Abstract
Purpose
CHIT1 is expressed by pulmonary macrophages, which is typically the site of entry for many environmental fungi that may increase the risk of pulmonary fungal infection and lead to hypersensitivity. The conserved expression of this gene in humans suggests its physiological importance in the mammalian lung.
Methods
The present study was conducted with a total of 964 subjects, including 483 healthy controls and 481 asthma patients. DNA samples were extracted from blood, and the genotyping was done using polymerase chain reaction method.
Results
Statistical analysis revealed that the 24 bp duplication in CHIT1 gene polymorphism shows highly significant association in heterozygous (wild/dup) genotype with OR 1.74, 95 % CI (1.29–2.36), and p = 0.000. However, the homozygous mutant genotype (dup/dup) was found to be non-significant with OR 1.06, 95 % CI (0.69–1.63), and p = 0.786. The combination of both wild/dup and dup/dup was also found to be highly significant with OR 1.57, 95 % CI (1.18–2.11), and p = 0.002.
Conclusions
This is the first study conducted in India which reports a significant association between 24 bp duplication in CHIT1 gene polymorphism and asthma in the studied North Indian population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Asthma is a major health problem worldwide and is characterized by chronic airway inflammation after interaction with allergens, environmental irritants, or infections [1]. In India, 0.2 % of all the deaths and 0.5 % of "National Burden of Diseases" are due to asthma [2]. Also, 44 % of asthma worldwide has an environmental impact [3]. This scenario is causing a considerable economic burden on healthcare system which has, therefore, intensified the research on asthma pathogenesis.
The CHIT1 gene, also known as Chitinase 1 or Chitotriosidase 1, is a 50-kDa mammalian chitotriosidase enzyme that is detected in the serum of both healthy and diseased individuals [4]. Enzymatically active chitinases cleave chitin, which is present in diverse organisms like fungal cell wall, exoskeleton of mites and arthropods, lining of the insect gut, and the microfilarial sheath of parasitic nematodes [5, 6]. These pathogens use chitin for their protection against the animal or plant host immune machinery, and absence of this chitin may lead to the death of the pathogen. Therefore, chitinase production is very important in the life cycle of any fungi or parasite for its survival in the host [7]. Chitinase breaks down the β-1,4-glycosidic bonds of chitin to release N-acetylglucosamine dimers, which are then acted upon by N-acetylglucosaminidase [8]. CHIT1 is produced, stored, and secreted by macrophages and neutrophils and plays a pivotal role in the activation of innate immunity against chitin-containing pathogens [9]. Also, they induce Th2-dominated immune responses which are associated with allergy, including the increased production of proinflammatory cytokines, serum IgE, and prominent tissue eosinophilia. These proinflammatory cytokines such as GM-CSF and TNFα stimulate the expression of chitotriosidase whereas IFNγ and IL4 inhibit its expression [10, 11] as observed in the previous data. However, recently it was found that the level of CHIT1 expression depends upon macrophages polarization. The IFNγ or LPS activate classical (M1) polarized macrophages and IL4 activate alternative (M2) polarized macrophages that indicates an increase level of this enzyme in specialized macrophages [12].
The human CHIT1 gene is located on chromosome 1q31–32 which contains 12 exons and extends up to 20 kb. A duplication of 24 base pair in exon 10 of CHIT1 gene was found by sequencing method that caused a recessive mutation. This duplication introduced a 3′ splice site in the coding region of CHIT1 that resulted in the deletion of subsequent 87 nucleotides. Loss of these nucleotides introduces a change of amino acid residues from 344 to 372 in the polypeptide chain of chitotriosidase which leads to the inactivation of chitinolytic activity [13]. Inactivation of this enzyme leads to the survival of pathogen which may play a role in the occurrence of asthma. It was also found that individuals having heterozygous allele for the duplication had a mild enzymatic activity [13–15].
The CHIT1, secreted by the activated macrophages, is detected in the plasma, was first discovered in patients suffering from Gaucher disease. Elevated levels of the enzyme activity are detectable by many techniques and acted as a biomarker in several diseases such as Niemann-Pick disease [16], β-thalassemia [17], sarcoidosis [18], multiple sclerosis [19], coronary artery disease [20], and in parasitic infections such as Plasmodium falciparum malaria [15].
