Abstract
Late-onset Alzheimer’s disease (AD) is a complex age-related neurodegenerative disorder that likely involves epigenetic factors. To better understand the epigenetic state associated with AD, we surveyed 420,852 DNA methylation (DNAm) sites from neurotypical controls (N = 49) and late-onset AD patients (N = 24) across four brain regions (hippocampus, entorhinal cortex, dorsolateral prefrontal cortex and cerebellum). We identified 858 sites with robust differential methylation collectively annotated to 772 possible genes (FDR < 5%, within 10 kb). These sites were overrepresented in AD genetic risk loci (p = 0.00655) and were enriched for changes during normal aging (p < 2.2 × 10−16), and nearby genes were enriched for processes related to cell-adhesion, immunity, and calcium homeostasis (FDR < 5%). To functionally validate these associations, we generated and analyzed corresponding transcriptome data to prioritize 130 genes within 10 kb of the differentially methylated sites. These 130 genes were differentially expressed between AD cases and controls and their expression was associated with nearby DNAm (p < 0.05). This integrated analysis implicates novel genes in Alzheimer’s disease, such as ANKRD30B. These results highlight DNAm differences in Alzheimer’s disease that have gene expression correlates, further implicating DNAm as an epigenetic mechanism underlying pathological molecular changes associated with AD. Furthermore, our framework illustrates the value of integrating epigenetic and transcriptomic data for understanding complex disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is an age-related neurodegenerative disease of complex etiology and is characterized by a progressive decline of memory and cognitive faculties. Although late-onset AD risk has a strong genetic component including the APOE locus [10] and several other loci identified through genome-wide association studies (GWAS) [32], disease risk is also influenced by lifestyle and environmental factors such as diet [61], sleep [35], education and literacy [29], history of head trauma [14], and level of physical activity [53]. Epigenetic marks such as DNA methylation (DNAm) represent molecular regulatory mechanisms through which environmental and lifestyle factors may modulate AD risk, including through interaction with underlying genetic risk. A better understanding of the epigenetic regulation of gene expression in late-onset Alzheimer’s disease (LOAD) could facilitate the discovery of more viable preventive and therapeutic strategies for this devastating disease.
While previous studies have identified global DNAm alterations in Alzheimer’s disease [6, 9, 44], recent landmark epigenome-wide association studies (EWAS) [3, 12, 40] have identified DNAm changes around specific genes. In a study using postmortem brain tissue from the dorsolateral prefrontal cortex (DLPFC), DNAm is robustly associated with AD neuropathology at 71 genome-wide sites, ultimately implicating seven genes within 50 kb with dysregulated expression: CDH23, DIP2A, RHBDF2, RPL13, SERPINF1, SERPINF2, and ANK1 [12]. A parallel study found that DNAm of a region near ANK1 strongly correlated with neuropathological measures in three cortical brain regions (entorhinal cortex, superior temporal gyrus, and prefrontal cortex), but not cerebellum, suggesting that some epigenetic perturbations in AD occur across multiple cortical regions [40]. Likewise, a 48 kb region within the HOXA gene cluster was differentially methylated in AD across multiple cortical regions [65]. Subsequent studies [59, 62, 64, 66, 69] have integrated DNAm and genetic evidence to highlight AD relevant genes. Thus far, however, these approaches have implicated only a few genes associated with DNAm variation in AD.
While these previous studies have successfully identified DNAm differences associated with AD, they have been limited in their ability to connect these epigenetic differences to corresponding gene expression changes. Previous analyses often used targeted qPCR for a small number of transcripts and did not comprehensively survey gene expression changes among all implicated DNAm loci. Moreover, the relationship between DNAm and gene expression has remained unclear from earlier studies because each data type originated from non-overlapping subjects. Integrating DNAm with an unbiased method for surveying the transcriptome—RNA sequencing (RNA-seq)—may therefore offer deeper insight into [20] epigenetically mediated transcription dysregulation associated with AD.
We, therefore, performed multi-stage analyses incorporating paired DNA methylation and gene expression data. Given the unique cellular composition and potentially distinct susceptibilities to AD neuropathology of different brain regions, we chose to study four regions from each brain donor: dorsolateral prefrontal cortex (DLPFC), entorhinal cortex (ERC), hippocampus (HIPPO) all three previously implicated in AD; and the cerebellar cortex (CRB). In our first stage, we compared the DNAm landscape of neurotypical controls to late-onset AD, with the Illumina’s Human Methylation 450k (HM450k) array. Then, for genes adjacent to DNAm loci identified from this analysis, we analyzed RNA-seq data for differential expression between AD cases and controls, as well as for correlation between DNAm and gene expression levels. This undertaking represents one of the most comprehensive integrations of epigenetic and transcriptomic data for late-onset AD in postmortem human brain tissue to date.
Results
Clinical characteristics of postmortem brain donors
The dataset used for our epigenome-wide scan consisted of 73 postmortem brain donors with DNA methylation (DNAm) data generated from four brain regions: entorhinal cortex (ERC), dorsolateral prefrontal cortex (DLPFC), hippocampus (HIPPO), and cerebellum (CRB) (Table S1). Our discovery cohort includes 49 neurotypical controls and 24 AD donors with a neuropathological diagnosis (see “Methods”) as estimated by standard Braak staging and CERAD scoring. Compared to controls, AD donors were older (p = 2.95 × 10−8, ttwo-tailed = 7.67) and had reduced overall brain mass (p = 9.33 × 10−6, ttwo-tailed = 5.24, Figure S1). APOE risk, defined here as the number of ε4 alleles, is a strong genetic factor underlying AD clinical risk [13, 38] and was indeed more common in our AD samples than our control samples (p = 2.99 × 10−5, Fisher’s Exact Test). Alzheimer’s disease was not significantly associated with differences in DNAm-estimated NeuN+ (neuronal) composition in any of the four brain regions in our sample (p > 0.05, Results S1, Figure S2a), in line with previous large epigenome-wide association studies [12]. These DNAm cell-type composition estimates are reflective of the majority of variance (principal component 1 ~ 67%) in the dataset (Pearson’s r = − 0.833, p < 2.2 × 10–16, Fig. S2b). Unlike DNAm-derived estimates, those based upon our transcriptome data showed reduced neuronal proportions in the ERC and HIPPO regions in AD samples compared to controls (Fig. S2c). Overall, there was moderate correlation between cell-type estimates derived from DNAm data with those derived from transcriptomic data (r = 0.283, p = 2.88 × 10−6, Fig. S2d). There was no significant association between epigenetic age acceleration and AD in any of the brain regions (p > 0.05 Results S2, Fig. S3a, b).
