Abstract
Mutations of isocitrate dehydrogenase 1 (IDH1) gene are most common in glioma, arguably preceding all known genetic alterations during tumor development. IDH1 mutations nearly invariably target the enzymatic active site Arg132, giving rise to the predominant IDH1R132H. Cells harboring IDH1R132H-heterozygous mutation produce 2-hydroxyglutarate (2-HG), which results in histone and DNA hypermethylation. Although exogenous IDH1R132H transduction has been shown to promote anchorage-independent growth, the biological role of IDH1R132H in glioma remains debatable. In this study, we demonstrate that heterozygous IDH1R132H suppresses but hemizygous IDH1R132H promotes anchorage-independent growth. Whereas genetic deletion of the wild-type allele in IDH1R132H-heterozygous cells resulted in a pronounced increase in neurosphere genesis, restoration of IDH1 expression in IDH1R132H-hemizygous cells led to the contrary. Conversely, anchorage-independent growth was antagonistic to the mutant IDH1 function by inhibiting gene expression and 2-HG production. Furthermore, we identified that in contrast to IDH1R132H-hemizygous neurosphere, IDH1R132H-heterozygous cells maintained a low level of reducing power to suppress neurosphere genesis, which could be bypassed, however, by the addition of reducing agent. Taken together, these results underscore the functional importance of IDH1 mutation heterozygosity in glioma biology and indicate functional loss of mutant IDH1 as an escape mechanism underlying glioma progression and the pathway of redox homeostasis as potential therapeutic targets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Somatic mutations of the isocitrate dehydrogenase 1 (IDH1) gene occur in > 70% of World Health Organization grade II and grade III gliomas as well as secondary glioblastoma. These missense mutations affect primarily Arg132, giving rise to the predominant type IDH1R132H [36, 59]. IDH1 encodes a cytosolic enzyme that converts isocitrate to 2-oxoglutarate (2-OG) concomitant with the production of reduced nicotinamide adenine dinucleotide phosphate (NADPH). Tumors lacking IDH1 mutations often have functionally equivalent mutations in IDH2, a mitochondrial gene in the citric acid cycle. Similar mutations have also been found in other types of cancer, including acute myeloid leukemia [34, 55] and intrahepatic cholangiocarcinoma [42].
Regarded as arguably the earliest genetic event in glioma development [6, 30, 48, 52, 56], IDH mutations are believed to be oncogenic and drive glioma development and progression [5]. Mutant IDH is best characterized by the gain of a neomorphic activity to produce 2-hydroxyglutarate (2-HG) from NADPH-dependent reduction of 2-OG [12, 19, 55]. 2-HG inhibits multiple 2-OG-dependent dioxygenases, including histone demethylases and 5-methylcytosine hydroxylases [11, 58]. Likewise, IDH1R132H transduction inhibits histone demethylation and induces DNA hypermethylation in cell culture and animal models, and, furthermore, blocks cell differentiation [32, 44, 51]. In addition, small-molecule targeting of mutant IDH induces tumor cell differentiation by blocking 2-HG production [41, 53], even though the effect on glioma growth remains controversial [41, 50].
Interestingly, IDH-mutant glioma patients are known to have a favorable outcome as compared with IDH-wildtype glioma patients [6, 36, 59]. In keeping with this, IDH1R132H expression in transduced astrocytes and glioblastoma cells inhibited rather than stimulated tumor growth [4, 8]. Comparative analyses of glioma samples also revealed attenuated oncogenic signaling, such as the phosphatidylinositol-3-kinase/protein kinase B (PI3K/AKT) pathway, glycolysis, and hypoxia, in IDH-mutant glioma compared with IDH-wildtype glioma [2, 4, 7, 9, 22]. We have, therefore, hypothesized that the biological consequence of IDH mutations in glioma is to ameliorate patient survival, at least in part, by inhibiting oncogenic signaling [22].
Apparently, this hypothesis is at odds with the key observations that suggest an oncogenic role of mutant IDH in glioma genesis. Specifically, immortalized human astrocytes, when transduced with IDH1R132H in comparison with wild-type IDH1, developed increased anchorage-independent growth in soft agar and neurosphere [1, 29, 51]; however, whether anchorage-independent growth maintains IDH1R132H transgene expression is not clear. It is noteworthy that IDH1 mutations are heterozygous in glioma [36, 59] and that wild-type IDH1 is obligatory for 2-HG production [27, 54]. Interestingly, loss of either mutant or wild-type IDH1 allele occurs in glioma patients, patient-derived xenograft growth, and ex vivo neurosphere culture [3, 9, 27], suggesting functional selection against mutant IDH1. In this study, we tested this possibility by investigating the role of wild-type IDH1 in neurosphere genesis using both loss-of-function and gain-of-function approaches in IDH1R132H-heterozygous and IDH1R132H-hemizygous glioma cells, respectively. We uncovered an unexpected relationship between IDH1R132H expression and anchorage-independent growth and, furthermore, provided a link between IDH1 mutation and redox homeostasis that dictates neurosphere growth.
Materials and methods
Plasmid construction and viral production
IDH1 and platelet-derived growth factor subunit B (PDGFB) cDNA were amplified from reverse-transcribed total RNA of normal human brain (BioChain). IDH1R132H was created using PCR-mediated site-directed mutagenesis. To coexpress the gene of interest with a fluorescent protein or firefly luciferase, we employed Gibson assembly (New England Biolabs) and engineered in pDONR221 (ThermoFisher Scientific) a marker gene and the gene of interest linked by P2A (Suppl. Figs. S1a, S2a). P2A allows stoichiometric expression after self-cleavage [28]. Through Gateway LR Clonase reactions, genes of interest were shuttled into the destination vector pLenti6.3/TO/V5-DEST [10] or RCASBP(A) [20, 31]. The inserts of these constructs were sequenced at the DNA Sequencing Core of University of Utah. Yellow fluorescent protein (YFP)* was a byproduct of YFP-IDH1R132H harboring a nonsense mutation at IDH1 Gly15.
Lentiviruses were produced in 293FT cells with the Viral Power HiPerform T-REx Vector kit (Invitrogen) according to the manufacturer’s protocol [10]. Retroviruses were generated according to the standard protocol [57]. Briefly, retroviral vectors were transfected into the DF-1 chicken embryonic fibroblasts using SuperFect transfection reagent (Qiagen) as per the manufacturer’s protocol. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 39 °C and in 5% CO2. Recombinant retroviruses were collected from condensed culture media 10 days after transfection. The recombinant viruses were purified through a 0.42-μm polyethersulfone syringe filter. Viral infection was performed in the presence of hexadimethrine bromide at a final concentration of 6 μg/mL.