Until now, very few case–control studies have been conducted worldwide to investigate the role of CHIT1 gene polymorphism in asthma. To the best of our knowledge, we have investigated the impact of 24 bp duplication in exon 10 of CHIT1 gene with asthma in the North Indian population, for the first time.
Materials and Methods
Ethical Clearance
Ethical clearance for this study was granted by the Ethics Committee, PGIMER, Chandigarh, India, vide Approval Memo no. PG-1Trg-10 on 21.9.2010 for conducting the research work on human blood samples. After doctors’ diagnosis, each patient gave a written consent, prior to inducting him/her in the study, and only the patients who fulfilled the criteria of global initiative for asthma (GINA) guidelines were recruited in the study.
Inclusion Criteria
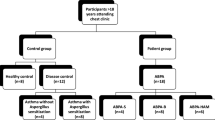
Patients from different states of North India viz. Chandigarh, Punjab, Haryana, Himachal Pradesh, Uttaranchal, Jammu and Kashmir, Rajasthan, Uttar Pradesh, and New Delhi were recruited for this study. A total of 481 patients were enrolled as cases visiting out patient department (OPD), Pulmonary Medicine at PGIMER, Chandigarh, India; while 483 age-matched healthy individuals without any symptoms of atopic, pulmonary disease, any other co-morbid disease, or smoking habits were recruited as controls.
Exclusion Criteria
Asthma patients with history of any other respiratory illness like COPD, tuberculosis, pneumonia, and bronchitis were excluded from this study. Apart from these, any other co-morbid illness such as diabetes mellitus, hypertension, or pregnant females were also not recruited as cases in this study.
Lung Function Test
Spirometry test was performed strictly in accordance with the Association of Respiratory Technician and Physiologists (ARTP) guidelines [21] for generating pneumotachographs which are helpful in assessing conditions such as asthma, COPD etc., using the device Spiro 233 (PK Morgan, Rainham, Kent, UK).
Out of the 481 asthma patients, spirometry was done for 377 asthmatics, which were further categorized according to their severity. The frequency of mild obstruction and heterozygous genotypes was found to be higher in the studied population.
Total IgE Measurement
Total IgE was measured using ImmunoCAPs with the device Phadia 100 IDM version 5.43 (Thermo Fisher Scientific Inc., USA) in serum samples of both control and asthma patients to screen allergy.
Skin Prick Test (SPT) and serum specific IgE against Aspergillus fumigatus were also done in some patients to distinguish asthma and ABPA (Allergic bronchopulmonary aspergillosis). Only negative SPT patients with specific IgE < 0.35 KUA/L were recruited in the study (Table 1).
Blood Collection and Processing for Genomic DNA
Approximately 5-ml blood was collected from each patient as well as control subjects in EDTA coated vials and stored at −80 °C until genomic DNA extraction was done. Genomic DNA was isolated from frozen whole blood samples using SSC Buffer method [22] and checked on 0.8 % agarose gel by electrophoresis before storage at −20 °C for further use.
Genotype Detection
The 24 bp duplication in CHIT1 gene was detected by polymerase chain reaction with the specific primers aligned around the duplication region [23]: CHIT1 F- 5′AGCTATCTGAAGCAGAAG-3′ and CHIT1 R- 5′GGAGAAGCCGGCAAAGTC-3′.
The PCR was carried out in a thermal cycler, in a total volume of 25 µl containing 10X PCR buffer, 3 mM MgCl2, 1 mg/ml BSA, 50 pmol of each primer, 10 mM of each dNTP, 0.125 U Taq polymerase, and 2 µl genomic DNA with the conditions initial denaturation at 94 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s with a final 72 °C extension for 5 min. The results were observed directly by electrophoresis on 2 % agarose gels stained with ethidium bromide and visualized by UV transillumination. Fragments of 75 and 99 bp were obtained, which indicated the wild and mutant alleles of Chit genes, respectively (Fig. 1).
Statistical Analysis
All the statistical analyses were performed using the SPSS software for Windows version 20.0 (SPSS, Inc., Chicago, IL, USA) and Epi Info version 3.4.7 (Centers for Disease Control and Prevention, Atlanta, GA, USA). Chi-square analysis was used to compare the genotype and allele frequency between asthma and control groups. Odds ratio (OR) and 95 % confidence interval (CI) were used for the assessment of risk factors, and p value <0.05 was considered significant.
