Abstract
Central neurocytomas are rare central nervous system neoplasms. Since the first description, approximately 500 cases of these tumors have been published to date. Nevertheless, only a limited number of genetic studies on these tumors have been reported. Here we investigated 20 “typical” central neurocytomas using array-based comparative genomic hybridization (array-CGH) with the GenoSensor Array 300. The functional significance of detected chromosomal aberrations harboring potent candidate genes was also examined at the mRNA expression level. Each tumor examined displayed DNA copy-number aberrations (CNAs), and mean number of CNAs per tumor was 38.1 ± 7.1 (range 19–53). Frequent gains were mapped at 2p24.1–22.1, 10q23.3–26.3, 11q23–25, and 18q21.3–qter. Frequent losses were identified at 1pter–36.3, 1p34.3, 6q13–21, 12q23–qter, 17p13.3, 17q11–23, and 20pter–12.3. There were 10 gained and 23 lost single DNA clones affecting ≥40% of samples tested. mRNA expression levels of 24 selected candidate genes harbored in these imbalanced clones were analyzed. MYCN, PTEN, and OR5BF1 were strongly overexpessed, whereas BIN1, SNRPN, and HRAS were found to be strongly underrepresented at the transcriptional level. Thus these data support that MYCN oncogene gain/overexpression accompanied by reduced expression of BIN1 tumor suppressor may contribute to central neurocytoma tumorigenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Central neurocytomas are rare central nervous system (CNS) neoplasms that occur predominantly in young adults [11, 27]. The tumors are usually found in the lateral ventricles occupying the septum pellucidum region. Pathologically, these tumors demonstrate neuronal differentiation as suggested by electron microscopy and immunohistochemistry findings [11]. Central neurocytomas are generally considered benign and are associated with favorable prognosis. However, certain neurocytomas display histological “atypia” and possess increasing proliferative activity, resulting in poor clinical outcomes [25, 27]. The histogenesis of central neurocytoma is still unclear. It remains a matter of discussion whether these neoplasms arise from cells committed to neuronal differentiation, or from multipotent progenitor cells in the periventricular matrix [11, 27].
Since the first description of central neurocytoma by Hassoun in 1982, approximately 500 cases have been published to date [25, 27]. Nevertheless, a limited number of genetic studies on these tumors have been conducted. Conventional cytogenetic analyses revealed normal or near-diploid karyotypes, while a few studies discovered either a gain of chromosome 7 or loss of chromosome 17 copy-number [4, 9, 30]. No TP53 mutation, EGFR and MYCN amplifications were found in central neurocytomas [6, 23, 31, 32]. Comparative genomic hybridization (CGH) analysis was used in ten samples and appreciable chromosomal alterations were revealed in six neurocytomas [32]. Gains at chromosomes 2p, 10q, 13q, and 18q were the more frequent genetic aberrations. Due to strong histological similarity between neurocytomas and oligodendrogliomas, analysis of the loss of heterozygosity (LOH) of 1p and 19q was also performed in these tumors. Tong et al. [31] observed interstitial deletions of 1p and 19q, although most of the informative markers were retained. Fujisawa et al. [6] were unable to found allelic losses on 1p and 19q in eight neurocytomas using FISH probes. Expression profiling of four central neurocytomas displayed their close genetic similarity to cerebellar liponeurocytomas and their marked differences relative to medulloblastoma transcriptional profiles [8]. Thus, the genetic background and molecular mechanisms underlying pathogenesis of central neurocytomas remain largely unknown, and consequently, the distinct nosologic position of these tumors is still uncertain.
In the present study we investigated the molecular pathogenesis of 20 central neurocytomas using a combined experimental design. Initially, we applied an array-based CGH method (or array-CGH) with the GenoSensor Array 300 to identify recurrent DNA copy-number imbalances in these tumors. The functional significance of detected chromosomal aberrations harboring potent candidate genes was further examined at the mRNA expression level.