CpG-site DNAm differences between AD subjects and unaffected controls
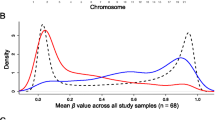
To identify differentially methylated sites with “shared effects” across multiple brain regions [40, 65], we adopted a powerful cross-brain region strategy (Results S3, Fig. S4, Table S2–S3, see “Methods”) where we analyzed all of our samples in conjunction (N = 269 total DNAm samples; Table S1). With this cross-region analysis, we identified 858 DNAm sites differentially methylated between AD and controls (FDR < 5%, Table S4). Collectively, these sites were within 10 kb of 1156 Gencode-annotated genes (v25), had a small median absolute difference in DNAm of 3.98%, and were relatively more methylated in subjects with AD compared to controls (N = 491, 57.2%, p = 2.30 × 10−5, Fig. 1a). The most prominent association between AD and DNAm within 10 kb of a protein-coding gene was with cg23703062 near ANKRD30B, a gene not previously implicated in AD (Fig. 1b). Other highly significant methylation sites replicated previously reported associations with cg19803550 (within 10 kb of WDR81 and SERPINF2, p = 3.48 × 10−1, 4th most significant) and cg05066959 (ANK1, p = 9.52 × 10−11), among several others suggesting that DNAm alterations in AD are replicable [12, 40]. However, the majority of our findings were novel associations (830 DNAm sites not presented in either of the landmark studies) indicating that there are several more DNAm loci associated with AD beyond those previously identified (Fig. S5). When we hierarchically clustered adjusted beta values of these 858 significant DMPs for all of our samples (N = 269), we found that samples clustered primarily by cerebellar vs. non-cerebellar brain region, then secondarily by AD diagnosis (Fig. 1c). Post hoc region-specific statistics were highly correlated across all four brain regions at these sites, confirming our expectation that AD has similar associations with DNAm at many of these sites across the different brain regions (Fig. S6).
a Histogram of effect sizes for significant differentially methylated probes (DMPs). 858 DNAm sites are significantly differentially methylated with the cross-region model (at false discovery rate, FDR < 5%). Of these 858 sites, 367 are less methylated (hypomethylated) and 491 are more methylated (hypermethylated) in AD patients compared to the unaffected controls. The greater number of hypermethylated sites constitutes statistically significant enrichment (p = 2.30 × 10−5). bANKRD30B differentially methylated probe (DMP) hypermethylation. Black points represent unaffected control samples and red points represent samples with a diagnosis of symptomatic Alzheimer’s disease (AD). Under the cross-region model, a DNAm site near ANKRD30B (cg23703062) was significantly more methylated in AD samples (red) than unaffected controls (black). Plotted beta values were adjusted for age, sex, ancestry, and estimates of technical variation using a linear model with logit transformation. c Heatmap of cross-region differentially methylated probes (DMPs). We adjusted beta-values of 858 differentially methylated probes identified by the cross-region model by regressing out covariates (age, sex, ancestry and estimates technical variance), then Z scaling the columns (DNAm sites). For hierarchical clustering and visualization purposes, Z values less than the 1st percentile or greater than the 99th percentile were set equal to these percentile Z values (i.e., thresholded) to limit the effect of extreme values. Brain samples (rows) and DNAm sites (columns) were clustered using Euclidean distance (dendrogram not shown for columns). dANKRD30B differentially methylated region (DMR). A series of neighboring probes overlapping the transcription start site (TSS) of ANKRD30B are more methylated in Alzheimer’s disease (red) than in unaffected controls (black). ANKRD30B ankyrin repeat domain 30B
We assessed the robustness of these findings using a series of sensitivity analyses. We found little effect of DNAm-estimated neuronal composition on these results, as adjusting for the estimated proportion of neurons resulted in almost all DNAm sites (sensitivity model 1, 842/858, 98.1%) remaining FDR significant with highly correlated global t statistics between both models (r = 0.954, p < 2.2 × 10−16, Fig. S7a). However, these results were somewhat expected given the lack of association between estimated neuronal composition estimates and diagnosis (p > 0.068) in our dataset. Furthermore, differential DNAm does not appear to be driven by the greater burden of APOE risk in our AD samples: t statistics were highly correlated with a model adjusting for APOE ε4 dosage (sensitivity model 2, r = 0.910, p < 2.2 × 10−16, Fig. S7b). Moreover, a slightly greater proportion of previously reported DMPs were consistent with the cross-region model than with the region-specific models of AD (Table S5). These analyses suggest that cross-region DMP findings from our discovery model are not confounded by cell-type composition or APOE risk burden differences and previously reported DMPs are reasonably consistent.
Regional DNAm differences between AD subjects and unaffected controls
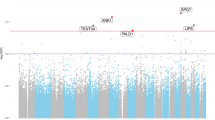
Next, we tested for regional differences in DNAm that could relate to additional regulatory mechanisms [24] via differentially methylated regions (DMRs) using a bump hunting strategy that jointly tests neighboring DNAm sites for differential methylation [26]. We found four DMRs between AD cases and controls under a cross-region model (at family-wise error rate, FWER < 0.05, Table S6). The most significant DMR was a hypermethylated region 1136 base pairs (bp) long that overlapped an exon 57 bp downstream from the DUSP22 transcription start site (TSS) and spans nine probes (FWER = 0, Fig. S8a). Post hoc region-specific analyses revealed DUSP22 hypermethylation in ERC (FWER = 0.053), HIPPO (FWER = 0.08), and DLPFC (FWER = 0.088) but not in CRB (FWER = 0.455). Another DMR overlaps the TSS of ANKRD30B (FWER = 0.027, 511 bp, nine contiguous probes, Fig. 1d). The third most significant region was hypomethylated and overlaps the promoter of JRK (384 bp from TSS, FWER = 0.029, 5 bp long, two contiguous probes, Fig. S8b) and the fourth DMR overlaps NAPRT (FWER = 0.047, 615 bp long, seven contiguous probes, Fig. S8c). These results suggest a more limited role of regional changes in DNAm associated with AD.
