Abstract
Purpose
The aim of this study was to evaluate in the vagina of 60 pre-menopausal women the detection of orally administered multispecies probiotic formulations showing anti-microbial properties in test in vitro.
Methods
A randomized, double-blind, three-arm parallel pilot study was carried out on 60 pre-menopausal women. Subjects were randomly divided in three groups (F_1, F_2, F_3). Each group received a daily oral administration of probiotic mixtures (for 14 days and at the day 21, 7 days after the wash-out) containing: Lactobacillus acidophilus and Lactobacillus reuteri (F_1), or Lactobacillus plantarum, Lactobacillus rhamnosus and Bifidobacterium animalis subsp. lactis (F_2), or placebo (F_3), respectively. Vaginal swabs were collected at four experimental times, at t0 and at t7, t14 and t21 days, and analyzed by qPCR. At the same time, the anti-microbial activity of the probiotic formulations was verified by assays in vitro against microorganisms as Escherichia coli and Candida albicans.
Results
L. acidophilus and L. reuteri as well as L. plantarum, L. rhamnosus and B. lactis were significantly increased on 7 days in the groups administered with F_1 and F_2, respectively, compared to group F_3. A similar significant trend was observed on 21 days, 7 days after the wash-out. F_1 and F_2 showed coherent anti-microbial properties.
Conclusion
Both probiotic formulations F_1 and F_2, chosen because of their anti-microbial activity against pathogens responsible for vaginal dysbiosis and infections, led to vaginal detection and enhancement of the amount of species of formulates when orally administered. This work provides the basis for further clinical investigations of the F_1 and F_2 capacity to prevent or treat uro-genital infections.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Probiotics are, according to the FAO and WHO’s definition, “live microorganisms which when administered in adequate amounts confer a health benefit on the host” [1]. Several health benefits of probiotics are known: their administration is beneficial in preventing and curing different types of diarrhea [2]; they are used in the treatment of inflammatory intestinal diseases [3] and it has been demonstrated that they help preventing from allergies [4]. Usually, the gut is the target organ for probiotic formulations, but recently other organs and tissues have been related to probiotics, such as skin, hair [5], the oral cavity [6] and the vagina [7], by showing their positive effects for human health. In the specific case of vagina, the microbial species play an important role in the maintenance of health and prevention from infections throughout several mechanisms: occupation of specific adhesion sites of the uro-vaginal epithelium, maintenance of a low pH and production of anti-microbial metabolites such as acids, bacteriocins, hydrogen peroxides and anti-adhesive polysaccharides [8]. In physiological conditions, the vagina hosts mainly Lactobacillus spp. which concurs to a healthy microbiota, hence preventing from colonization of pathogenic bacteria and fungi [9, 10]. On the contrary, the depletion of vaginal lactobacilli facilitates the overgrowth of diverse species such as Gardnerella vaginalis, Atopobium vaginae, Candida albicans, Escherichia coli, responsible for vaginal dysbiosis and uro-genital infections [9–12]. Different probiotics were reported [13–15] to restore the normal vaginal homeostasis by colonization of lactobacilli, when administered topically.
Recent works [16, 17] also suggest that oral administration of lactobacilli and bifidobacteria would colonize both the intestinal and vaginal mucosal surfaces. This opens a new prospect for probiotic therapy: the challenge is to develop probiotics formulation to treat the uro-genital infections and to reduce the recurrences of disease in time, thanks to the capacity to control the pathogenicity of other microbes and to restore the normal ecological balance in vagina. However, these studies did not describe the efficacy of oral administration in term of presence and persistence of probiotic at the vaginal mucosa.
In this context, the aim of the present work is to evaluate in the vagina of 60 pre-menopausal healthy women the detection of orally administered multispecies probiotic formulations showing anti-microbial properties in test in vitro. A formulation F_1 containing L. acidophilus PBS066 and L. reuteri PBS072 and the other F_2 composed by L. plantarum PBS067, L. rhamnosus PBS070 and B. animalis subsp. lactis PBS075 were compared with the placebo (F_3). Data on these two mixtures supported the anti-microbial activity exerted by the above-mentioned single strains by assays in vitro using cell-free supernatants against microorganisms as E. coli and C. albicans.