Cell culture
IMA mut/IDH1 and IMA mut/− cells were cultured as described previously [27]. Briefly, adherent IMA cells were maintained in a complete growth medium consisting of 45% stem cell medium (knockout minimum essential medium with 20 ng/mL epidermal growth factor (EGF), 50 ng/mL basic fibroblast factor (bFGF), 1 × B-27 (Life Technologies), 1 × GlutaMAX (Life Technologies), 1 × non-essential amino acids solution (Life Technologies), 10 ng/mL leukocyte inhibitory factor, 2 μg/mL heparin), 45% DMEM, and 10% FBS. The IDH1R132H heterozygous and hemizygous status in IMA mut/IDH1 and IMA mut/− cells, respectively, was verified by DNA sequencing.
BT142 mut/− cells were purchased from American Type Culture Collection and maintained initially as neurosphere in culture medium as described [33]. Briefly, BT142 mut/− cells were cultured in DMEM/F12 (1:1) with 0.9% glucose, 4 mM l-glutamine, 25 µg/mL insulin, 100 µg/mL transferrin, 20 nM progesterone, 15 µM putrescine, 30 nM selenite, 20 ng/mL EGF, 100 ng/mL PDGF-AA, 20 ng/mL FGF, and 2 µg/mL heparan sulfate. BT142 mut/− cells were subsequently grown under adherent conditions in the IMA complete growth medium described above for lentiviral transduction at 2 multiplicities of infection to generate BT142 mut/YFP* and BT142 mut/YFP-IDH1. These resultant YFP-expressing cells were enriched by flow cytometry, and their IDH1R132H statuses were verified by DNA sequencing.
Astrocytes were extracted from brains of less than 2-day-old Nes-tva;Cdkn2afl/fl pups [40]. Brains were minced on ice and dissociated with 0.25% trypsin–EDTA for 15 min at 37 °C with agitation. The reaction was stopped by the addition of a growth medium (Roswell Park Memorial Institute medium supplemented with 10% FBS and 1 × l-glutamine/penicillin/streptomycin) and centrifuged to remove tissue debris. After overnight culture, cells were immortalized with Cre recombinase-mediated deletion of Cdkn2a. The resultant NA1 line was transduced first with a retrovirus expressing luc-PDGFB or mCherry-PDGFB and then with one expressing YFP-IDH1, YFP*, or YFP-IDH1R132H. Infection was conducted 3 times a day for a total of 3 consecutive days. The resultant cells with fluorescent signals were enriched by flow cytometry, and the IDH1R132H status was verified by DNA sequencing.
Gene expression and DNA copy number
Total RNA was extracted with the E.Z.N.A. total RNA kit I (Omega Biotek) or in tandem with genomic DNA extraction with ZR-Duet DNA/RNA Miniprep Kit (ZymoResearch) and converted with GoScript reverse transcriptase (Promega) according to the manufacturer’s protocol. Gene expression was determined by PCR with PDGFB primer set (forward 5′-TTGGCTCGTGGAAGAAGG-3′ and reverse 5′-GAGTTTGGGGCGTTTTGG-3′) at 55 °C for 30 cycles, with IDH1 primer set (forward 5′-GAGAAGAGGGTTGAGGAG-3′ and reverse 5′-CTTTTGGGTTCCGTCACT-3′) at 56 °C for 30 cycles, with YFP-IDH1 primer set (forward 5′-GGCCGACAAGCAGAAGAA-3′ and reverse 5′-GGCCCAGGAACAACAAAA-3′) at 57 °C for 30 cycles, with ACTB primer set (forward 5′-GCGAGAAGATGACCCAGA-3′ and reverse 5′-GGAAGGAAGGCTGGA AGA-3′) at 56 °C for 28 cycles, and with Actb primer set (forward 5′-TGCTATGTTGCTCTAGACTT-3′ and reverse 5′-CTCATCGTACTCCTGCTT-3′) at 55 °C for 28 cycles.
Genomic DNA from transduced NA1 cells was extracted as described above. PCR amplification was conducted for quantification of PDGFB and IDH1R132H transgenes with primer sets and conditions described above and mouse endogenous Il2 with the primer set (5′-CTAGGCCACAGAATTGAAAGATCT-3′ and reverse 5′-GTAGGTGGAAATTCTAGCATCATCC-3′) at 52 °C for 30 cycles.
Whole-cell extracts were prepared essentially as described previously [21] in a lysis buffer containing 20 mM HEPES pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 20% glycerol freshly supplemented with 0.5 mM dithiothreitol and protease inhibitor cocktail (Roche) [10]. Western blot analysis was performed as described previously [10]. Primary antibodies and corresponding dilutions were as follows: 1:5000 anti-β-actin (Sigma A5441), 10 μg/mL anti-FLAG (Sigma F3165), 1:500 anti-HA (Abcam ab13834), and 0.5 μg/mL anti-V5 (Sigma V8012). Secondary antibodies were 1:10,000 anti-mouse HRP (JIR 115-035-146) and 1:10,000 anti-rabbit HRP (JIR 711-035-152).
2-HG analysis
Approximately 3 × 107 BT142 and 1 × 108 NA1 cells were washed with phosphate-buffered saline, pelleted, flash frozen in liquid nitrogen, and stored at 80 °C prior to gas chromatography–mass spectrometry analysis at the Metabolomics Core of University of Utah using an Agilent 5977B GC–MS with HES source and an Agilent 7693A automatic liquid sampler. Collected data in triplicate were normalized by mean centering to the internal standard d4-succinate.
2-HG concentrations in adherent and neurosphere cultures were determined using D-2-Hydroxyglutarate (D2HG) Colorimetric Assay Kit (BioVision), as per manufacturer recommendations. For each replicate approximately 1 × 105 cells were split into three wells designated as sample, 5 nmol D2HG spiked sample, and sample background. D2HG in each cell type was determined in triplicate, calculated according to the manufacturer’s protocol, and expressed as nmol/mg protein.
Neurosphere growth and quantitative assay
Neurosphere growth was observed by seeding 10,000 cells per well in a 24-well plate with neural stem-cell media consisting of Neurobasal media (Invitrogen) supplemented with B-27, 10 ng/mL bFGF, and 20 ng/mL EGF (Invitrogen) for all cell types analyzed. Bright-field microscopy images were acquired on day 7 for comparison. For quantitative assays, each cell type was seeded in a 96-well plate at 5 cells per well for 48 wells in triplicate in the same medium. The media was replenished with the addition of 20 µL of neural stem-cell medium at day 4, 7, and 10. Neurosphere genesis was determined by counting spheres of ≥ 50 µm on day 14. Each replicate was converted to percentage in reference to the total number of cells seeded, and unpaired t tests were performed to determine two-tailed p values.
Flow cytometry of fluorescent cells
Neurosphere cultures were collected and sifted with a 40-µm cell strainer and a 100-µm cell strainer to remove single cells and cell aggregates. Adherent and neurosphere cells were then dissociated enzymatically and analyzed in triplicate by flow cytometry (BD FACSCanto, BD Biosciences) with BD FACSDiva Software (BD Biosciences) and FlowJo (FlowJo). Populations of YFP+ and/or mCherry+ cells were obtained and the mean fluorescent intensities were analyzed.