Brain region-dependent differential methylation
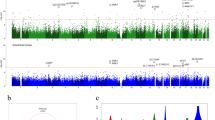
Given that some brain regions, such as cerebellar cortex, are putatively less susceptible to AD pathology than other brain regions, we next assessed whether Alzheimer’s disease is associated with DNAm in a brain region-dependent manner (i.e., an interaction model, brain region by diagnosis). We found 11,518 DNAm sites with region-dependent AD effects (at FDR < 5%, Table S7, Fig. S9). The most significant region-dependent effect is seen for a DNAm site near ANK1, which is more methylated in AD subjects for DLPFC, ERC, and HIPPO but less methylated for CRB (cg11823178, p = 3.41 × 10−21, Fig. 2a). Another example of a region-dependent effect can be seen for a CpG near CSNK1G2 (cg01335597, p = 3.04 × 10−15, Fig. 2b). Region-dependent sites were mostly distinct from the cross-region DMPs identified above, though a subset of 130 sites was significant in both models (Fig. 2c). These results demonstrate the power of combining data across multiple brain regions into joint statistical models and help further partition DNAm differences in AD by brain region.
a Example region-dependent DMP at cg11823178. Black points represent unaffected control samples and red points represent samples with a diagnosis of symptomatic Alzheimer’s disease (AD). The DNAm site cg11823178 is within 10 kb of protein-coding gene ANK1. The effect of Alzheimer’s disease (AD) upon DNAm at this site is brain region dependent, with cortical brain regions, DLPFC, hippocampus (HIPPO), and entorhinal cortex (ERC), having a large difference between AD cases and unaffected controls, whereas little difference is observed in cerebellum (CRB). ANK1 ankyrin 1. b Example region-dependent DMP at cg01335597. The DNAm site cg01335597 is within 10 kb of CSNK1G2, and is less methylated in CRB but more methylated in DLPFC, HIPPO, and ERC, in AD cases relative to unaffected controls. CSNK1G2 casein kinase 1 gamma 2. c Venn diagram of cross-region and region-dependent sites. The number of overlapping and distinct cross-region and region-dependent DNAm sites is shown (at FDR < 5%)
Normal aging vs. AD-associated DNAm changes
Older age is a strong risk factor for Alzheimer’s disease [36] and indeed our AD donors were older than our controls (p = 2.95 × 10−8). We, therefore, sought to disentangle the effects of normal aging on DNAm from the effects of AD on DNAm by assessing the association between DNAm and age in the subset of neurotypical controls (N = 187, age range 52–83 years) and comparing these age-related associations with AD associations. Cross-region AD t statistics were significantly associated with age-related t statistics, but the absolute correlation was weak (Pearson’s r = 0.089, Fig. S10a), suggesting that on the genome-wide scale, cross-region DNAm differences in AD are generally distinct from the DNAm signature of normal aging, suggesting some Alzheimer’s disease specific epigenetic mechanisms. Nevertheless, 18.1% of the significant AD-associated DNAm sites (154/858) exhibited similar age-related changes in unaffected controls compared to only 1.66% of DNAm sites not associated with AD (6967/419,994; at p < 1×10−3 and same directionality; Odds Ratio OR = 13.0, p < 2.2 × 10−16). DNAm changes in normal aging, therefore, further support the biological relevance of some of the most promising DNAm association, including CpG sites near the genes: WDR81 (cg19803550, paging = 1.31 × 10−13), ANK1 (cg05066959, paging = 4.85 × 10−13), MYO1C (cg14462670, paging = 4.39 × 10−4), and BIN1 (cg22883290, paging = 1.00 × 10−10). In contrast, p values from the region-dependent model were highly correlated on a global scale with p values from a region-dependent normal aging model (r = 0.752, p < 2.2 × 10−16, Fig. S10b). We, therefore, focused further analyses on cross-region differential methylation, which appeared to be less susceptible to confounding by age (see Results S4 for additional region-dependent analyses).
Biological relevance of genes near differential DNAm
We sought to better characterize the genomic and biological correlates of the DNAm changes identified under our cross-region model. Differentially methylated sites were enriched for CpG shelves and shores (Fig. S11), suggesting that these genomic features may be more dynamic in AD. Then, we tested whether genes near differentially methylated sites were enriched in known pathways and ontologies while accounting for the biased distribution of CpGs per gene. Overall, DMPs were preferentially in close proximity to genes involved in processes related to cell adhesion, immunity, and calcium binding (at FDR < 5%, Table S8a). Further analysis suggests that hypermethylated DNAm sites are driving these enrichments rather than hypomethylated sites, which did not appear to be enriched in any particular process (Tables S8b, c). Thus, the gain of DNA methylation at these cross-region sites implicates genes involved in cell adhesion, immunity, and calcium binding in AD.
AD genetic risk loci are enriched for differential methylation
Given that AD risk has a strong genetic component, we investigated whether differential methylation was located preferentially within AD risk loci from a large GWAS meta-analysis [32]. While only a small number of differentially methylated sites were present in AD risk loci (5/858, 0.58%), it represents statistically significant enrichment when compared to the background overlap with risk loci (562/419,432, 0.13%; OR = 4.37, p = 0.00655, Fisher’s exact test, Table S9). This enrichment was fully driven by hypermethylated sites (N = 5, OR = 7.74, p = 0.000588). When we relaxed our stringency for differential methylation (nominal p < 0.01), the enrichment in GWAS loci remained statistically significant (OR = 1.90, p = 0.00423). Of note, probes within AD risk loci were more often dropped during quality control than those outside the loci (192 probes dropped, OR = 2.21, p < 2.2 × 10−16; see Appendix for more details). Dropped probes were located mostly within the HLA-DRB5 locus (index SNP: rs9271192;131/192, 68.2%) that was previously reported [69] to be differentially methylated, so our analysis may underestimate the relative enrichment in GWAS loci. These results indicate that differentially methylated probes overlap putative AD genetic risk loci more than expected, which can hopefully be expanded upon and further refined through larger GWAS efforts in AD.
Replication with an independent cohort
To assess the replicability of these cross-region DNAm differences, we re-analyzed processed, public AD DNAm data from four brain regions, DLPFC, ERC, CRB, and superior temporal gyrus (STG), under a similar model (Ncontrol = 94 samples, NAD = 239 samples from Lunnon et al. [40]; see “Methods” for further details). We found 28.0% of cross-region differentially methylated sites were consistent in this independent dataset (with p < 0.05 and the same direction of effect; 240/858, OR = 3.84, p < 2.2 × 10−16, Table S10a). Notably, a site near ANKRD30B was more methylated in AD samples than controls (cg23703062, Preplication = 0.000355). These moderate replication rates are similar to those we observed previously (Table S5), suggesting reasonable concordance between AD DNAm differences from different brain cohorts.