The pilot study developed in this paper represents an example to investigate how oral consumption of probiotics formulations can lead to increased levels of the consumed species in the vagina of the women recruited in the study that showed anti-microbial activity against microorganisms potentially involved in uro-genital infections.
Materials and methods
Strains, probiotic formulations and culture conditions
This study comprised five strains of Lactobacillus spp. and Bifidobacterium spp. supplied from a private collection (Principium Europe Srl) (Table 1).
Unless otherwise specified, Lactobacillus spp. strains were cultured in deMan, Rogosa and Sharpe (MRS) medium. For Bifidobacterium spp. strains the MRS medium was supplemented with 0.3 g/L l-cysteine hydrochloride monohydrate (cMRS) (Sigma-Aldrich). The cultures were incubated at 37 °C under microaerophilic or anaerobic conditions using anaerobic atmosphere generation bags (Anaerogen, Oxoid).
Two different formulations containing lactobacilli and bifidobacteria of the study (mix F_1 and mix F_2) or placebo (mix F_3) were prepared (Table 2). The composition of the probiotic mix F_1 was as follows: 5 × 109 CFU L. acidophilus PBS066 (40 mg as lyophilized), 5 × 109 CFU L. reuteri PBS072 (30 mg as lyophilized), 320 mg inulin, 5 mg silica, 5 mg talc. The F_2 composition was as follows: 5 × 109 CFU L. plantarum PBS067 (12 mg as lyophilized), 5 × 109 CFU L. rhamnosus PBS070 (20 mg as lyophilized), 5 × 109 CFU B. animalis subsp. lactis PBS075 (60 mg as lyophilized), 298 mg inulin, 5 mg silica, 5 mg talc. Placebo (F_3) composition was as follows: 390 mg inulin, 5 mg silica, 5 mg talc.
Both single strains and formulations were tested for their anti-microbial activity [18]. As antagonistic microorganisms for anti-microbial activity assays, E. coli ATCC 25922 and C. albicans ATCC 10231 were employed. With the exception of C. albicans, maintained in Sabouraud (Oxoid) agar (1.5% w/v Agar Technical, Oxoid) medium, the antagonists were cultured in Tryptic Soy Agar (TSA; Oxoid) at 37 °C in aerobiosis.
Assessment of the anti-microbial activity
Inhibitory activity of formulates not-neutralized cell-free supernatant on the growth of antagonistic microorganisms
Over-night MRS (or cMRS) cultures were centrifuged at 12,000×g at 4 °C for 10 min. The pHs of the supernatants were recorded and measured. An aliquot of the not-neutralized (NN) supernatant fractions was filtered with 0.22 µm pore filter membranes to remove any residual bacterial cell.
The antimicrobial activities were detected by measuring the growth inhibition of E. coli ATCC 25922 and C. albicans ATCC 10231 in liquid cultures in the presence of the NN cell culture supernatants. Single colonies from freshly streaked plates of the antagonists were resuspended in saline solution at a concentration of 106 CFU/mL. The assay was performed in 96-well plates. Each well contained: double concentrated TSB, 10% (v/v) cell suspension of antagonistic strains, 25% (v/v) of NN cell culture supernatants and distilled water up to 150 µL as final volume.
Positive controls were prepared by substituting to NN cell culture supernatants an equal volume of not-inoculated MRS medium. Bifidobacterium sp. strain NN supernatant was used as Ref. [18]. Plates were incubated at 37 °C in aerobiosis for 24 h. At regular times, the cultures were sampled and the growth was evaluated by count plate technique. Experiments were performed three times with a standard error around ±10%.