Fluorescent microscopy
Adherent and neurosphere cultures were stained with 10 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) for 30 min and 1 h, respectively, prior to fluorescent microscopy using an Axiovert 200 inverted microscope with Axiovision microscopy software (Carl Zeiss, Germany) or a Nikon A1R confocal microscope with NIS-Elements confocal software (Nikon Instruments). The images of mCherry, YFP, CellROX Deep Red, and DAPI were converted with an open-source image analysis platform (Fiji) [46] and presented in single colors and merged.
Redox study
Oxidative stress was measured using CellROX Deep Red Reagent (Invitrogen) as per manufacturer protocol. Adherent cells were plated at 20% confluence in a 4-well culture chamber slide and cultured for 3 days prior to the assay. Neurosphere culture was done for 7 days prior to the assay. After staining with the CellROX reagent, cells were fixed with 3.7% paraformaldehyde and stained with 10 μg/mL DAPI. Fluorescent microscopy images were obtained using a Nikon A1R confocal microscope with NIS-Elements confocal software (Nikon Instruments).
GSH/GSSG-Glo Assay (Promega) was used according to the manufacturer’s recommendations to assess GSH/GSSG ratio in adherent and neurosphere samples. For each replicate, 2 × 104 cells were used to assess in triplicate total GSH and GSSG. GSH/GSSG ratios and the GSH and GSSG concentrations were calculated from GSH standard curves. Total NAD(P)H levels were analyzed using NAD(P)H-Glo Detection System (Promega) according to the manufacturer’s recommendations. Briefly, 1 × 105 cells were lysed with 0.2 N NaOH prior to the addition of the detection reagent.
Intracellular redox state was altered by the continuous treatment with 30 µM hydrogen peroxide or 1 mM NAC at cell seeding, day 3, and day 6 before microscopy and flow cytometry analyses. For quantitative neurosphere assays, cells were incubated with NAC at a final concentration of 1 mM at seeding (day 0). The media was replenished by the addition of 20 μL of neural stem-cell medium containing 1 mM NAC at days 4, 7, and 10.
Statistical analysis
Students t tests and one-way ANOVA were performed using the GraphPad Prism 7 to determine means and standard deviations along with statistical significance of experimental data in replicates of three or as indicated. Significance was defined as *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Results
Mutant IDH1 markedly suppresses neurosphere genesis in a wild-type IDH1-dependent fashion
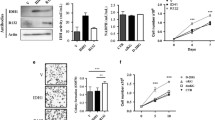
The anaplastic astrocytoma cell line IMA mut/IDH1 is IDH1R132H heterozygous [27], whereas the anaplastic oligoastrocytoma cell line BT142 mut/− is IDH1R132H hemizygous, derived from the heterozygous BT142 [33]. Interestingly, when cultured under neurosphere conditions, BT142 mut/− cells developed robust and much larger neurosphere than IMA mut/IDH1 cells (Fig. 1a, left). To quantify neurosphere genesis, we seeded cells in 96-well plates at 5 cells per well, which had been determined empirically for neurosphere formation at ≤ 1 per well. The quantitative assay revealed a greater than sevenfold difference between BT142 mut/− and IMA mut/IDH1 cells (5.83 versus 0.83%) (Fig. 1a, right), which apparently suggested an inhibitory effect of wild-type IDH1 on neurosphere genesis.
Mutant IDH1 suppresses neurosphere genesis in a wild-type allele-dependent fashion. a Comparison of BT142 mut/− and IMA mut/IDH1 neurosphere growth in micrographs (left) and quantitative assays (right). Scale bars: 100 μm. b Significant reduction of neurosphere genesis in BT142 mut/− with exogenous wild-type IDH1 in reference to YFP*. c Significant increase of neurosphere genesis in IMA mut/IDH1 (n = 4) after deletion of endogenous wild-type IDH1 (n = 3). d Suppression of neurosphere genesis by expression of mutant IDH1 (mIDH1) but not wild-type IDH1 (IDH1) in luc-PDGFB transduced mouse astrocytes. ***p < 0.001; ****p < 0.0001
To test this notion in the same genetic background, we introduced wild-type IDH1 into BT142 mut/− cells using a lentivirus that expressed nuclear YFP, P2A, and IDH1 in tandem to establish the BT142 mut/YFP-IDH1 line (Suppl. Fig. S1a). A control line BT142 mut/YFP* was created with another lentivirus expressing the same YFP-IDH1 transcript but with a nonsense mutation at IDH1 Gly15. After fluorescent enrichment by flow cytometry, the BT142 mut/YFP-IDH1 line was authenticated by the expression of YFP-IDH1 mRNA and hemagglutinin (HA)-tagged IDH1 protein, the presence of both wild-type and mutant IDH1 sequences at Arg132, and a greater than sixfold increase in 2-HG (Suppl. Fig. S1b–e). Importantly, restoration of IDH1 expression in BT142 mut/− resulted in significant reduction of neurosphere growth compared with BT142 mut/YFP* (Suppl. Fig. S1f). Further quantitative analysis showed a 55% reduction in neurosphere genesis with the introduction of wild-type IDH1 (2.78% in BT142 mut/YFP-IDH1 versus 6.11% in BT142 mut/YFP*, Fig. 1b). Conversely, deletion of wild-type IDH1 allele from IMA mut/IDH1, which resulted in an 87-fold decrease in 2-HG levels [27], increased neurosphere genesis by more than threefold (3.05% in IMA mut/− versus 0.83 in IMA mut/IDH1) (Fig. 1c). Taken together, these results support the involvement of wild-type IDH1 in suppressing neurosphere genesis but also imply the role of mutant IDH1 in promoting neurosphere growth, as reported previously [1, 24, 51].
An alternative interpretation of the above results, however, is that mutant IDH1 requires a wild-type allele for the inhibition of neurosphere genesis because of its functional dependence on wild-type IDH1 to produce 2-HG [27, 54]. To distinguish these two possibilities, we investigated whether expression of wild-type or mutant IDH1 suppressed neurosphere growth in a wild-type IDH1 background. To that end, we employed a mouse astrocyte line NA1 that had been transformed with an avian retrovirus, RCAS/luc-PDGFB, expressing firefly luciferase (luc) and PDGFB (Fig. S2a), and was further transduced with RCAS/YFP-mIDH1 (expressing YFP-IDH1R132H) or RCAS/YFP-IDH1. After validation of transgene expression, sequencing confirmation, and 2-HG production in YFP-mIDH1 cells (Fig. S2b–e), quantitative analysis confirmed an approximately two-thirds reduction of neurosphere genesis by YFP-mIDH1 in comparison with YFP-IDH1 (0.88 versus 2.4%) (Fig. 1d). Thus, these results support the alternative hypothesis that mutant IDH1 markedly suppresses neurosphere genesis in a wild-type allele-dependent fashion, whereas loss of the wild-type allele promotes neurosphere growth.