Materials and methods
Patient population and pathological analysis
In total, 20 adult patients who were treated for newly diagnosed central neurocytomas at the Burdenko Neurosurgical Institute from January 1, 1995, to January 1, 2004 were included in this study. The mean age was 27 years (range 18–44 years) and there were 11 women and 9 men among them. In all cases the tumor was located in both lateral ventricles involving the septum pellucidum. All patients underwent open surgery with gross total (12 cases) and subtotal (8 cases) tumor resection. After the operation, all patients experienced a stable clinical course without signs of disease progression.
Histological diagnoses were performed according to the current World Health Organization histologic classification [11]. All tumors were composed of sheets of uniform, round cells with perinuclear haloes intermingled with fine background neuropil and acellular fibrillary zones. Additionally, all samples displayed strong immunoreactivity for synaptophysin and Neu-N antibodies. Also, 11 tumors were examined with an electron microscope and “classic” neurocytoma patterns, such as dendrites, synapses and neurosecretory granules were found. There were no signs of histological atypia (mitotic activity, microvascular proliferation and necrosis) in these 20 neurocytomas. Immunohistochemistry with a Ki-67 (MIB1) antibody was performed on all samples and the Ki-67 labeling indices varied from 0.12 to 1.89% (mean 0.93 ± 0.56%). Approval to link laboratory data to clinical data was obtained by the Institutional Review Board.
Sample processing
Samples from each tumor were collected at the time of initial operation, immediately frozen in liquid nitrogen, and stored at −80°C until nucleic acid isolation and analysis. To confirm the presence of a viable tumor (>90% of neoplastic cells), the cryosections of each manually dissected neurocytoma sample were stained with H&E and reviewed before DNA and RNA extraction. DNA was isolated according to a protocol that applies the DNeasy tissue kit (Qiagen, Hilden, Germany). Total RNA was extracted from 12 available tumor samples according to a protocol that applies TRIzol reagent and RNeasy midi spin columns (Qiagen). DNA and total RNA concentration and quality were determined by absorption spectrophotometry and their integrity and purity were analyzed on 1% agarose gel. Also, the integrity of each RNA sample was monitored with the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
DNA labeling, hybridization, and analysis of the GenoSensor Array 300
Pretreatment and analysis of the microarrays was performed as described previously [15]. Briefly, approximately 100 ng of DNA from tumor samples (test DNA) and sex-mismatched normal reference human DNA were labeled by a random priming reaction according to the protocols recommended by the manufacturer (Vysis, Inc., Abbott Laboratories S.A., Downers Grove, IL, USA). Aliquots of labeled DNA were mixed with the hybridization buffer and the hybridization mixture was then introduced to the genomic DNA microarray slides. We used the commercially available GenoSensor Array 300 (Vysis, Inc.), which contained 287 genomic targets cloned from P1 and BAC libraries, and printed in triplicate. This microarray platform includes cancer amplicons/oncogenes, tumor suppressor genes, loci of insertions/deletions/duplications, telomeres and markers added to reduce gaps (for detailed information see http://www.vysis.com). The microarray slides were incubated for 72 h at 37°C, washed in formamide/standard saline citrate solutions, counterstained with DAPI and covered with coverslip glasses. The microarray slides were then analyzed using the GenoSensor Reader System (Vysis, Inc.) according to the manufacturer’s instructions. Based on previous control experiments using the GenoSensor Array 300 [1, 15, 21], we determined losses, gains and amplifications of target DNA sequence copy-number by a green/red ratio <0.8, >1.2, and >2.0, respectively.
Fluorescence in situ hybridization (FISH)
Thirty-five paraffin-embedded samples of central neurocytomas (including all samples applied for microarray experiments) were examined by a two-color interphase FISH assay that was performed on 5 μm-thick sections. The following fluorochrome-labeled probes were applied (all produced by Vysis, Inc., besides 2p11–q11): centromere 2p11–q11 probe (RPZD, Berlin, Germany) and locus-specific 2p24(MYCN) probe, 10p11–q11/10q23(PTEN) dual-color probe set, and locus-specific 17p13(LIS1)/17q11(RARA) dual-color probe set. Pretreatment of slides, hybridization, posthybridization processing, and signal detection were performed as described previously [14].