Anti-microbial activity of formulates vs antagonistic microorganisms by using living cells
The antimicrobial activity of the probiotic mixtures containing lactobacilli and bifidobacteria living cells against the antagonists was evaluated by the overlay method, using the protocol described by Presti et al. [18]. A suspension of each mix F_1 and mix F_2 or placebo (mix F_3) was cultured into MRS broth until the O.D.600nm was 0.2. Then, 50 µL of each culture was spread by forming a stripe 2 cm wide across the MRS (or cMRS) agar plates. The plates were incubated in anaerobiosis at 37 °C for 24 h. After strain growth, plates were overlaid with 10 mL of melted TSA or MYPG (3 g/L Malt extract, Difco; 3 g/L Yeast extract, Biolife; 3 g/L Bacto Peptone, Difco; 2 g/L glucose, Sigma-Aldrich; pH, 6.2) soft agar (8 g/L) media. E. coli ATCC 25922 and C. albicans ATCC 10231 colonies from freshly streaked plates were resuspended into 0.9% (w/v) saline solution at a concentration of 108 CFU/mL. Cell suspensions were streaked over TSA (or MYPG in the case of C. albicans) surface with a cotton swab. The plates were incubated at 37 °C for 24 h in aerobic conditions. The anti-microbial ability of the examined lactobacilli and bifidobacteria strains was semi-quantitatively evaluated in terms of absent (−), moderate (+) and strong (++) growth inhibition of the antagonist, depending on the dimension of the inhibition halos.
DNA extraction and manipulation
DNA from microbial cultures was extracted by the “InstaGene Matrix” (Biorad): 1 mL of culture (109 CFU/mL) was centrifuged at 4000 rpm and 12 °C for 15 min. DNA was extracted from the pellet following the protocol provided by the manufacturer. DNA was also extracted from tenfold dilutions of the 109 CFU/mL culture up to 102 CFU/mL to set up qPCR analysis.
DNA from microbial cultures was used as template to set up primers and PCR conditions to apply for the detection of bacteria of the formulates by qPCR. Primers were designed as follows. The two DNA regions, pre-16S rRNA and IS 16S/23S rRNA sequences [19, 20] were used to identify species-specific DNA markers. For each probiotic, the obtained sequence served as a query to perform BLAST searches against publically available nucleotide databases (http://blast.ncbi.nlm.nih.gov). We collected the top BLAST hits, with a query cover >70% and identity >80%, and aligned them with the query sequence by using Clustal Omega [21] at http://www.ebi.ac.uk/Tools/msa/clustalo/. Forward and reverse primers were manually designed with the least conserved nucleotide sequences. Selected primers were firstly tested in silico on “Primer-BLAST” at the NCBI BLAST web site (http://www.ncbi.nlm.nih.gov/tools/primer-blast) running a default BLAST search against the entire database with the designed primers as queries.
PCR analyses were performed by using the identified primers and by using 10 ng of DNA extracted from each microbial culture. PCRs were performed with by puReTaq Ready-To-Go PCR beads (GE HealthCare Biosciences, Buckinghamshire, UK) in a 25 μL reaction according to the manufacturer’s instructions. PCR cycles consisted of an initial denaturation step for 7 min at 94 °C, 35 cycles of denaturation (45 s at 94 °C), annealing (30 s at 55 °C), extension (1 min at 72 °C) and a final extension at 72 °C for 7 min.
The efficacy of amplification was verified by agarose gel electrophoresis and ethidium bromide staining. In addition, each PCR product was sequenced to verify the correspondence of the selected region. Primer synthesis and DNA sequencing were supplied by Primm, Milan, Italy.
qPCR: primers validation and standard curves
Semi-quantitative analyses of probiotics were set up using qPCR. The 10 μL qPCR mix was assembled with 5 μL of “SsoFast EvaGreen Supermix with Low ROX” (BIO-RAD) and 0.5 μL of 10 μM primer forward and 10 μM primer reverse and 4 μL of DNA template. qPCR was performed in triplicate in the ABI 7500 Real-Time PCR system (Applied Biosystems) using standard cycling conditions: 2 min at 50 °C, 10 min at 95 °C, 40 cycles of 15 s at 95 °C and 1 min at 60 °C. This qPCR program was followed by a dissociation step to verify specificity.
Tenfold dilutions of DNA extracted from microbial cultures were tested in order to verify the detection limit of the qPCR reaction and hence select an optimum number of cycles for the reaction.
Standard curves were constructed for each strains using DNA extracted from microbial cells.
Cells were prepared using tenfold dilutions ranging from 109 to 102 CFU/mL.