Results
Study Population Characteristics
For characterizing the impact of 24 bp duplication in CHIT1 gene in the North Indian population, genotyping of a total of 964 subjects was done, including 483 healthy controls and 481 asthma patients. Other parameters such as gender, age, disease duration, atopic status, total serum IgE level, smoke exposure, family history, spirometry diagnosis, cough, occurrence, severity, BMI, etc., were also examined. The mean age for asthma patients was found to be 37.22 ± 14.1 years, and the mean age for healthy adults was 34.29 ± 12.2 years. Also the females outnumbered the males in both cases and controls. Mean disease duration was more than 9 years, and 28 % asthmatic patients had a family history of asthma (Table 1).
An association study was performed between CHIT1 and asthma in which we found that the control groups were in Hardy–Weinberg Equilibrium. Statistics for the allelic frequencies in both the groups were calculated (Table 2) in which we found that the wild allele was marginally more prevalent among the control subjects (58.4 %) than in the asthma patients (55.7 %), while the mutant allele was more prevalent among the asthmatics (44.30 %) compared to the controls (41.6 %), with OR 1.12, 95 % CI (0.93–1.34), and a non-significant p = 0.237.
While comparing the genotypic frequencies for 24 bp duplication, the homozygous genotype (wild/wild) was more prevalent among the controls (33.3 %) compared to the asthmatics (24.1 %). The heterozygous genotype (wild/dup) was significantly more prevalent among the asthma patients (63.2 %) compared to the controls (50.1 %) which conferred the increased risk toward asthma in the studied population with OR 1.74, 95 % CI (1.29–2.36), and highly significant value of p = 0.000. Also, the combination of wild/dup + dup/dup genotype revealed a highly significant p value of p = 0.002 with OR 1.57 and 95 % CI (1.18–2.11). However, the homozygous mutant genotype (dup/dup) was more prevalent in the control subjects (16.6 %) than in the asthma patients (12.7 %), with OR 1.06, 95 % CI (0.69–1.63), and p = 0.786 (Table 2).
Asthma patients were further categorized on the basis of the phenotypic characteristics of the disease (Table 3), as obtained from their detailed pro forma, such as gender (male/female), occurrence (seasonal/throughout), severity (wheezing on exertion/wheezing at rest), family history (positive/nil), rhinitis (positive/nil), allergy to at least two provoking factors (positive/nil), smoking status (non-smoker/ever-smoker), and longstanding cough (positive/nil) where no association was observed between the CHIT1 polymorphism and asthma patients (p > 0.05).
Discussion
This is the first study conducted in a North Indian population, which reveals the impact of 24 bp duplication in the CHIT1 gene on asthma. Although we confirmed that the heterozygous genotype (wild/dup) and combination of both the heterozygous and the mutant revealed an increased risk toward asthma in the studied population, we were unable to demonstrate a significant effect of this recessive mutation of CHIT1 gene on the susceptibility to asthma. Phenotypic characteristics also do not show any significant difference between asthma and control groups.
Allergic diseases like asthma occur after the interaction with allergens and are associated with a heightened release of Th2 cytokine, IgE levels, and eosinophilia [24]. Th2 immune response is also observed in parasitic infections [25]. It has been reported that the productions of chitinases and Chitinase-Like Proteins in mammals are associated with the upregulated expression of Th2 cells and possibly contribute to the inflammatory diseases such as bronchial asthma, rhinitis, and allergen-induced inflammation [26]. In addition to this, CHIT1 is expressed by pulmonary macrophages of lungs, which is typically the site of entry for many environmental fungi, which may increase the risk of pulmonary fungal infection that leads to hypersensitivity or complications in lung diseases [27].
In the present study, we found a highly significant association between the heterozygous genotype and asthma with p = 0.000, and the statistical analysis reveals that maximum individuals have heterozygous genotype. This finding is consistent with a study in which Boot et al. [13] concluded that in most ethnic groups, ~35 % population has a carrier genotype and 5 % is homozygous to the 24 bp duplication and homozygous individuals for this mutation, lack a functional chitotriosidase.
There are some studies that confirm an important role of chitinases in asthma [28–30], but we found a lack of association between dup/dup genotype and asthma. Our finding is consistent with a recent study conducted on a population size of 422 subjects in American children which showed lack of association of CHIT1 polymorphism with asthma and concluded that asthma exacerbations in children are due to environmental fungal burden and not due to mutation in the CHIT1 gene [31]. In another pediatric study conducted in a Caucasian population, with 322 asthmatic children and 270 randomly chosen adult controls, it was found that 24 bp duplication in CHIT1 does not play a major role in the development of bronchial asthma in the population [32].