Using gene expression to functionally validate differential methylation
We aimed to functionally validate differences in DNAm associated with AD and, therefore, tested for AD case–control gene expression differences. We generated transcriptome data for mostly the same samples used in the DNAm analysis, from the same brain regions: HIPPO, DLPFC, ERC and CRB (Ndonors: 50 controls, 26 AD cases; Nsamples: 196 controls, 92 AD cases). Then, we investigated expression of genes near differentially methylated sites (i.e., 645 DNAm sites corresponding to 772 genes within 10 kb; 218 DNAm sites were not within 10 kb of a Gencode v25 annotated gene and were not considered for this analysis). The majority of these genes were nominally differentially expressed between AD cases and controls in at least one brain region (p < 0.05, 52.7% of genes, 407/772, Table S11). The significance of only two genes, PLEC and CNPY2, survived stringent correction for multiple testing (threshold pbonferroni = 1.62 × 10−5, Ntests = 772 × 4 = 3088), but at a slightly less conservative threshold, six additional genes remained significant: ANKRD30B, ATP9B, SYTL2, PCDHGB1, ENSG00000242687.2, and ADAMTS2 (threshold pbonferroni = 6.48 × 10−5, Ntests = 772). Interestingly ANKRD30B, which was hypermethylated in AD at both spatial resolutions (DMP and DMR), was under-expressed in AD patients compared to controls in ERC (log2 fold change, LFC = − 1.50, p = 3.71 × 10−5) and HIPPO (LFC = − 1.70, p = 0.00242) but not DLPFC (LFC = − 0.434, p = 0.198) or CRB (LFC = − 0.304, p = 0.665; Fig. 3a). Other genes such as WDR81 and MYO1C that were previously [12] implicated via differential DNAm were differentially expressed here (Fig. 3b, c).
aANKRD30B differential expression. ANKRD30B is significantly less expressed in AD samples compared to controls in hippocampus and entorhinal cortex brain regions (ERC: p = 3.71 × 10−5, LFC = − 1.50; HIPPO: p = 0.00242, LFC = − 1.70), but not DLPFC or cerebellum (DLPFC: p = 0.198; LFC = − 0.434; CRB: p = 0.665, LFC = − 0.304). ANKRD30B ankyrin repeat domain 30B. bWDR81 differential expression. WDR81 is significantly more expressed in AD cases than in unaffected controls within ERC (CRB: p = 0.915, LFC = 0.0148; DLPFC: p = 0.111, LFC = 0.215; ERC: p = 0.00754, LFC = 0.295; HIPPO: p = 0.101, LFC = 0.293). WDR81 WD repeat domain 81. cMYO1C differential expression. MYO1C is significantly more expressed in AD cases than in unaffected controls within cortical brain regions (CRB: p = 0.826, LFC = 0.0299; DLPFC: p = 0.00836, LFC = 0.503; ERC: p = 0.0448, LFC = 0.382; HIPPO: p = 0.0324, LFC = 0.628). MYO1C myosin IC. d Greater DNAm at cg23703062 associates with reduced ANKRD30B expression in DLPFC, ERC, and HIPPO. Hypermethylation of DNAm site cg23703062, annotated to protein-coding gene ANKRD30B, associates with reduced gene expression in DLPFC, ERC, and HIPPO but not in CRB (CRB: p = 0.910, β = 0.00755; DLPFC: p = 0.0710, β = − 0.719; ERC: p = 0.0233, β = − 1.25; HIPPO: p = 0.101, β = − 0.792). ANKRD30B ankyrin repeat domain 30B
To further assess the functional link between DNAm and gene expression, we directly tested whether DNAm associates with gene expression for the subset of our samples that had both DNAm and RNA-seq data (Ndonors: 49 controls, 24 AD; Nsamples: 182 controls, 82 AD). We focused on the 407 genes with a nearby cross-region DMP and evidence of possible differential expression. We found DNAm associated with expression levels of 130 genes in at least one brain region (31.9%, at nominal p < 0.05, Table S12). This integrated approach refined our list of candidate genes implicated by a nearby differentially methylated site (within 10 kb), improving the biological resolution of our epigenome-wide scan (before: 39.1% of CpGs with multiple genes, after: 8.23% of CpGs with multiple genes; χ2 = 55.069, non-parametric p value = 1.6 × 10−8, 1 × 109 bootstraps). Overall, DNAm was inversely correlated with gene expression (79/130, 60.8%, p = 0.01406; using the strongest CpG-gene association), in line with the promoter-focused design of this microarray platform. Of note, hypermethylation of a site corresponding to ANKRD30B was associated with reduced gene expression in ERC, HIPPO, and DLPFC but not CRB (Fig. 3d). Other associations between DNAm probes and local expression of MYO1C (Fig. S12a) and DUSP22 (Fig. S12b) illustrate the heterogenous patterns of gene expression associated with DNAm and suggest additional brain region-specific factors contribute to gene expression regulation. Together, these findings further implicate DNAm as a potential mechanism underlying dysregulated gene expression in AD.
Discussion
DNA methylation (DNAm) is an epigenetic factor that is disrupted in Alzheimer’s disease (AD), and the effects of this epigenetic disruption have already been linked to several genes. To further validate previously reported genes and identify novel AD-relevant genes, we undertook a cross-brain region analysis of DNAm in Alzheimer’s disease (AD). Differential methylation analysis implicated biological processes previously hypothesized to underlie AD pathology—cell adhesion [39] and calcium ion homeostasis [41]—and were enriched for AD genetic risk loci [32]. Then, by linking DNAm data with corresponding transcriptome data, we were able to prioritize 130 genes that were differentially expressed between cases and controls and for which DNAm associated with expression. Together these findings further validate several previous, prominent DNAm associations such as those with ANK1 [12, 40] and DUSP22 [56] (Discussion S1), and implicate novel epigenetically dysregulated genes.
One promising gene implicated by our DNAm and gene expression evidence is ANKRD30B. Although little is known about the function of this protein-coding gene, it is expressed most strongly in breast, testis, and brain tissue. Furthermore, ANKRD30B is expressed in several cortical tissues affected by AD (DLPFC, ERC, HIPPO), and not cerebellum. Thus, while ANKRD30B is hypermethylated in all four brain regions, its expression is associated with DNAm and was reduced only in cortical regions and not cerebellum. The ANKRD30B protein product is predicted to contain an ankyrin repeat domain, a motif important for protein–protein interactions [27]. Interestingly, other genes previously implicated by DNAm evidence also encode proteins that contain ankyrin repeat domains (ANK1, ANKRD11).
Studying multiple brain regions is crucial to convert knowledge of epigenetic changes into insight of molecular risk mechanisms, because each region has a distinct cellular composition and regulatory landscape that contributes to neurophysiological function [11, 19, 25, 47, 57]. By studying the epigenome and transcriptome of four brain regions in parallel, three cortical regions susceptible to AD, and cerebellum, a region that is relatively protected, we were able to cast a wide net to capture AD-related differences. While we focused upon “cross-region” differential methylation—changes concordant across multiple brain regions—we also identified and characterized a large number of region-dependent differentially methylated sites. Moreover, even if differential methylation is shared across cerebellar and cortical brain regions, the gene expression correlates may not be. By studying the unique epigenetic profiles and their transcriptional correlates of multiple brain regions, we may glean deeper insight into the molecular mechanisms underlying differential susceptibility of brain regions to AD neuropathology. However, our study has several limitations. First and foremost, we cannot distinguish DNAm and gene expression differences that are causal in AD from those that relate to epiphenomena and secondary disease processes, such as neurodegeneration (Discussion S2). Indeed, the DNAm signature of other neurodegenerative diseases such as Lewy body dementia and Parkinson’s disease resembles that of Alzheimer’s disease at both the genome-wide [60] and single-gene level [63]. Nevertheless, some of the DNAm differences reported here may relate to AD etiology, because a subset of the differences was enriched within known late-onset AD genetic risk loci and correlated with gene expression changes. Further mechanistic studies to better untangle the timing of the co-occurring DNAm and expression changes are necessary to more confidently determine which of these DNAm changes are causative versus an epiphenomenon in the brains of patients with AD.