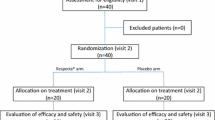
Pilot study design
This study consisted in a randomized, double-blind, three-arm parallel group, and placebo-controlled pilot study. It involved 60 pre-menopausal women aged between 18 and 50 years old not suffering from vaginal or urinary tract infections during the 12 months prior to the date of their enrollment in the study. All the study procedures were approved by the Independent Ethics Committee for Non-Pharmacological Clinical Investigations and were carried out according to World Medical Association’s (WMA) Helsinki Declaration and its amendments (Ethical Principles for Medical Research Involving Human Subjects, adopted by the 18th WMA General Assembly Helsinki, Finland, June 1964).
All subjects provided a written informed consent before initiation of any study-related procedures. The study took place at Farcoderm Srl facilities. Farcoderm Srl is an independent testing laboratory for in vitro and in vivo safety and efficacy assessment of cosmetics, food supplements and medical devices.
Depending on a randomization plan, the 60 volunteers were divided into three groups of treatment; each subject was treated, for 14 days, respectively, with a dietary supplement containing F_1 or F_2 or placebo F_3 (Table 2). Vaginal swabs of the enrolled subjects were collected within first 4–5 cm of vagina, to take a sample of vaginal secretion, at four experimental times: the day before first intake of probiotics, after 7, 14 and 21 days from the first intake (day 21 represents seven days after the end of the treatment). Vaginal swabs were stored at −20 °C until total DNA isolation.
The tested products consisted of food supplements (capsules) containing lactobacilli and bifidobacteria (Principium Europe Srl, Solaro, MI, Italy) (Table 1). The composition of the probiotic mix F_1, F_2, and F_3 is reported above (Table 2).
DNA analysis of vaginal samples
Total DNA was extracted from the vaginal swabs by the “InstaGene Matrix” (Biorad) according to the following protocol: 120 μL were taken from each swab and centrifuged at 13,000 rpm at 10 °C for 10 min; 200 μL of “InstaGene Matrix” were added to the pellet; after an incubation at 56 °C and 1000 rpm for 30 min and 8 min at 100 °C, the solution was centrifuged at 12,000 rpm for 3 min. The supernatant containing DNA was collected and stored at −20 °C. DNA quality was verified by agarose gel electrophoresis.
DNA extracted from swabs was analyzed in triplicate, with each one of the five primer sets, with qPCR mix and qPCR protocol described before. We chose to analyze 4 μL of 1:10 dilutions of isolated DNA in order to reduce the effect of possible PCR inhibitors.
Each qPCR run contained multiple Non-Template Controls (NTC).
Statistical methods
To examine the effect of the administration of different probiotic formulations, we used a linear mixed model (LM). The different probiotics, the capsule (treatment: F_1, F_2, F_3) and the time point at four different experimental times (timepoint: t0, t7, t14, t21 days) were used as the explanatory variables. Moreover, we considered volunteers (sample) as random effect.
To apply generalized linear mixed model (GLMM) under Poisson-lognormal error to account for higher variation at the lower end of target abundance, MCMC.qpcr R package was used to convert Ct data in bacterial counts. The conversion to approximate counts uses the following formula:
where E is the efficiency of amplification and Ct1 is the number of qPCR cycles required to detect a single target molecule.
Markov Chain Monte Carlo (MCMC) algorithm implemented in the package is used to sample from the joint posterior distribution over all model parameters, in order to estimate the effects of all experimental factors on the levels of specific microbial species. GLMM was used to test whether the levels of the different microbial species in different formulation groups (F_1, F_2, F_3) differed between the baseline (t0) and the subsequent time points (t7, t14, t21 days).
The experimental design is incorporated into the following model:
where the logarithm of bacterial counting rate is the response variable and the fixed factors are Formulation and Time (baseline and subsequent time points). The three remaining factors: sample (different subjects of the study), species:sample and species: residual are defined as random factors, accounting for the variation in quality and quantity of biological material among samples.
In particular, lmer package [22] lmerTest package [23] was used to perform linear mixed models and do statistical tests. Plots are performed using ggplot2 package [24].