Mammalian chitinase acts as an immune stimulator, was first discovered and extensively explored in the middle to late 1980s [33]. Chitinase cleaves chitin, which is the second most abundant polymer on earth, in both plants and animals. An experiment done for the quantification of chitinase mRNA in humans revealed that the highest level of CHIT1 mRNA was in human lungs, followed by spleen, fetal liver, and thymus. In the lungs, CHIT1 protects against chitin-containing pathogens and the expression of this gene is conserved in humans as well as mice, which suggests its physiological importance in the mammalian lung [34]. These observations led to the proposal that chitinases are important mediators in allergic diseases; however, additional studies are needed to define the complex interactions between host chitinases, fungal exposures, and subsequent inflammatory responses in the airway. Thus, we propose that mutation in the CHIT1 gene results in decreased chitinolytic activity that may increase the susceptibility to fungal infections and persistent fungal burden in the lungs, leading to conditions such as asthma.
Conclusions
The genetic findings of this study aim to shed light on the association of 24 bp duplication in exon 10 of CHIT1 gene with the susceptibility to asthma in the studied North Indian population. Our results show highly significant association of asthma in heterozygous genotype but the phenotype remains unaffected. Therefore, further studies are required to understand the immune regulatory effects of chitin on asthma severity.
References
March ME, Sleiman PMA, Hakonarson H (2013) Genetic polymorphisms and associated susceptibility to asthma. Int J Gen Med 6:253–265
Smith KR (2002) Indoor air pollution in developing countries: recommendations for research. Int J Indoor Environ Health 12:1–7
Pruss-Ustun A, Corvalan C (2006) Preventing disease through health environments. Towards an estimate of the environmental burden of disease, World Health Organisation
Juarez-Rendon KJ, Lara-Aguilar RA, Garcia-Ortiz JE (2012) 24-bp duplication on CHIT1 gene in Mexican population. Rev Med Inst Mex Seguro Soc 50(4):375–377
Araujo AC, Souto-Padron T, de Souza W (1993) Cytochemical localization of carbohydrate residues in microfilariae of Wuchereria bancrofti and Brugia malayi. J Histochem Cytochem 41:571–578
Debono M, Gordee RS (1994) Antibiotics that inhibit fungal cell wall development. Annu Rev Microbiol 48:471–497
Elias JA, Homer RJ, Hamid Q, Lee CG (2005) Chitinases and chitinase-like proteins in T(H)2 inflammation and asthma. J Allergy Clin Immunol 116(3):497–500
Tachu B, Pillai S, Lucius R, Pogonka T (2008) Essential role of chitinase in the development of the filarial nematode Acanthocheilonema viteae. Infect Immun 76(1):221–228
Kanneganti M, Kamba A, Mizoguchi E (2012) Role of chitotriosidase (chitinase1) under normal and disease conditions. J Epithel Biol Pharmacol 5:1–9
Malaguarnera L, Musumeci M, Di Rosa M, Scuto A, Musumeci S (2005) Interferon-γ, tumor necrosis factor-α, and lipopolysaccharide promote chitotriosidase gene expression in human macrophages. J Clin Lab Anal 19(3):128–132
van Eijk M, van Roomen CP, Renkema GH et al (2005) Characterization of human phagocyte-derived chitotriosidase, a component of innate immunity. Int Immunol 17:1505–1512
Di Rosa M, Malaguarnera G, De Gregorio C, D’Amico F, Mazzarino MC, Malaguarnera L (2013) Modulation of chitotriosidase during macrophage differentiation. Cell Biochem Biophys 66(2):239–247. doi:10.1007/s12013-012-9471-x
Boot RG, Renkema GH, Verhoek M et al (1998) The human chitotriosidase gene. Nature of inherited enzyme deficiency. J Biol Chem 273:25680
Canudas J, Cenarro A, Civeira F et al (2001) Chitotriosidase genotype and serum activity in subjects with combined hyperlipidemia: effect of the lipid-lowering agents, atorvastatin and bezafibrate. Metabolism 50:447–450
