-00727-3