Results
Anti-microbial activity of the probiotic formulates
The anti-microbial activity of formulates F_1 and F_2 was tested against C. albicans and E. coli, as an example of microorganisms responsible for uro-genital infections, by growth inhibition with non-neutralized cell-free supernatants and by overlay assay on living cells. Both probiotics mixtures showed a strong inhibition rate against E. coli mediated by microbial culture supernatants during 24 h, while no influence was detected vs C. albicans. The inhibition degree of each probiotic formulation compared to the corresponding single strain is reported in Fig. 1. The curve of activity of formulates is in line with the expected profile from single strain performance. The inhibition capacity of the mixtures F_1 and F_2 was further investigated by direct contact of their cultures against the same microorganisms. The growth inhibition halos around the multiple colonies were coherent with the anti-microbial inhibition profiles detected for the single strains. F_1 showed a moderate activity against both C. albicans and E. coli according to a better performance of L. acidophilus contrary to L. reuteri, while a stronger inhibition capacity vs both pathogens was observed for F_2. The results are showed in Fig. 2.
Effect of non-neutralized culture supernatants of L. acidophilus (square), L. plantarum (cross), L. rhamnosus (triangle), B. animalis subsp. lactis (filled square), L. reuteri (filled triangle), mixture F_1 (diamond), mixture F_2 (filled diamond) on the growth of E. coli and C. albicans: A1 mixture F_1 vs E. coli; A2 mixture F_1 vs C. albicans; B1 mixture F_2 vs E. coli; B2 mixture F_2 vs C. albicans
Set up of DNA molecular tools for probiotics identification
To identify each probiotic, polymorphic DNA regions were identified among the pre-16S rRNA and IS 16S/23S sequences [19, 20]. BLAST analysis performed against the entire default database (nr/nt), excluding species of interest, allowed to identify the most polymorphic regions (data not shown).
For L. rhamnosus and L. plantarum the DNA marker regions were found in the 16S/23S IS, while for the other three probiotics the pre-16S partial sequence resulted more polymorphic and suitable to identify marker regions. For each selected marker, species-specific primer pairs were identified (Table 3).
For each primer combination, standard qPCR curves were constructed by using DNA of pure culture of each strain. Results are reported in Table 4, together with the R 2 value, the slope and the efficiency of the amplification. The values obtained show how the qPCR reaction set up is optimal. The R 2 values are high (>0.99) and the efficiencies are between 80 and 115%, limits necessary for a reliable standard curve [25]. The analysis performed on different culture dilutions showed a linear dynamic range to be between 109 CFU/mL of culture and 102 CFU/mL.
Evaluation of vaginal probiotic amount in the subjects of the study
The selected primers were used to estimate the presence and persistence of each probiotic species in vagina from swab samples of the enrolled subjects by qPCR. PCR analyses were performed on DNA extracted from vaginal swabs at time t0, t7, t14 and t21 days.
The qPCR analysis demonstrated that the species-specific sequences associated with the probiotics of the formulations were detected only in vaginal DNA from subjects treated with the formulations F_1 and F_2 and not with the formulation F_3. Figure 3 shows the abundance of the different probiotic species for each treatment group at the different experimental times.
Ratio of probiotics of formulations (F_1 and F_2, vs F_3) by qPCR of species-specific sequences at the different times of treatment vs the amount at the baseline time point, expressed as bacterial counts. Upon the bars is reported the statistical analysis between treatments (***p < 0.001; **p < 0.01; *p < 0.05)
The subjects treated with F_1 showed an increase in the level of both L. acidophilus and L. reuteri compared with F_3. This has been observed at all the times compared with the t0, including t21, 7 days after the follow-up from the last probiotics administration. The same trend was observed for L. rhamnosus, L. plantarum and B. animalis subsp. lactis in women treated with F_2 formulation. Differently, the amount of all five strains remained constant throughout the study in the vaginal DNA of women treated with the placebo F_3.