Selection against IDH1R132H transgene during neurosphere but not adherent culture
Previous studies showed that mutant IDH1 promoted anchorage-independent growth of immortalized human astrocytes after IDH1R132H transduction [1, 24, 29, 51]; however, whether the expression of mutant IDH1 was maintained in those conditions was unclear. To visualize transgene expression within neurosphere, we employed fluorescent markers by replacing luc-PDGFB with mCherry-PDGFB (Suppl. Fig. S2a) and compared YFP-mIDH1 expression levels in reference to YFP*. After confirmation of transgene expression in NA1 astrocytes (Suppl. Fig. S3a), we observed an approximately sevenfold decrease in neurosphere growth of YFP-mIDH1 cells in reference to YFP* cells (Fig. S3b). Importantly, quantification of fluorescent microscopy images revealed only 35% of YFP+ cells within YFP-mIDH1 neurosphere in contrast to 80% of YFP+ within YFP* neurosphere (Fig. 2a, b), despite equivalent population of YFP+ cells in adherent culture between the two cell types (Fig. 2c). These data suggested specific selection against IDH1R132H transgene in neurosphere culture.
Suppression of YFP signal in PDGFB-transformed NA1 neurosphere expressing YFP-mIDH1. a Fluorescent microscopy of mCherry-PDGFB transformed neurosphere expressing YFP* (i, ii) and YFP-mIDH1 (iii, iv) demonstrating a marked suppression of YFP signal in YFP-mIDH1 neurosphere in comparison with YFP* neurosphere. b The YFP+ cell percentages were quantified in reference to DAPI-stained nuclei in YFP* neurosphere (n = 5) and YFP-mIDH1 neurosphere (n = 8). The analysis revealed significant reduction of YFP+ cell percentage in YFP-mIDH1 neurosphere. Unpaired t test was performed with two-tailed p value. ***p < 0.001. c Fluorescent microscopy of adherent NA1 expressing mCherry-PDGFB and YFP* (top) and mCherry-PDGFB and YFP-mIDH1 (bottom) showing equivalent population of YFP+ cells. Scale bar, 50 μm
To corroborate this finding, we determined the distributions of mCherry+YFP+ and mCherry+YFP− cells within the neurosphere with flow cytometry. Whereas ~ 90% of adherent YFP* and YFP-mIDH1 cells were mCherry+YFP+, in neurosphere culture only 28% of YFP-mIDH1 versus 83% of YFP* cells were double positive (Fig. 3a). Statistical analysis confirmed that in YFP-mIDH1 neurosphere less than one-third of cells were mCherry+YFP+ versus > 82% in YFP* neurosphere; however, in adherent culture almost 90% adherent YFP-mIDH1 cells were mCherry+YFP+ (Fig. 3b), consistent with the notion that mutant IDH1 transgene is selected against in neurosphere. Further analysis showed a marked reduction of log2 ratio of YFP+/YFP− population to − 1.20 in YFP-mIDH1 neurosphere compared with 2.37 in YFP* neurosphere and > 40% loss of YFP fluorescent intensity in YFP-IDH1 neurosphere (Fig. 3c, d).
Quantitative measurement of YFP+ cell population in PDGFB-transformed NA1 neurosphere expressing YFP-mIDH1. a Representative images of flow cytometry of the NA1 cells as described above cultured under adherent (top) and neurosphere (bottom) conditions. YFP and mCherry signals were plotted along x and y axes, respectively. The percentage of cells in each quadrant is indicated. b Statistical comparison of mean percentages of mCherry+YFP− and mCherry+YFP+ between YFP* and YFP-mIDH1 cells cultured under adherent (top) and neurosphere (bottom) conditions. c Comparison of YFP* and YFP-mIDH1 cells in neurosphere showing extremely significant reduction in YFP+/YFP− log2 ratio. d Comparison of YFP-mIDH1 neurosphere with YFP* neurosphere showing a significant decrease of YFP fluorescent intensity. Unpaired t tests were performed with two-tailed p values. **p < 0.01; ***p < 0.001; ****p < 0.0001
To obtain direct evidence for the suppression of IDH1R132H transgene in neurosphere, we detected significant reduction of IDH1R132H but not PDGFB expression by RT-PCR (Fig. 4a), which was further confirmed by using YFP-IDH1R132H as the amplicon for the elimination of cross-amplification of endogenous Idh1 (Suppl. Fig. S3c). The suppression of IDH1R132H expression was not a result of loss of DNA copy number (Fig. 4b). We conclude, therefore, that IDH1R132H transgene is incompatible with anchorage-independent growth.
Reduction of YFP-mIDH1 transcript levels in PDGFB-transformed NA1 neurosphere. a RT-PCR (left) and statistical analysis (right) showing differential expression of IDH1R132H between YFP* neurosphere and YFP-mIDH1 neurosphere. b Genomic DNA amplification (left) and statistical analysis (right) showed no statistical differences in genomic copy number between YFP* neurosphere and YFP-mIDH1 neurosphere. Endogenous β-actin (Actb) and interleukin 2 (Il2) were used as controls. Unpaired t tests were performed and are presented with two-tailed p values. ***p < 0.001
Selection against IDH1R132H function in IDH1R132H-heterozygous neurosphere but not adherent cells
In light of the requirement of wild-type IDH1 for the suppression of neurosphere growth, we speculated that wild-type IDH1 transgene is likely selected against in BT142 mut/YFP-IDH1 neurosphere owing to its obligatory role for mutant IDH1 function [27, 54]. Indeed, fluorescent microscopy revealed a marked reduction of fluorescent signal and YFP+ cells in YFP-IDH1 neurosphere compared with YFP* cells (Fig. 5a). Flow cytometry analysis revealed that less than one-third of the cells in YFP-IDH1 neurosphere were YFP+ in contrast to almost three-quarters in YFP* neurosphere, whereas equivalent percentages of YFP+ cells were observed under adherent growth conditions between the two cell types (Figs. 5b, c, 6a). Further analyses of log2 ratio of YFP+/YFP− population and YFP fluorescent intensity corroborated the extremely significant suppression of YFP signal specific to YFP-IDH1 neurosphere but not adherent culture (Fig. 5d, e). Furthermore, a marked reduction of YFP-IDH1 transcripts in neurosphere was observed (Fig. 6c).