In a similar vein, these DNAm changes can be regulated by other components of the genome and epigenome. For example, DNAm differences in AD can reflect underlying genetic risk, as was the case with a differentially methylated region overlapping the PM20D1 promoter [59]. Likewise, DNAm changes may interact with other epigenetic factors that regulate gene expression [31]. Current human brain epigenome datasets now include post-translational histone modifications and higher order chromatin interactions, which have been shown to significantly alter gene expression both, within specific genetic loci and across the genome in human postmortem brain tissue [4, 50, 67]. Recent studies implicate widespread changes in the distribution of histone modifications in AD [16, 17]. While in some cases histone modifications may act independently of DNAm changes to generate AD risk [43], in other scenarios there is likely epigenetic cross-talk between DNAm and histone modifications [58]. Intriguingly, whereas a study of H4K16 acetylation, an active chromatin associated modification, identified AD-related differences weakly inversely associated with age-related changes [49], we observed the opposite correlation with DNAm, albeit the association was weak. The canonical roles of DNAm as a transcriptionally repressive epigenetic mark and histone acetylation as an activating one may reconcile these conflicting patterns. Disruption of multiple epigenetic factors may converge via regulating transcriptional networks [46, 48, 70] that are disrupted in AD. Our study suggests that disrupted DNAm plays a role within this complex gene regulatory framework. We identified 130 genes that had evidence of differential expression between cases and controls and for which expression associated with nearby DNAm that differed in AD. These differentially expressed genes may involve cell type-specific dysregulation. For instance, a recent study of cell-sorted cortical tissue found DNAm differences existed in both neurons and glia that associated with AD neuropathology (Braak stage) [15]. Likewise, a study of laser captured brain tissue suggests that microglia are responsible for differential expression of the epigenetically dysregulated gene, ANK1 [45]. Understanding the transcriptional output of these epigenetic changes in AD across multiple brain regions, which have different susceptibilities to AD neuropathology, will be a challenging, but rewarding endeavor. Our publicly available dataset contributes unique matched RNA-seq and DNAm data that can be used for development of genomic integration statistical methods.
The concordance between prior differential methylation studies and ours was modest (~ 28%), albeit similar to previous levels of concordance among epigenome-wide association studies. One factor potentially limiting concordance with prior studies is our use of neuropathological diagnosis as an outcome, as opposed Braak staging and CERAD scores. Additionally, our study used a modest number of AD cases (N = 24), as it included four brain regions and corresponding RNA-sequencing data. Aside from increasing sample sizes through additional data collection, jointly analyzing data from multiple existing studies via a meta-analysis framework may yield new, robust clues into the epigenetic fingerprint of AD. Meta-analyses of other genomic data types have offered valuable insights into the genetic [28, 32, 42] and transcriptional [51] correlates of Alzheimer’s disease. Likewise, a meta-analysis of DNAm studies would improve power to detect DNAm differences associated with AD and may also help reveal factors underlying study heterogeneity.
Although our study provides novel evidence supporting the role of DNAm in gene dysregulation, the extent to which DNAm contributes to AD pathogenesis remains unclear. While the DNAm array used here (HM450k) offers base-level resolution of DNAm differences, it is a biased sample of the human methylome and focuses on CpG DNAm in a promoter context. Indeed, DNAm in non-CpG contexts has been recently suggested to be more dynamic than DNAm in CpG contexts within human brain tissue and is likely relevant to neuropsychiatric diseases [52, 55]. Furthermore, we found that CpGs differentially methylated between AD cases and controls were preferentially located within regions of the genome sparsely sampled in the HM450k array. Thus, further studies with complementary methodologies (e.g., whole genome bisulfite sequencing) and meta-analysis of existing publicly available HM450k data will better resolve the relative contribution of DNAm to AD pathogenesis.
In summary, we used paired DNAm and transcriptome data from four brain regions to link methylation differences in AD to local gene dysregulation. These results support the role of DNAm as an epigenetic mechanism underlying gene dysregulation and support novel genes involved in AD. More generally, they illustrate the value of integrating epigenetic and transcriptomic data to study complex disease.
Methods
Postmortem brain tissue dissections
DLPFC (Brodmann areas 9 and 46), hippocampal formation, entorhinal cortex at the level of the anterior hippocampus and cerebellar cortex were dissected from frozen postmortem brains using a hand-held visually guided dental drill (Cat #UP500-UG33, Brasseler, Savannah, GA) as previously reported [37]. In addition to demographic matching between AD control subjects, the subjects included in the control group had no clinical or neurological history, or history of alcohol or substance abuse, or positive toxicology screens for illicit substances.
Alzheimer’s neuropathology diagnosis
All postmortem brains of subjects (controls and AD) have been sampled for neuropathology and specific Alzheimer’s lesions: beta-amyloid plaques, neurofibrillary tangles and tau-positive neurites. Samples were also genotyped for the apolipoprotein E gene (APOE). The sampled tissue sections were fixed in 10% buffered formalin and paraffin embedded into blocks for microscopic analysis (10 micron thickness). Sections included the superior frontal gyrus, middle and superior temporal gyri, inferior parietal cortex, occipital cortex, amygdala, hippocampus and entorhinal cortex, anterior thalamus, midbrain, pons, medulla, and cerebellum (including cerebellar cortex and deep cerebellar nuclei). Sections were silver-stained using the Hirano method [68] and immune stained using antibodies against ubiquitin, phosphorylated anti-tau (PHF-1) and beta-amyloid protein (6E10). Microscopic preparations were examined using conventional light microscopy. Alzheimer’s neuropathology ratings include the Braak [5] staging schema evaluating tau neurofibrillary tangle burden, and the CERAD scoring system as a measure of senile plaque burden (neuritic and diffuse). An Alzheimer’s likelihood diagnosis was then performed based on the published consensus recommendations for postmortem diagnosis of Alzheimer’s disease [23] as in prior publications [8].