The increase of microbial cell number and the permanence of probiotic species at higher levels after the end of the oral administration (t21) indicate that these species are actually more abundant in the vaginal microbiota. This increase was observed with statistical significance (p value <0.05) since 7 days after the beginning of treatment for L. reuteri and L. acidophilus of the F_1 group and for L. rhamnosus and B. animalis subsp. lactis of the F_2 group. Instead, the increase of L. plantarum resulted statistically significant (p value <0.05) since 14 days from the beginning of the treatment. The permanence of bacteria was observed for all the species studied until the t21, 7 days from the last probiotics intake. In fact, the differences of t14 and t21 from t7 are not statistically significant (p value >0.05) for all probiotics, with the exception of L. plantarum.
For F_3 formulation there were not so variations in the amount of selected probiotics for the entire 21-day period.
Discussion
The microbial species that inhabit the vaginal tract play an important role in the maintenance of health and prevention of infections. In particular, the presence of high numbers of lactic acid bacteria in the vagina is often equated with an health status [26]. It appears evident that the balance between a healthy and diseased state involves an equilibrium which can depends on different factors, such as hormone levels, douching, sexual practices, as well as bacterial interactions, and host defenses [27, 28]. A way to increase the level of vaginal lactobacilli is through the use of probiotics; two ways are commonly applied: direct application through vagitories or indirect application by oral consumption of probiotics. Several probiotics have been found to both increase the overall level of vaginal lactobacilli and to aid in the treatment of bacterial vaginosis [26]. Moreover, the capacity of probiotics to exert beneficial effects in human health is recognized to be strain-specific and a multi-species formulation could take the advantage of combining a greater spectrum of activities.
In this context, the present work allowed us to assess in the vagina of 60 pre-menopausal healthy women of a pilot study, the detection of orally administered multispecies probiotic formulations showing anti-microbial properties in test in vitro. First, we tested the antagonistic capacity against E. coli and C. albicans of the formulation F_1 and F_2 through cell supernatants and through the overlay contact between probiotic formulations and pathogens. Both formulations showed a strong anti-microbial activity against E. coli in both the conditions, as expected by the average trend of the single strains included in the formulations, while inhibition against C. albicans occurred only in the overlay assay, likewise the tested single strains. This is probably due to the reliable inhibitory activity of lower pH upon the E. coli growth, while the same conditions are not effective against yeasts. Moreover, it is reported that bacteriocins produced by probiotics are active only at certain pH ranges but can be neutralized at different pHs; this could be the reason for not having observed any inhibitory effect on the pathogens growth by using the neutralized cell supernatants [18].
Probiotics can reach the target of vagina when orally consumed, as an alternative way to a direct local administration; so they can locally exert their effect by competition–displacement of pathogens, reducing the infection relapses and related symptoms [26, 29].
Although previous studies [30–32] had already suggested this possibility, it is not so expected and the mechanism is still unclear. The current pilot study was aimed to investigate the vaginal detection after oral consumption of mixtures of probiotics would lead to increased levels of the consumed multispecies in the vagina of the enrolled subjects.
Our analysis showed an increased levels of all probiotics species (p value <0.05) detected in the vaginal swabs of women consuming the formulates F_1 and F_2 in comparison to women of the placebo group. The only exception was for L. plantarum, which was higher starting from 7 to 21 days of administration. This might suggest that: (1) L. plantarum is not as efficient as other probiotics, in colonizing vaginal mucosa from the intestinal region, (2) L. plantarum is already present at high levels in vaginal mucosa of healthy women and its abundance is not easily perturbed by the oral administration of probiotics. For this reason, L. plantarum might need a longer treatment or may require a higher concentration in administered capsules.
The abundance of probiotic strains in the vaginal DNA was assessed for both formulations until the last day of the experiment (t21 days), 7 days after the last intake (p value <0.05). This indicates that probiotics can actually colonize the vaginal microbiota in a short time. It would be particularly interesting to demonstrate if these can persist also after the menses. Indeed, the set-up of this study was purposely planned to match menstrual cycle of volunteers, with the first swab collected after menses and the last 21 days later, before the next menses. This was done to prevent from having blood traces in the vaginal swabs, so possibly altering the results.
The DNA markers used allowed to monitor the assessment and persistence of each probiotic species in the vaginal microbiota of healthy pre-menopausal women during the treatment and after the follow-up period. Our findings suggested that the five selected DNA markers can detect the increasing level and persistence of the studied bacteria into the vagina by using qPCR methods.