Functional suppression of mutant IDH1 in IDH1R132H-heterozygous neurosphere. a Fluorescent microscopy of BT142 mut/− neurosphere expressing YFP* (left) and YFP-IDH1 (right) showed a marked suppression of YFP signal in BT142 mut/YFP-IDH1 neurosphere in comparison with BT142 mut/YFP* neurosphere. Statistical comparison of mean percentages of mCherry+YFP− and mCherry+YFP+ between BT142 mut/YFP* and BT142 mut/YFP-IDH1 cells cultured under adherent (b) and neurosphere (c) conditions. Comparison of BT142 mut/YFP-IDH1 neurosphere with BT142 mut/YFP* neurosphere showing extremely significant reduction of YFP+/YFP− log2 ratio (d) and YFP fluorescent intensity (e). Unpaired t tests were performed with two-tailed p values. One-way ANOVA of 2-HG concentrations in adherent and neurosphere conditions showing extremely significant decreases in BT142 mut/YFP-IDH1 (f) and IMA mut/IDH1 (g) neurosphere. **p < 0.01; ***p < 0.001; ****p < 0.0001
Lower levels of ROS in IDH1R132H-heterozygous cells. a Fluorescent microscopy of BT142 mut/YFP* (i, ii) and BT142 mut/YFP-IDH1 (iii, iv) cells cultured under adherent (i, iii) and neurosphere (ii, iv) conditions showing marked reduction of YFP signal in BT142 mut/YFP-IDH1 neurosphere in comparison with BT142 mut/YFP*. ROS levels are indicated by the intensity of Deep Red, as also shown in merged images with DAPI. b Flow cytometry analysis of ROS in adherent (red) and neurosphere (blue) culture of BT142 mut/YFP* (top) and BT142 mut/YFP-IDH1 (bottom). c RT-PCR showing pronounced reduction of YFP-IDH1 transcripts in BT142 mut/YFP-IDH1 neurosphere compared with BT142 mut/YFP* neurosphere. d Comparison of ROS levels between BT142 mut/− (i) and IMA mut/IDH1 (ii) and between luc-PDGFB transduced NA1/YFP* (iii) and NA1/YFP-mIDH1 (iv) cultured under adherent conditions
To provide evidence for functional inhibition of IDH1R132H in BT142 mut/YFP-IDH1 neurosphere, we compared the 2-HG concentration in adherent culture with that in neurosphere. In keeping with IDH1 transgene inhibition, the 2-HG concentration in BT142 mut/YFP-IDH1 neurosphere was reduced approximately by two-thirds in comparison with adherent culture; however, no such change was observed in BT142 mut/− cells (Fig. 5f). Moreover, we confirmed this finding in the native IDH1R132H-heterozygous IMA mut/IDH1 cells, where the 2-HG concentration in neurosphere was reduced by more than 53% (Fig. 5g). Taken together, these results indicate that anchorage-independent growth selects against mutant IDH1 function by targeting either wild-type or mutant IDH1 transgene, which is in accordance with the eventual loss of wild-type IDH1 allele in BT142 and secondary glioblastomas and of mutant IDH1 allele in brain tumor stem-cell cultures [9, 27].
Increased reactive oxygen species in IDH1R132H-hemizygous cells
Anchorage-independent growth is associated with elevated levels of reactive oxygen species (ROS) when malignant cells detach from an extracellular matrix for deregulated proliferation [26, 45]. Accordingly, we utilized the CellROX Deep Red reagent to assess ROS levels in neurosphere compared with adherent culture. Fluorescent microscopy showed increased ROS levels, as indicated by Deep Red fluorescent signal, in the neurosphere of both BT142 mut/YFP* and BT142 mut/YFP-IDH1 cells in comparison with their respective adherent counterparts (Fig. 6a). This finding was confirmed by a more than fourfold increase of fluorescent signal in neurosphere by flow cytometry (Fig. 6b).
Interestingly, we observed an approximately twofold increase of ROS in BT142 mut/YFP* cells compared with BT142 mut/YFP-IDH1 cells, which suggested that loss of the wild-type allele in IDH1R132H heterozygous cells increases ROS levels, as shown similarly in Idh1-deficient hepatocytes [23]. In keeping with this, the IDH1R132H-hemizygous BT142 mut/− cells showed much higher levels of ROS than the IDH1R132H-heterozygous IMA mut/IDH1 cells (Fig. 6d). Furthermore, deletion of IDH1 allele from IMA mut/IDH1 cells markedly increased ROS levels (Fig. 8e). To provide evidence that IDH1R132H requires a wild-type allele to keep ROS levels low, we observed that IDH1R132H expression decreased ROS levels in PDGFB-transduced NA1 (Fig. 6d). Taken together, these results not only confirmed the increased ROS levels in anchorage-independent growth, but, more importantly, indicated the loss of wild-type allele as a means of elevating ROS levels and promoting neurosphere genesis in IDH1-mutant glioma cells.
IDH1R132H-heterozygous cells maintain low levels of reducing power to suppress neurosphere growth
To explore the possibility that redox homeostasis differs between IDH1R132H-hemizygous and IDH1R132H-heterozygous cells, we determined their reduced glutathione-to-oxidized glutathione (GSH/GSSG) ratio and the GSH and GSSG concentrations. BT142 mut/YFP* neurosphere had a GSH/GSSG ratio of 19 compared with a ratio of 4 in BT142 mut/YFP-IDH1 neurosphere, in contrast to their respective ratios of 7.6 and 6.4 in adherent culture (Fig. 7a). The low GSH/GSSG ratio in BT142 mut/YFP-IDH1 neurosphere was attributable to the modest increase of GSH concentration despite 88% increase of GSSG concentration compared with adherent conditions (Fig. 7b, c). By contrast, BT142 mut/YFP* neurosphere gained a high ratio owing to 54% increase of GSH concentration and 40% decrease of GSSG concentration. Similar results were obtained in IMA mut/− and IMA mut/IDH1 cells (Fig. 7e–g). In addition to the lack of GSH increase in IDH1R132H-heterozygous neurosphere, no NAD(P)H increase was observed in reference to adherent culture, whereas IDH1R132H-hemizygous neurosphere increased NAD(P)H levels by an average of twofold (Fig. 7d, h). We speculated, therefore, that it is the reducing power rather than ROS alone that determines neurosphere genesis, i.e., cells capable of producing sufficient reducing power can meet the demand of increased ROS for anchorage-independent growth.