Quantitative real-time polymerase chain reaction with genomic DNA (QRT-PCR)
QRT-PCR with genomic DNA was performed on a 7300 real-time PCR system (Applied Biosystems, Foster City, CA, USA) using TaqMan single exon-containing probes to PTEN, HIC1, OR5BF1, and ZNF439 (all pre-made probes from Applied Biosystems) and intron-containing TaqMan probe to MYCN obtained from the Institute of Molecular Biology (Moscow, Russia). All 20 DNA neurocytoma samples applied for array-CGH were examined, and PCR amplification was performed in duplicate (aliquots of 50 ng) for each sample. We quantified the tumor DNA by comparing the target locus to the reference 18S rRNA probe. The relative target copy-number level was also normalized to normal human genomic DNA. Conditions for the quantitative PCR reaction and calculation of copy-number changes of target gene relative to the endogenous control and calibrator were performed as described previously [22]. Based on these calculations, copy numbers < 1.58 were considered to be a loss of DNA targets whereas those >2.53 were considered to be gains.
Quantitative reverse transcription real-time polymerase chain reaction (QRT-RT-PCR)
Levels of mRNA for 24 selected candidate genes (see Tables 3, 4) were also measured using a real-time fluorescence detection method. Single-stranded cDNA was generated from total RNA using the High Capacity cDNA Archive Kit (Applied Biosystems). QRT-RT-PCR was performed in duplicate for each sample with the 7300 system according to the manufacturer’s instructions, using TaqMan gene expression assays with pre-made gene primers (all Applied Biosystems). Amplification products were verified by agarose gel electrophoresis. To standardize the amount of sample cDNA, endogenous control amplicon (GAPDH) was used as a housekeeping gene. For tissue-specific normalization, a reference pool of human brain, cerebral cortex total RNA (Cat. No. 636561; BD Bioscience, Heidelberg, Germany) was applied as a calibrator tissue control. The relative quantification of each target was calculated using sequence detection SDS software (version 1.3.1.) according to a previously published algorithm based on threshold criteria of ±2.0 [13].
Allelic discrimination testing using real-time polymerase chain reaction analysis of single nucleotide polymorphism (SNP) on 17p13.3 chromosomal region
Real-time PCR SNP analysis of 14 intragenic SNP markers spanning of 17p13.3 chromosome locus was performed using predeveloped TaqMan SNP genotyping assay reagents (see Table 5) according to the manufacturer’s instructions. Order and allocation of the markers relative to 17p13.3 locus were determined according to the SNP Browser Software. Eight blood DNA samples, isolated from healthy individuals, were used a control. Additionally, we studied three DNA samples obtained from medulloblastomas with isochromosome 17q, which was detected by high-resolution array-CGH analysis [18].
Immunohistochemistry with antibody to LIS1 protein
Immunohistochemical analysis was performed on 5 μm-thick paraffin sections mounted on poly-l-lysine-coated slides. The sections were microwaved in antigen unmasked solution (BD Bioscience, San Jose, CA, USA). Goat polyclonal antibody to LIS1 (N-19; sc 7577, 1:200; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was applied and the sections were incubated overnight at 4°C. Immunostain visualization was achieved with standard streptavidin–biotin–peroxidase technique (ImmunoCruz TM Staining System; sc-2053; Santa Cruz Biotechnology). The slides were stained with 3,3′-diaminobenzidine, counterstained with hematoxylin, and mounted. For the negative control procedure, primary antibody was substituted according to commercially produced control reagents. For the positive control procedure, sections were obtained from normal non-tumoral human brain tissue.
Statistical analysis
Intraclass correlation analysis was used to assess the degree of linear associations between pairs of variables. The Chi-square test and Fishers’ exact test were performed to analyze relationships between clinical variables, pathological features and genetic alterations. A significant correlation between two parameters was taken at the 95% confidence interval. Probability (P) values <0.05 were considered significant.