Malaguarnera L, Simpore J, Prodi DA et al (2003) A 24-bp duplication in exon ten of human chitotriosidase gene from the sub-Saharan to the Mediterranean area: role of parasitic diseases and environmental conditions. Genes Immun 4:570–574
Brinkman J, Wijburg FA, Hollak CE, Groener JE, Verhoek M (2005) Plasma chitotriosidase and CCL18: early biochemical surrogate markers in type B Niemann-Pick disease. J Inherit Metab Dis 28:13–20
Altarescu G, Rudensky B, Abrahamov A et al (2002) Plasma chitotriosidase activity in patients with beta-thalassemia. Am J Hematol 71(1):7–10
Boot RG, Hollak CE, Verhoek M et al. (2010) Plasma chitotriosidase and CCL18 as surrogate markers for granulomatous macrophages in sarcoidosis. Clin Chim Acta 411(1–2):31–36. doi: 10.1016/j.cca.2009.09.034. Epub 2009 Oct 4
Comabella M, Domınguez C, Rio J et al (2009) Plasma chitotriosidase activity in multiple sclerosis. Clin Immunol 131:216–222
Karadag B, Kucur M, Isman FK, Hacibekiroglu M, Vural VA (2008) Serum chitotriosidase activity in patients with coronary artery disease. Circ J 72(1):71–75
Miller MR, Hankinson J, Brusasco V et al (2005) Standardisation of spirometry. ATS/ERS Task Force. Eur Respir J 26(2):319–338
Roe BA, Crabtree JS, Khan AS (1996) DNA isolation and sequencing.In: Rickwood D (ed) Essential techniques series, John Wiley and Sons, New York, p 85–86, 116–117
Ales M, Igor M, Barbara S, Marjeta Z, Borut P (2010) The role of chitotriosidase duplication gene polymorphism in the susceptibility to sarcoidosis. Zdr Vestn 79:837–842
Barnes PJ (2008) The cytokine network in asthma and chronic obstructive pulmonary disease. J Clin Invest 118(11):3546–3556. doi:10.1172/JCI36130
McKenzie AN (2000) Regulation of T helper type 2 cell immunity by interleukin-4 and interleukin-13. Pharmacol Ther 88:143–151
Sutherland TE, Maizels RM, Allen JE (2009) Chitinases and chitinase-like proteins: potential therapeutic targets for the treatment of T-helper type 2 allergies. Clin Exp Allergy 39(7):943–955. doi:10.1111/j.1365-2222.2009.03243.x
Vicencio AG, Chupp GL, Tsirilakis K et al (2010) CHIT1 mutations: genetic risk factor for severe asthma with fungal sensitization? Pediatrics 126:e982
Zimmermann N, King NE, Laporte J et al (2003) Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest 111(12):1863–1874
Zhu Z, Zheng T, Homer RJ et al (2004) Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science 304(5677):1678–1682
Zhao J, Zhu H, Wong CH, Leung KY, Wong WS (2005) Increased lungkine and chitinase levels in allergic airway inflammation: a proteomics approach. Proteomics 5(11):2799–2807
Wu AC, Lasky-Su J, Rogers CA, Klanderman BJ, Litonjua AA (2010) Fungal exposure modulates the effect of polymorphisms of chitinases on emergency department visits and hospitalizations. Am J Respir Crit Care Med 182(7):884–889
Bierbaum S, Superti-Furga A, Heinzmann A (2006) Genetic polymorphisms of chitotriosidase in Caucasian children with bronchial asthma. Int J Immunogenet 33:201–204. doi:10.1111/j.1744-313X.2006.00597.x
Lee CG, Da Silva CA, Lee JY, Hartl D, Elias JA (2008) Chitin regulation of immune responses: an old molecule with new roles. Curr Opin Immunol 20(6):684–689. doi:10.1016/j.coi.2008.10.002
Ohno M, Togashi Y, Tsuda K et al (2013) Quantification of chitinase mRNA levels in human and mouse tissues by real-time PCR: species-specific expression of acidic mammalian chitinase in stomach tissues. PLoS ONE 8(6):e67399. doi:10.1371/journal.pone.0067399
Acknowledgment
J. Singh is highly grateful to the Department of Science & Technology, New Delhi under the Ministry of Science & Technology, India, for providing research Grant (SR/FT/LS-018/2008) for the study.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sinha, S., Singh, J., Jindal, S.K. et al. Association of 24 bp Duplication of Human CHIT1 Gene with Asthma in a Heterozygous Population of North India: A Case–Control Study. Lung 192, 685–691 (2014). https://doi.org/10.1007/s00408-014-9605-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00408-014-9605-6