Redox homeostasis controls neurosphere genesis. GSH/GSSG ratios were significantly increased in IDH1R132H-hemizygous BT142 mut/YFP* (a) and IMA mut/− (e) neurosphere in contrast to their heterozygous controls. The increased ratio was attributable to the increase of GSH concentration and the concomitant decrease of GSSG concentration in IDH1R132H-hemizygous BT142 mut/YFP* (b, c) and IMA mut/− (f, g) neurosphere. The decreased GSH/GSSG ratios in BT142 mut/YFP-IDH1 and IMA mut/IDH1 resulted primarily from increased GSSG concentrations (c, g). NAD(P)H levels were increased only in IDH1R132H-hemizygous neurosphere (d, h). One-way ANOVA was performed. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
In support of this theory, continuous treatment of BT142 mut/YFP* with hydrogen peroxide (H2O2) at 30 μM resulted in a pronounced reduction of neurosphere growth, whereas treatment of BT142 mut/YFP-IDH1 cells with 1 mM reducing N-acetyl cysteine (NAC) markedly increased neurosphere genesis (Fig. 8a). Similar results were obtained with H2O2 treatment of IMA mut/− and NA1/YFP* cells and NAC treatment of IMA mut/IDH1 and NA1/YFP-mIDH1cells (Suppl. Fig. S4a, b). Furthermore, quantitative analysis of NAC treatment revealed a nearly twofold increase of neurosphere genesis for BT142 mut/YFP-IDH1 cells (from 2.78 to 5.28%) and a > 3.5-fold increase for IMA mut/IDH1 cells (from 0.83 to 3.06%) (Fig. 8b). Likewise, NAC treatment also enhanced BT142 mut/YFP* neurosphere growth, whereas H2O2 treatment had no significant effect on BT142 mut/YFP-IDH1 neurosphere growth (Fig. 8c). These results suggest that NAC promotes neurosphere genesis by providing reducing power to meet the requirement of increased ROS. Therefore, we expected that treated IDH1R132H-heterozygous neurosphere would contain increased, rather than decreased, ROS. Indeed, a striking increase of Deep Red signal was observed in NAC-treated neurosphere in both BT142 mut/YFP-IDH1 and IMA mut/IDH1 cells (Fig. 8d, e; Suppl. Fig. S4c). By contrast, H2O2 treatment failed to elevate ROS further. Taken together, these results not only indicate the importance of reducing power in controlling neurosphere genesis but also suggest “shedding” of the wild-type allele as a means of gaining reducing power for IDH1R132H glioma progression.
Redox reagents alter neurosphere genesis. a Comparison of BT142 mut/YFP* neurosphere growth with and without H2O2 treatment and BT142 mut/YFP-IDH1 neurosphere growth with and without NAC treatment as specified. Scale bar 100 μm. b Quantitative analysis of neurosphere genesis of BT142 mut/YFP-IDH1 and IMA mut/IDH1 in response to NAC treatment. Unpaired t tests were performed with two-tailed p values. **p < 0.01; ***p < 0.001. c Comparison of BT142 mut/YFP* neurosphere growth with and without NAC treatment and BT142 mut/YFP-IDH1 neurosphere growth with and without H2O2 treatment as specified. Scale bar 100 μm. Increased ROS levels, as indicated by Deep Red signal, after NAC treatment of BT142 mut/YFP-IDH1 (d) and IMA mut/IDH1 (e) cells in comparison with untreated. No obvious increase of ROS after H2O2 treatment of IDH1R132H-hemizygous cells
Discussion
This study provides cogent evidence for the absolute requirement of a wild-type allele for mutant IDH1 to suppress anchorage-independent growth through the maintenance of redox homeostasis. In response to a need for increased intracellular ROS for anchorage-independent growth, IDH1R132H-heterozygous cells inhibit growth by limiting the production of reducing power, whereas IDH1R132H-hemizygous cells bypass the redox checkpoint to generate more reducing power. In particular, we demonstrated that restoration of wild-type IDH1 expression in IDH1R132H-hemizygous cells suppressed reducing power generation and neurosphere genesis; conversely, deletion of the wild-type allele in IDH1R132H-heterozygous cells gave rise to the contrary. Our results are consistent with the genetic and functional characteristics of IDH1 mutations being heterozygous and dependent on a wild-type allele for 2-HG production [27, 36, 54, 59].
It should be noted that previous studies reported increased neurosphere growth or colony formation using IDH1R132H transgene in comparison with wild-type IDH1 [1, 24, 29, 51]; however, whether these conditions maintained IDH1R132H expression was not examined. We found selection against either wild-type or mutant IDH1 transgene in neurosphere, but not adherent, culture and marked reduction of 2-HG concentrations in neurosphere, which indicate the functional incompatibility between heterozygous IDH1R132H mutation and anchorage-independent growth. Pusch et al. reported that human gliomas select against rare IDH1 mutations such as R132G, R132C, and R132S because of their higher 2-HG levels in comparison with the prevalent R132H [39]. The ‘toxic” effect of high levels of 2-HG on glioma biology seems to be in agreement with the growth-suppressive function of 2-HG achieved by extending the lifespan of Caenorhabditis elegans [15]. Although the heterozygous state of rare IDH1 mutations in glioma remain to be determined, our results shed light on the perplexing loss of either wild-type or mutant IDH1 allele during neurosphere culture and tumor progression [3, 9, 27] and may also account for the lack of neurosphere growth of murine Idh1R132H neural progenitor cells [1]. It remains to be seen, however, whether our finding is relevant to other IDH-mutant cancer types such as acute myeloid leukemia and intrahepatic cholangiocarcinoma [14, 34, 42, 55] because, unlike IDH-mutant glioma, no IDH mutation-associated survival benefits have been identified in these cancers [17, 37]. Additionally, it seems unlikely that our study is relevant to cancers harboring IDH2 mutations because mutant IDH2 produces 2-HG independent of wild-type allele [54].
Our results provide a mechanistic understanding of how redox homeostasis plays an important role in mediating the suppression of neurosphere growth in heterozygous IDH1R132H cells. Although adherent IDH1R132H-hemizygous and IDH1R132H-heterozygous cells shared similar profiles of redox homeostasis, under neurosphere conditions the two cell types manifested remarkable differences in GSH/GSSG ratio and NAD(P)H level (Fig. 7), which were also observed between IDH1-wildtype and IDH1-mutant cells upon ionizing radiation or endotoxin exposure [23, 35]. Although loss of wild-type IDH1 allele resulted in marked reduction of 2-HG levels (Fig. 5f, g) [27, 54], whether IDH1R132H-hemizygous cells have decreased 2-OG levels is unclear; however, metabolic reprogramming for maintaining 2-OG levels through mitochondrial IDH2 and conversion from glutamine would be expected [38]. Therefore, loss of IDH1R132H heterozygosity enables IDH1R132H-hemizygous cells to produce reducing power for anchorage-independent growth. Likewise, the reducing agent NAC promotes such growth by allowing IDH1R132H-heterozygous cells to circumvent the redox checkpoint.
The importance of reducing power in coping with ROS increases in anchorage-independent growth [26, 45] is consistent with NADPH production as one of the emerging hallmarks of cancer metabolism [38]. Reductive carboxylation supports redox homeostasis during anchorage-independent growth [26], but IDH1-mutant cells exhibit decreased reductive glutamine metabolism concomitant with increased oxidative metabolism of the citric acid cycle [18]. The divergence between oxidative metabolism in heterozygous IDH1-mutant cells and reductive metabolism in anchorage-independent growth may pose a metabolic conflict, which is consistent with the suppression of neurosphere genesis and conversely the selection against mutant IDH1 function. Gaining an understanding of how heterozygous IDH1-mutant glioma cells maintain a low level of redox homeostasis should help elucidate the mechanism by which hemizygous IDH1-mutant cells elevate the level of redox homeostasis to promote anchorage-independent growth. Owing to its dependence on wild-type IDH1 [26], reductive glutamine metabolism seems a less likely mechanism because hemizygous IDH1-mutant cells showed increased NAD(P)H levels. Generation of reducing power involves several major metabolic pathways, including the 5′ adenosine monophosphate-activated protein kinase (AMPK)–fatty acid oxidation pathway, the pentose phosphate pathway, the serine–glycine one-carbon pathway, and the malic enzyme-mediated conversion of malate to pyruvate [13, 25, 38, 47]. Investigation of these pathways is warranted to identify the one that is regulated by mutant IDH1.