As lactobacilli and bifidobacteria have a fundamental role on the vaginal well-being, and more specifically an anti-microbial activity, we would highlighted that the anti-microbial effect detected against several pathogen microorganisms suggests that the five selected strains could similarly exert the antagonistic activity in vivo.
Conclusions
In conclusion, this study reports that the oral intake of two probiotic multi-species mixtures leads to an evident colonization of vagina of 60 volunteers of probiotic bacteria showing in vitro anti-microbial activity against pathogens involved in uro-genital infections. The adopted molecular tool represents a valid instrument to be used in future clinical trials to correlate the eventual clinical outcomes with the effective colonization in the treatment or prevention of vaginal dismicrobism and uro-genital infections.
References
Joint FAO/WHO (2001) Expert consultation on evaluation of health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria
Saavedra JM, Bauman NA, Oung I, Perman JA, Yolken RH (1994) Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhea and shedding of rotavirus. Lancet 344(8929):1046–1049
Meijer BJ, Dieleman LA (2011) Probiotics in the treatment of human inflammatory bowel diseases: update 2011. J Clin Gastroenterol 45(Suppl):S139–S144
Kuitunen M (2013) Probiotics and prebiotics in preventing food allergy and eczema. Curr Opin Allergy Clin Immunol 13(3):280–286
Levkovich T, Poutahidis T, Smillie C, Varian BJ, Ibrahim YM, Lakritz JR et al (2013) Probiotic bacteria induce a ‘Glow of Health’. PLoS One 8(1):e53867
Haukioja A (2010) Probiotics and oral health. Eur J Dent 4(3):348–355
Borchert D, Sheridan L, Papatsoris A, Faruquz Z, Barua JM, Junaid I et al (2008) Prevention and treatment of urinary tract infection with probiotics: review and research perspective. Indian J Urol 24(2):139–144
Coman MM, Verdenelli MC, Cecchini C, Silvi S, Orpianesi C, Caspani M et al (2015) In vitro evaluation on HeLa cells of protective mechanisms of probiotic lactobacilli against Candida clinical isolates. J Appl Microbiol 119(5):1383–1390 (Epub 2015/10/05)
Abad CL, Safdar N (2009) The role of lactobacillus probiotics in the treatment or prevention of urogenital infections–a systematic review. J Chemother 21(3):243–252
Martinez RC, Franceschini SA, Patta MC, Quintana SM, Candido RC, Ferreira JC (2009) Improved treatment of vulvovaginal candidiasis with fluconazole plus probiotic Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14R. Lett Appl Microbiol 48(3):269–274 (Epub 2009/02/02)
Martius J, Krohn MA, Hillier SL, Stamm WE, Holmes KK, Eschenbach DA (1988) Relationship of vaginal Lactobacillus species, cervical Chlamydia trachomatis, and bacterial vaginosis to preterm birth. Obstet Gynecol 71(1):89–95
Aldunate M, Srbinovski D, Hearps AC, Latham CF, Ramsland PA, Gugasyan R et al (2015) Antimicrobial and immune modulatory effects of lactic acid and short chain fatty acids produced by vaginal microbiota associated with eubiosis and bacterial vaginosis. Front Physiol 6:164
Stapleton AE, Au-Yeung M, Hooton TM, Fredricks DN, Roberts PL, Czaja CA et al (2011) Randomized, placebo-controlled phase 2 trial of a Lactobacillus crispatus probiotic given intravaginally for prevention of recurrent urinary tract infection. Clin Infect Dis 52(10):1212–1217 (Epub 2011/04/14)
Falagas M, Betsi GI, Athanasiou S (2007) Probiotics for the treatment of women with bacterial vaginosis. Clin Microbiol Infect 13(7):657–664
Palmeira-de-Oliveira R, Palmeira-de-Oliveira A, Martinez-de-Oliveira J (2015) New strategies for local treatment of vaginal infections. Adv Drug Deliv Rev 92:105–122 (Epub 2015/07/02)