Processing DNA methylation (DNAm) data
Human Methylation 450k (HM450k) arrays were run as specified by the manufacturer upon DNA extracted from each brain region. After generating array data for 398 postmortem brain tissue samples, resulting idat files were imported and then rigorously preprocessed with minfi [2]. We did not observe batch effects with several different quality control metrics. After removing low-quality samples (N = 5), we normalized the data with stratified quantile normalization [2]. We removed 64,660 probes that met one or more of the following exclusion criteria: (a) poor quality, (b) cross-reactive, (c) included common genetic variants, (d) mapped to a sex chromosome, and (e) did not map to hg38. We dropped 13 samples for one or more of the following reasons: (i) DNAm-predicted sex did not match phenotypic sex, (ii) 450k genotype clustered inappropriately, (iii) 450k genotype did not match SNP-chip genotype, and (iv) clustered inappropriately on principal component analysis. After these conservative quality control steps, 420,852 high-quality probes available for 377 samples remained for further analysis. Postmortem donors not meeting our diagnostic criterion for controls or AD were not included for analysis, leaving 269 tissue samples (Ndonors: 24 AD, 49 controls). We estimated DNAm cell-type compositions using a deconvolution algorithm [22] with a flow-sorted DLPFC reference [18] and estimated age acceleration with Horvath’s clock[21]. For additional details about DNAm processing, see Supplemental Methods (Figs. S13–S18).
Brain-region stratified analyses
We tested for differences between AD cases and controls in predicted neuronal proportion and age acceleration, stratified by brain region, with linear models. We included age, sex, and ancestry as covariates in these models. Likewise, we tested for case-control differences in RNA-seq estimated neuronal proportions using a seperate linear model for each brain region while adjusting for age, sex, race, mitochondrial mapping rate, gene assignment rate, and RIN.
Cross-region differential methylation analysis
In our “cross-region” model for AD case–control differences (N = 269 samples), we tested beta values from 420,852 probes for differential methylation between Alzheimer’s disease cases and controls using limma [54] with the model:
for DNAm site i and donor j and binary disease status Diagnosisj adjusting for brain region, age, sex, the first multidimensional scaling (MDS) component of genotype data (ancestry), and the first two negative control principal components (PCs). Afterward, we tested an interaction model, i.e., an F test of the estimated AD effect from each brain region, while adjusting for the same covariates as above. Both of these models incorporated multiple brain regions from each donor, and thus repeated measures, so we used the generalized least squares technique to estimate statistical parameters to appropriately account for the repeated biological measures. We estimated a consensus correlation with limma’s duplicateCorrelation function in which donors are blocks and different brain regions are repeated observations. Then, we incorporated this consensus correlation while estimating statistics for each DNAm site using the generalized least squares technique via limma’s lmFit function. We performed sensitivity analyses to assess how robust our results were to potential confounds (see Supplemental Methods for additional details).
Differential methylated region analysis
To assess whether regions of the genome were differentially methylated between Alzheimer’s disease cases and controls, we used bumphunter with the same models as above [26]. We implemented the bumphunter method with the minfi Bioconductor package’s bumphunter function using 1000 bootstrap iterations, a cutoff of 0.05 for defining continuous probes, smoothing done with the locfitByCluster function, and otherwise default parameters [2]. We controlled for multiple testing using the family wise error rate (FWER) to determine statistical significance.
Processing RNA sequencing (RNA-seq) data
RNA sequencing was performed with protocols described at [7]. In summary, TruSeq Stranded Total RNA Library Preparation kit with Ribo-Zero ribosomal RNA depletion (Illumina) libraries were generated and sequenced on an Illumina HiSeq 2000 at the LIBD Sequencing Facility, producing a mean of 130.6 million 100-bp paired-end reads per sample. Raw sequencing reads were quality checked with FastQC [1]. Quality checked reads were mapped to the hg38/GRCh38 human reference genome with splice-aware aligner HISAT2 version 2.0.4 [30]. Gene-level quantification based on GENCODE release 25 (GRCh38.p7) annotation was run on aligned reads using featureCounts (subread version 1.5.0-p3) [34] with a mean 34.9% (SD = 4.6%) of mapped reads assigned to genes. Quality controls metrics such as gene assignment rate, mitochondrial mapping rate, and RNA integrity number (RIN) were significantly associated with brain region and case–control status (Fig. S19a–c). These quality control metrics were, therefore, included as covariates in downstream analyses (see below).
Differential expression of differentially methylated genes and association with DNAm
DNAm sites were annotated to all Gencode v25 genes within 10 kb upstream or downstream (20 kb region) using the GenomicRanges Bioconductor package [33]. These genes were then tested for corresponding AD-case–control differential gene expression. Analyses were stratified by brain region and used 196 control and 92 AD RNA-seq samples processed as described in the previous section. We normalized counts for 25,587 Gencode annotated genes that were expressed in at least one brain region with limma’s voom function then tested for AD case–control differences under an empirical Bayesian framework [54] while adjusting for RNA Integrity Number (RIN), age, sex, race, mitochondrial mapping rate, and gene assignment rate.
We tested then the association between DNAm at each differentially methylated site and log-transformed, normalized gene expression (log2[RPKM + 1]) for all Gencode (v25) annotated genes within 10 kb with at least partial evidence of differential expression (nominal p < 0.05 in at least one brain region). This analysis used 182 control and 82 AD samples with matching DNAm and RNA-seq data. We tested for these associations in R using a linear model (lm function) that adjusted for the covariates above and was stratified by brain region.
Availability of data and materials
Raw and processed DNA methylation data is available through GEO accession GSE125895. The computer code used to generate results is available on GitHub: https://github.com/LieberInstitute/Alzheimers_DNAm.