Results from this study corroborate the theory that IDH1 mutations in glioma are anti-oncogenic [22], which may account for the lack of glioma development in Idh1R132H knock-in mice despite robust production of 2-HG [1, 43] and conversely the shortened survival of xenografted animals treated with an IDH1-mutant inhibitor [49]. Of note, despite the observation of cellular and molecular features associated with glioma genesis surrounding the subventricular zone of the Idh1R132H knock-in mice, no tumor was detected [1]. The anti-oncogenic theory is also consistent with the observations that IDH1 mutations are an early, rather than late, event of glioma development [6, 56] and glioma progression is consistently associated with functional loss, but not gain, of IDH1 mutations [3, 9, 27].
Together with previous studies, this study not only underscores the functional importance of the heterozygosity of IDH1 mutations but also suggests inactivation/attenuation of mutant IDH1 function as an escape mechanism underlying glioma progression through genetic changes of IDH1 locus and/or downstream epigenetic changes, including global DNA demethylation [7] and resultant IGFBP2 expression [16, 22, 60]. A greater understanding of the escape mechanisms may provide novel therapeutic targets for the prevention of glioma progression.
References
Bardella C, Al-Dalahmah O, Krell D, Brazauskas P, Al-Qahtani K, Tomkova M et al (2016) Expression of Idh1R132H in the murine subventricular zone stem cell niche recapitulates features of early gliomagenesis. Cancer Cell 30:578–594. https://doi.org/10.1016/j.ccell.2016.08.017
Birner P, Pusch S, Christov C, Mihaylova S, Toumangelova-Uzeir K, Natchev S et al (2014) Mutant IDH1 inhibits PI3K/Akt signaling in human glioma. Cancer 120:2440–2447. https://doi.org/10.1002/cncr.28732
Borodovsky A, Salmasi V, Turcan S, Fabius AWM, Baia GS, Eberhart CG et al (2013) 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft. Oncotarget 4:1737–1747
Bralten LBC, Kloosterhof NK, Balvers R, Sacchetti A, Lapre L, Lamfers M et al (2011) IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Ann Neurol 69:455–463. https://doi.org/10.1002/ana.22390
Cairns RA, Mak TW (2013) Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov 3:730–741. https://doi.org/10.1158/2159-8290.CD-13-0083
Cancer Genome Atlas Research Network (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372:2481–2498. https://doi.org/10.1056/NEJMoa1402121
Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA et al (2016) Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164:550–563. https://doi.org/10.1016/j.cell.2015.12.028
Chen R, Nishimura MC, Kharbanda S, Peale F, Deng Y, Daemen A et al (2014) Hominoid-specific enzyme GLUD2 promotes growth of IDH1R132H glioma. Proc Natl Acad Sci USA 111:14217–14222. https://doi.org/10.1073/pnas.1409653111
Chesnelong C, Chaumeil MM, Blough MD, Al-Najjar M, Stechishin OD, Chan JA et al (2014) Lactate dehydrogenase A silencing in IDH mutant gliomas. Neurooncology 16:686–695. https://doi.org/10.1093/neuonc/not243
Choi H, Gillespie DL, Berg S, Rice C, Couldwell S, Gu J et al (2015) Intermittent induction of HIF-1α produces lasting effects on malignant progression independent of its continued expression. PLoS ONE 10:e0125125. https://doi.org/10.1371/journal.pone.0125125
Chowdhury R, Yeoh KK, Tian Y-M, Hillringhaus L, Bagg EA, Rose NR et al (2011) The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep 12:463–469. https://doi.org/10.1038/embor.2011.43
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM et al (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462:739–744. https://doi.org/10.1038/nature08617
Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD (2014) Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510:298–302. https://doi.org/10.1038/nature13236
Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ et al (2010) DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 17:13–27. https://doi.org/10.1016/j.ccr.2009.11.020
Fu X, Chin RM, Vergnes L, Hwang H, Deng G, Xing Y et al (2015) 2-Hydroxyglutarate inhibits ATP synthase and mTOR signaling. Cell Metab 22:508–515. https://doi.org/10.1016/j.cmet.2015.06.009
Fuller GN, Rhee CH, Hess KR, Caskey LS, Wang R, Bruner JM et al (1999) Reactivation of insulin-like growth factor binding protein 2 expression in glioblastoma multiforme: a revelation by parallel gene expression profiling. Cancer Res 59:4228–4232
Goyal L, Govindan A, Sheth RA, Nardi V, Blaszkowsky LS, Faris JE et al (2015) Prognosis and clinicopathologic features of patients with advanced stage isocitrate dehydrogenase (IDH) mutant and IDH wild-type intrahepatic cholangiocarcinoma. Oncologist 20:1019–1027. https://doi.org/10.1634/theoncologist.2015-0210
Grassian AR, Parker SJ, Davidson SM, Divakaruni AS, Green CR, Zhang X et al (2014) IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res 74:3317–3331. https://doi.org/10.1158/0008-5472.CAN-14-0772-T
Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG et al (2010) Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 207:339–344. https://doi.org/10.1084/jem.20092506
Holmen SL, Williams BO (2005) Essential role for Ras signaling in glioblastoma maintenance. Cancer Res 65:8250–8255. https://doi.org/10.1158/0008-5472.CAN-05-1173
Huang LE, Arany Z, Livingston DM, Bunn HF (1996) Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem 271:32253–32259
Huang LE, Cohen AL, Colman H, Jensen RL, Fults DW, Couldwell WT (2017) IGFBP2 expression predicts IDH-mutant glioma patient survival. Oncotarget 8:191–202. https://doi.org/10.18632/oncotarget.13329
Itsumi M, Inoue S, Elia AJ, Murakami K, Sasaki M, Lind EF et al (2015) Idh1 protects murine hepatocytes from endotoxin-induced oxidative stress by regulating the intracellular NADP(+)/NADPH ratio. Cell Death Differ 22:1837–1845. https://doi.org/10.1038/cdd.2015.38
Izquierdo-Garcia JL, Viswanath P, Eriksson P, Cai L, Radoul M, Chaumeil MM et al (2015) IDH1 mutation induces reprogramming of pyruvate metabolism. Cancer Res 75:2999–3009. https://doi.org/10.1158/0008-5472.CAN-15-0840
Jeon S-M, Chandel NS, Hay N (2012) AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature 485:661–665. https://doi.org/10.1038/nature11066
Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA et al (2016) Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 532:255–258. https://doi.org/10.1038/nature17393
Jin G, Reitman ZJ, Duncan CG, Spasojevic I, Gooden DM, Rasheed BA et al (2013) Disruption of wild-type IDH1 suppresses D-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res 73:496–501. https://doi.org/10.1158/0008-5472.CAN-12-2852
Kim JH, Lee S-R, Li L-H, Park H-J, Park J-H, Lee KY et al (2011) High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE 6:e18556. https://doi.org/10.1371/journal.pone.0018556
Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S et al (2012) Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483:484–488. https://doi.org/10.1038/nature10898