Strus M, Chmielarczyk A, Kochan P, Adamski P, Chełmicki Z, Chełmicki A et al (2012) Studies on the effects of probiotic Lactobacillus mixture given orally on vaginal and rectal colonization and on parameters of vaginal health in women with intermediate vaginal flora. Eur J Obstet Gynecol Reprod Biol 163(2):210–215 (Epub 2012/06/19)
Reid G, Charbonneau D, Erb J, Kochanowski B, Beuerman D, Poehner R et al (2003) Oral use of Lactobacillus rhamnosus GR-1 and L. fermentum RC-14 significantly alters vaginal flora: randomized, placebo-controlled trial in 64 healthy women. FEMS Immunol Med Microbiol 35(2):131–134
Presti I, D’Orazio G, Labra M, La Ferla B, Mezzasalma V, Bizzaro G et al (2015) Evaluation of the probiotic properties of new Lactobacillus and Bifidobacterium strains and their in vitro effect. Appl Microbiol Biotechnol 99(13):5613–5626 (Epub 2015/03/07)
Ward LJ, Timmins MJ (1999) Differentiation of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus by polymerase chain reaction. Lett Appl Microbiol 29(2):90–92
Sul SY, Kim HJ, Kim TW (2007) Kim HY (2007) Rapid identification of Lactobacillus and Bifidobacterium in probiotic products using multiplex PCR. J Microbiol Biotechnol 17(3):490–495
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Bates D (2005) Fitting linear mixed models in R. R News 5(1):27–30
Kuznetsova A, Brockhoff PB, Christensen RHB (2013) lmerTest: Tests for random and fixed effects for linear mixed effect models (lmer objects of lme4 package). R package version 1.2
Wickham H (2011) ggplot2. Wiley Interdiscip Rev Comput Stat 3(2):180–185
Zhang T, Fang HH (2006) Applications of real-time polymerase chain reaction for quantification of microorganisms in environmental samples. Appl Microbiol Biotechnol 70(3):281–289 (Epub 2006/02/10)
Borges S, Silva J, Teixeira P (2014) The role of lactobacilli and probiotics in maintaining vaginal health. Arch Gynecol Obstet 289(3):479–489 (Epub 2013/11/30)
Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ (2012) Understanding vaginal microbiome complexity from an ecological perspective. Transl Res 160(4):267–282 (Epub 2012/03/06)
Cribby S, Taylor M, Reid G (2008) Vaginal microbiota and the use of probiotics. Interdiscip Perspect Infect Dis 2008:256490
Wagner RD, Johnson SJ (2012) Probiotic lactobacillus and estrogen effect on vaginal epithelial gene expression responses to Candida albicans. J Biomed Sci 19:58–65
Jespers V, Menten J, Smet H, Poradosú S, Abdellati S, Verhelst R et al (2012) Quantification of bacterial species of the vaginal microbiome in different groups of women, using nucleic acid amplification tests. BMC Microbiol 12:83
De Alberti D, Russo R, Terruzzi F, Nobile V, Ouwehand AC (2015) Lactobacilli vaginal colonisation after oral consumption of Respecta® complex: a randomised controlled pilot study. Arch Gynecol Obstet 292(4):861–867 (Epub 2015/04/09)
Vujic G, Jajac Knez A, Despot Stefanovic V, Kuzmic Vrbanovic V (2013) Efficacy of orally applied probiotic capsules for bacterial vaginosis and other vaginal infections: a double-blind, randomized, placebo-controlled study. Eur J Obstet Gynecol Reprod Biol 168(1):75–79 (Epub 2013/02/07)
Acknowledgements
The research was conducted through the Probioplus4Food Project, funded by MIUR and Lombardy Region, Italy. We thank Principium Europe Srl for supplying the bacterial strains and oral probiotic capsules. We thank Dr. Anna Sandionigi for supporting the statistical analyses.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
An erratum to this article is available at http://dx.doi.org/10.1007/s00404-016-4280-x.
Rights and permissions
About this article
Cite this article
Mezzasalma, V., Manfrini, E., Ferri, E. et al. Orally administered multispecies probiotic formulations to prevent uro-genital infections: a randomized placebo-controlled pilot study. Arch Gynecol Obstet 295, 163–172 (2017). https://doi.org/10.1007/s00404-016-4235-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00404-016-4235-2