References
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD et al (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30:1363–1369. https://doi.org/10.1093/bioinformatics/btu049
Bakulski KM, Dolinoy DC, Sartor MA, Paulson HL, Konen JR, Lieberman AP et al (2012) Genome-wide DNA methylation differences between late-onset Alzheimer’s disease and cognitively normal controls in human frontal cortex. J Alzheimers Dis 29:571–588. https://doi.org/10.3233/JAD-2012-111223
Bharadwaj R, Peter CJ, Jiang Y, Roussos P, Vogel-Ciernia A, Shen EY et al (2014) Conserved higher-order chromatin regulates NMDA receptor gene expression and cognition. Neuron 84:997–1008. https://doi.org/10.1016/j.neuron.2014.10.032
Braak H, Braak E (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathologica 82(4):239–259
Chouliaras L, Mastroeni D, Delvaux E, Grover A, Kenis G, Hof PR et al (2013) Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer’s disease patients. Neurobiol Aging 34:2091–2099. https://doi.org/10.1016/j.neurobiolaging.2013.02.021
Collado-Torres L, Burke EE, Peterson A, Shin JH, Straub RE, Rajpurohi A et al (2018) Regional heterogeneity in gene expression, regulation and coherence in hippocampus and dorsolateral prefrontal cortex across development and in schizophrenia. bioRxiv. https://doi.org/10.1101/426213
Conejero-Goldberg C, Hyde TM, Chen S, Herman MM, Kleinman JE, Davies P et al (2015) Cortical Transcriptional Profiles in APOE4 Carriers with Alzheimer's disease: patterns of protection and degeneration. J Alzheimers Dis 48(4):969–978
Coppieters N, Dieriks BV, Lill C, Faull RL, Curtis MA, Dragunow M (2014) Global changes in DNA methylation and hydroxymethylation in Alzheimer’s disease human brain. Neurobiol Aging 35:1334–1344. https://doi.org/10.1016/j.neurobiolaging.2013.11.031
Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW et al (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261:921–923
Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S et al (2012) Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol 13:R43. https://doi.org/10.1186/gb-2012-13-6-r43
De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L et al (2014) Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci 17:1156–1163. https://doi.org/10.1038/nn.3786
Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R et al (1997) Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278:1349–1356
Fleminger S (2003) Head injury as a risk factor for Alzheimer's disease: the evidence 10 years on; a partial replication. J Neurol Neurosurg Psychiatry 74(7):857–862
Gasparoni G, Bultmann S, Lutsik P, Kraus TFJ, Sordon S, Vlcek J et al (2018) DNA methylation analysis on purified neurons and glia dissects age and Alzheimer’s disease-specific changes in the human cortex. Epigenet Chromatin 11:41. https://doi.org/10.1186/s13072-018-0211-3
Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai LH et al (2015) Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 518:365–369. https://doi.org/10.1038/nature14252
Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM et al (2012) An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature 483:222–226. https://doi.org/10.1038/nature10849
Guintivano J, Aryee MJ, Kaminsky ZA (2013) A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics 8:290–302. https://doi.org/10.4161/epi.23924
Gutierrez-Arcelus M, Ongen H, Lappalainen T, Montgomery SB, Buil A, Yurovsky A et al (2015) Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLoS Genet 11:e1004958. https://doi.org/10.1371/journal.pgen.1004958
Hasin Y, Seldin M, Lusis A (2017) Multi-omics approaches to disease. Genome Biol 18:83. https://doi.org/10.1186/s13059-017-1215-1
Horvath S (2013) DNA methylation age of human tissues and cell types. Genome Biol 14:R115. https://doi.org/10.1186/gb-2013-14-10-r115
Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH et al (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform 13:86. https://doi.org/10.1186/1471-2105-13-86
Hyman BT, Trojanowski JQ (1997) Consensus recommendations for the postmortem diagnosis of Alzheimer disease from the National Institute on Aging and the Reagan Institute Working Group on diagnostic criteria for the neuropathological assessment of Alzheimer disease. J Neuropathol Exp Neurol 56(10):1095–1097
Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P et al (2009) The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41:178–186. https://doi.org/10.1038/ng.298
Jaffe AE, Irizarry RA (2014) Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol 15:R31. https://doi.org/10.1186/gb-2014-15-2-r31
Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP et al (2012) Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol 41:200–209. https://doi.org/10.1093/ije/dyr238
Jager D, Stockert E, Gure AO, Scanlan MJ, Karbach J, Jager E et al (2001) Identification of a tissue-specific putative transcription factor in breast tissue by serological screening of a breast cancer library. Cancer Res 61:2055–2061
Jansen I, Kiddle SJ, Posthuma D. Genetic meta-analysis identifies 9 novel loci and functional pathways for Alzheimer’s disease risk. Nature Genet. https://doi.org/10.17863/CAM.35461
Kaup AR, Nettiksimmons J, Harris TB, Sink KM, Satterfield S, Metti AL et al (2015) Cognitive resilience to apolipoprotein E epsilon4: contributing factors in black and white older adults. JAMA Neurol 72:340–348. https://doi.org/10.1001/jamaneurol.2014.3978
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. https://doi.org/10.1038/nmeth.3317
Klein HU, Bennett DA, De Jager PL (2016) The epigenome in Alzheimer’s disease: current state and approaches for a new path to gene discovery and understanding disease mechanism. Acta Neuropathol 132:503–514. https://doi.org/10.1007/s00401-016-1612-7
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45:1452–1458. https://doi.org/10.1038/ng.2802
Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R et al (2013) Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. https://doi.org/10.1371/journal.pcbi.1003118
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/bioinformatics/btt656
Lim AS, Kowgier M, Yu L, Buchman AS, Bennett DA (2013) Sleep fragmentation and the risk of incident Alzheimer’s disease and cognitive decline in older persons. Sleep 36:1027–1032. https://doi.org/10.5665/sleep.2802
Lindsay J, Laurin D, Verreault R, Hebert R, Helliwell B, Hill GB et al (2002) Risk factors for Alzheimer’s disease: a prospective analysis from the Canadian Study of Health and Aging. Am J Epidemiol 156:445–453
Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM et al (2006) Critical factors in gene expression in postmortem human brain: focus on studies in schizophrenia. Biol Psychiatry 60:650–658. https://doi.org/10.1016/j.biopsych.2006.06.019
Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G (2013) Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 9:106–118. https://doi.org/10.1038/nrneurol.2012.263
Liu G, Jiang Y, Wang P, Feng R, Jiang N, Chen X et al (2012) Cell adhesion molecules contribute to Alzheimer’s disease: multiple pathway analyses of two genome-wide association studies. J Neurochem 120:190–198. https://doi.org/10.1111/j.1471-4159.2011.07547.x
Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M et al (2014) Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat Neurosci 17:1164–1170. https://doi.org/10.1038/nn.3782
Magi S, Castaldo P, Macri ML, Maiolino M, Matteucci A, Bastioli G et al (2016) Intracellular calcium dysregulation: implications for Alzheimer’s disease. Biomed Res Int 2016:6701324. https://doi.org/10.1155/2016/6701324
Marioni RE, Harris SE, Zhang Q, McRae AF, Hagenaars SP, Hill WD et al (2018) GWAS on family history of Alzheimer’s disease. Transl Psychiatry 8:99. https://doi.org/10.1038/s41398-018-0150-6