Lai A, Kharbanda S, Pope WB, Tran A, Solis OE, Peale F et al (2011) Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol 29:4482–4490. https://doi.org/10.1200/JCO.2010.33.8715
Loftus SK, Larson DM, Watkins-Chow D, Church DM, Pavan WJ (2001) Generation of RCAS vectors useful for functional genomic analyses. DNA Res 8:221–226
Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O et al (2012) IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483:474–478. https://doi.org/10.1038/nature10860
Luchman HA, Stechishin OD, Dang NH, Blough MD, Chesnelong C, Kelly JJ et al (2012) An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neurooncology 14:184–191. https://doi.org/10.1093/neuonc/nor207
Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K et al (2009) Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361:1058–1066. https://doi.org/10.1056/NEJMoa0903840
Molenaar RJ, Botman D, Smits MA, Hira VV, van Lith SA, Stap J et al (2015) Radioprotection of IDH1-mutated cancer cells by the IDH1-mutant inhibitor AGI-5198. Cancer Res 75:4790–4802. https://doi.org/10.1158/0008-5472.CAN-14-3603
Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P et al (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321:1807–1812. https://doi.org/10.1126/science.1164382
Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Krönke J, Bullinger L et al (2010) IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol 28:3636–3643. https://doi.org/10.1200/JCO.2010.28.3762
Pavlova NN, Thompson CB (2016) The emerging hallmarks of cancer metabolism. Cell Metab 23:27–47. https://doi.org/10.1016/j.cmet.2015.12.006
Pusch S, Schweizer L, Beck A-C, Lehmler J-M, Weissert S, Balss J et al (2014) D-2-Hydroxyglutarate producing neo-enzymatic activity inversely correlates with frequency of the type of isocitrate dehydrogenase 1 mutations found in glioma. Acta Neuropathol Commun 2:19. https://doi.org/10.1186/2051-5960-2-19
Robinson JP, Vanbrocklin MW, Guilbeault AR, Signorelli DL, Brandner S, Holmen SL (2010) Activated BRAF induces gliomas in mice when combined with Ink4a/Arf loss or Akt activation. Oncogene 29:335–344. https://doi.org/10.1038/onc.2009.333
Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C et al (2013) An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 340:626–630. https://doi.org/10.1126/science.1236062
Saha SK, Parachoniak CA, Ghanta KS, Fitamant J, Ross KN, Najem MS et al (2014) Mutant IDH inhibits HNF-4α to block hepatocyte differentiation and promote biliary cancer. Nature 513:110–114. https://doi.org/10.1038/nature13441
Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio IIC et al (2012) D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev 26:2038–2049. https://doi.org/10.1101/gad.198200.112
Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brüstle A et al (2012) IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 488:656–659. https://doi.org/10.1038/nature11323
Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY et al (2009) Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461:109–113. https://doi.org/10.1038/nature08268
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T et al (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. https://doi.org/10.1038/nmeth.2019
Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M et al (2013) Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496:101–105. https://doi.org/10.1038/nature12040
Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y et al (2015) Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet 47:458–468. https://doi.org/10.1038/ng.3273
Tateishi K, Wakimoto H, Iafrate AJ, Tanaka S, Loebel F, Lelic N et al (2015) Extreme Vulnerability of IDH1 mutant cancers to NAD+ depletion. Cancer Cell 28:773–784. https://doi.org/10.1016/j.ccell.2015.11.006
Turcan S, Fabius AWM, Borodovsky A, Pedraza A, Brennan C, Huse J et al (2013) Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT inhibitor decitabine. Oncotarget 4:1729–1736
Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E et al (2012) IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483:479–483. https://doi.org/10.1038/nature10866
Wakimoto H, Tanaka S, Curry WT, Loebel F, Zhao D, Tateishi K et al (2014) Targetable signaling pathway mutations are associated with malignant phenotype in IDH-mutant gliomas. Clin Cancer Res 20:2898–2909. https://doi.org/10.1158/1078-0432.CCR-13-3052
Wang F, Travins J, Delabarre B, Penard-Lacronique V, Schalm S, Hansen E et al (2013) Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 340:622–626. https://doi.org/10.1126/science.1234769
Ward PS, Lu C, Cross JR, Abdel-Wahab O, Levine RL, Schwartz GK et al (2013) The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem 288:3804–3815. https://doi.org/10.1074/jbc.M112.435495
Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA et al (2010) The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17:225–234. https://doi.org/10.1016/j.ccr.2010.01.020
Watanabe T, Nobusawa S, Kleihues P, Ohgaki H (2009) IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 174:1149–1153. https://doi.org/10.2353/ajpath.2009.080958
von Werder A, Seidler B, Schmid RM, Schneider G, Saur D (2012) Production of avian retroviruses and tissue-specific somatic retroviral gene transfer in vivo using the RCAS/TVA system. Nat Protoc 7:1167–1183. https://doi.org/10.1038/nprot.2012.060
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H et al (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19:17–30. https://doi.org/10.1016/j.ccr.2010.12.014
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W et al (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773. https://doi.org/10.1056/NEJMoa0808710
Zheng S, Houseman EA, Morrison Z, Wrensch MR, Patoka JS, Ramos C et al (2011) DNA hypermethylation profiles associated with glioma subtypes and EZH2 and IGFBP2 mRNA expression. Neurooncology 13:280–289. https://doi.org/10.1093/neuonc/noq190
Acknowledgements
We wish to thank Kristin Kraus for editorial assistance and Rodney Stewart for providing mCherry cDNA. We acknowledge the Metabolomics Core, the Fluorescence Microscope Core, and the Flow Cytometry Facility at the University of Utah for providing services. This work was supported in part by a National Institutes of Health Grant (CA084563) from the National Cancer Institute, the Department of Neurosurgery at the University of Utah, funds in conjunction with Grant P30CA042014 awarded to Huntsman Cancer Institute and to the Nuclear Control Program at Huntsman Cancer Institute, and an award from the Foreign Science and Technology Cooperation Plan of Jiangxi Province (20151BDH80009).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tiburcio, P.D.B., Xiao, B., Berg, S. et al. Functional requirement of a wild-type allele for mutant IDH1 to suppress anchorage-independent growth through redox homeostasis. Acta Neuropathol 135, 285–298 (2018). https://doi.org/10.1007/s00401-017-1800-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-017-1800-0