Marzi SJ, Leung SK, Ribarska T, Hannon E, Smith AR, Pishva E et al (2018) A histone acetylome-wide association study of Alzheimer’s disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat Neurosci 21:1618–1627. https://doi.org/10.1038/s41593-018-0253-7
Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD (2009) Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer’s disease. PLoS One 4:e6617. https://doi.org/10.1371/journal.pone.0006617
Mastroeni D, Sekar S, Nolz J, Delvaux E, Lunnon K, Mill J et al (2017) ANK1 is up-regulated in laser captured microglia in Alzheimer’s brain; the importance of addressing cellular heterogeneity. PLoS One 12:e0177814. https://doi.org/10.1371/journal.pone.0177814
Miller JA, Guillozet-Bongaarts A, Gibbons LE, Postupna N, Renz A, Beller AE et al (2017) Neuropathological and transcriptomic characteristics of the aged brain. Elife. https://doi.org/10.7554/elife.31126
Montano CM, Irizarry RA, Kaufmann WE, Talbot K, Gur RE, Feinberg AP et al (2013) Measuring cell-type specific differential methylation in human brain tissue. Genome Biol 14:R94. https://doi.org/10.1186/gb-2013-14-8-r94
Mostafavi S, Gaiteri C, Sullivan SE, White CC, Tasaki S, Xu J et al (2018) A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat Neurosci 21:811–819. https://doi.org/10.1038/s41593-018-0154-9
Nativio R, Donahue G, Berson A, Lan Y, Amlie-Wolf A, Tuzer F et al (2018) Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat Neurosci 21:497–505. https://doi.org/10.1038/s41593-018-0101-9
Ng B, White CC, Klein HU, Sieberts SK, McCabe C, Patrick E et al (2017) An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat Neurosci 20:1418–1426. https://doi.org/10.1038/nn.4632
Patel H, Dobson RJB, Newhouse SJ (2018) Meta-analysis of Alzheimer’s disease brain transcriptomic data. bioRxiv. https://doi.org/10.1101/480459
Price AJ, Collado-Torres L, Ivanov NA, Xia W, Burke EE, Shin JH et al (2018) Divergent neuronal DNA methylation patterns across human cortical development: critical periods and a unique role of CpH methylation. bioRxiv. https://doi.org/10.1101/428391
Radak Z, Hart N, Sarga L, Koltai E, Atalay M, Ohno H et al (2010) Exercise plays a preventive role against Alzheimer’s disease. J Alzheimers Dis 20:777–783. https://doi.org/10.3233/JAD-2010-091531
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Rizzardi LF, Hickey PF, DiBlasi VR, Tryggvadóttir R, Callahan CM, Idrizi A et al (2019) Neuronal brain-region-specific DNA methylation and chromatin accessibility are associated with neuropsychiatric trait heritability. Nature Neurosci 22(2):307–316
Sanchez-Mut JV, Aso E, Heyn H, Matsuda T, Bock C, Ferrer I et al (2014) Promoter hypermethylation of the phosphatase DUSP22 mediates PKA-dependent TAU phosphorylation and CREB activation in Alzheimer’s disease. Hippocampus 24:363–368. https://doi.org/10.1002/hipo.22245
Sanchez-Mut JV, Aso E, Panayotis N, Lott I, Dierssen M, Rabano A et al (2013) DNA methylation map of mouse and human brain identifies target genes in Alzheimer’s disease. Brain 136:3018–3027. https://doi.org/10.1093/brain/awt237
Sanchez-Mut JV, Graff J (2015) Epigenetic alterations in Alzheimer’s disease. Front Behav Neurosci 9:347. https://doi.org/10.3389/fnbeh.2015.00347
Sanchez-Mut JV, Heyn H, Silva BA, Dixsaut L, Garcia-Esparcia P, Vidal E et al (2018) PM20D1 is a quantitative trait locus associated with Alzheimer’s disease. Nat Med 24:598–603. https://doi.org/10.1038/s41591-018-0013-y
Sanchez-Mut JV, Heyn H, Vidal E, Moran S, Sayols S, Delgado-Morales R et al (2016) Human DNA methylomes of neurodegenerative diseases show common epigenomic patterns. Transl Psychiatry 6:e718. https://doi.org/10.1038/tp.2015.214
Scarmeas N, Stern Y, Tang MX, Mayeux R, Luchsinger JA (2006) Mediterranean diet and risk for Alzheimer’s disease. Ann Neurol 59:912–921. https://doi.org/10.1002/ana.20854
Smith AR, Mill J, Smith RG, Lunnon K (2016) Elucidating novel dysfunctional pathways in Alzheimer's disease by integrating loci identified in genetic and epigenetic studies. Neuroepigenetics 6:32–50
Smith AR, Smith RG, Burrage J, Troakes C, Al-Sarraj S, Kalaria RN et al (2018) A cross-brain regions study of ANK1 DNA methylation in different neurodegenerative diseases. Neurobiol Aging 74:70–76. https://doi.org/10.1016/j.neurobiolaging.2018.09.024
Smith AR, Smith RG, Condliffe D, Hannon E, Schalkwyk L, Mill J et al (2016) Increased DNA methylation near TREM2 is consistently seen in the superior temporal gyrus in Alzheimer’s disease brain. Neurobiol Aging 47:35–40. https://doi.org/10.1016/j.neurobiolaging.2016.07.008
Smith RG, Hannon E, De Jager PL, Chibnik L, Lott SJ, Condliffe D et al (2018) Elevated DNA methylation across a 48-kb region spanning the HOXA gene cluster is associated with Alzheimer’s disease neuropathology. Alzheimers Dement. https://doi.org/10.1016/j.jalz.2018.01.017
Watson CT, Roussos P, Garg P, Ho DJ, Azam N, Katsel PL et al (2016) Genome-wide DNA methylation profiling in the superior temporal gyrus reveals epigenetic signatures associated with Alzheimer’s disease. Genome Med 8:5. https://doi.org/10.1186/s13073-015-0258-8
Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK et al (2016) Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538:523–527. https://doi.org/10.1038/nature19847
Yamamoto T, Hirano A (1986) A comparative study of modified Bielschowsky, Bodian and thioflavin S stains on Alzheimer's neurofibrillary tangles. Neuropathol Appl Neurobiol 12(1):3–9
Yu L, Chibnik LB, Srivastava GP, Pochet N, Yang J, Xu J et al (2015) Association of brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol 72:15–24. https://doi.org/10.1001/jamaneurol.2014.3049
Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA et al (2013) Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153:707–720. https://doi.org/10.1016/j.cell.2013.03.030
Funding
This work was supported by the funding from Lieber Institute for Brain Development and the Maltz Research Laboratories.
Author information
Authors and Affiliations
Contributions
SAS performed analyses and led the writing of the manuscript. RAB assessed neuropathology, contributed to biological study design and interpretation, and writing of the manuscript. LCT processed RNA-seq data and contributed to its analysis. RT performed APOE4 genotyping. JHS performed data generation. AD performed clinical reviews. JRW contributed to the analysis of the DNAm data. DRW contributed to the study design, interpretation of the results, and writing of the manuscript. TMH performed tissue dissections, contributed to the study design, interpretation of the results, and writing of the manuscript. JEK oversaw tissue collection and contributed to the study design, interpretation of the results, and writing of the manuscript. AEJ and VSM co-led the study, including the design, statistical analyses, interpretation, and writing of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval and consent to participate
Postmortem human brain tissue was obtained by autopsy primarily from the Offices of the Chief Medical Examiner of the District of Columbia, and of the Commonwealth of Virginia, Northern District, all with informed consent from the legal next of kin (protocol 90-M-0142 approved by the NIMH/NIH Institutional Review Board).
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Semick, S.A., Bharadwaj, R.A., Collado-Torres, L. et al. Integrated DNA methylation and gene expression profiling across multiple brain regions implicate novel genes in Alzheimer’s disease. Acta Neuropathol 137, 557–569 (2019). https://doi.org/10.1007/s00401-019-01966-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-019-01966-5