Results
Genomic imbalances in central neurocytomas detected by array-CGH
Each tumor examined displayed DNA copy-number aberrations (CNAs) and the mean number of CNAs per tumor was 38.1 ± 7.1 (range 19–53). The mean number of low-level gains was 14.7 ± 4.1 (range 7–26) and the mean number of copy-number losses was 20.4 ± 3.8 (range 11–31). There were no samples with high-level copy-number gains/amplifications in these 20 tumors. More frequent gains involving at least two neighboring clones were mapped at 2p24.1–22.1 (85%), 10q23.3-26.3 (70%), 11q23–25 (60%), and 18q21.3–qter (50%). Correspondingly, more frequent losses were identified in 1pter-36.3 (70%), 1p34.3 (60%), 6q13–21 (80%), 12q23–qter (60%) 17p13.3 (80%), 17q11–23 (70%), and 20pter–12.1 (60%). The frequently gained and lost single DNA clones affecting ≥40% of samples tested are outlined in Tables 1 and 2, respectively.
Correlation analysis of CNAs by chromosomal location revealed a significant association between imbalances in neighboring DNA clones along the same chromosomes and an inverse correlation between loss of BIN1 locus and gain of MYCN (r = −0.63, P = 0.0005).
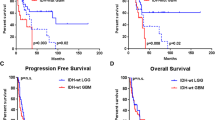
For validation of array-CGH findings, we compared the results of this study with data obtained by FISH (Fig. 1) and QRT-PCR analyses. There was a high level of concordance between the data from all these techniques (see Tables 1, 2). Also, results obtained by all these methods were correlated strongly for each individual case (P < 0.05).
Array-CGH (a) validation by FISH (b, c). Array-based cytogenetic profile of central neurocytoma (female; 26 years) displays 44 copy-number aberrations including gain of MYCN (b; numerous red signals), loss of BIN1, gain of PTEN (c; numerous red signals), and loss of 10p11–q11 (c; single green signals). In addition, numerous losses on both arms of chromosome 17 could be seen
We were unable to find any association between array-CGH data and known clinico–pathological variables, including patient age (<30 or >30 years), gender, and Ki-67 labeling index (<1 vs. >1%).
mRNA expression level for selected candidate genes detected by QRT-RT-PCR
To evaluate the potential effect of defined genomic CNAs for central neurocytoma molecular pathogenesis, we analyzed mRNA expression of 24 selected candidate genes harbored in imbalanced regions of genomic DNA (see Tables 3, 4). Compared with the human cerebral cortex total RNA, MYCN, PTEN, and OR5BF1 genes were strongly overexpessed in all 12 samples tested, based on threshold ratio ±2.0. In contrast, BIN1 and SNRPN genes were found to be strongly underrepresented at the transcriptional level in all neurocytomas examined, whereas the HRAS gene was also down-regulated in 10 of 12 samples. For these six genes, the level of over- or underexpression tended to be greater in tumors with gain or loss of the corresponding DNA clones. However, these differences were not significant. No consistent gene dosage effects between DNA copy-number status and relative gene expression level for the remaining 18 respective candidates were observed.
Allelic discriminations analysis of 17p13.3 chromosomal region
In as much as 17p13.3 was found to be frequently deleted in central neurocytomas, we performed analysis of allelic status for 14 intragenic SNP markers spanning this region. All tumors showed at least one monoallelic loss on this chromosomal locus with a more frequent breakpoint involving 3′ untranslated region of LIS1 gene (Table 5, Fig. 2). Eight control blood DNA specimens disclosed diploid profile for all SNP markers tested, whereas three medulloblastoma specimens showed monoallelic losses for all probes examined.
Graphic summary of the results from the allelic discrimination analysis of 17p13.3 locus using 14 intragenic SNP markers. Case numbers are given on the top of the panel and the investigated SNP markers are given on the left of the panel. Order and allocation of the markers relative to 17p13.3 locus were determined according to the SNP Browser Software. Filled squares SNP loci with monoallelic loss (homozygosity), open squares heterozygous SNP loci with diploid (balanced) alleles; LIS1(i) intron SNP marker, LIS1(u) UTR3′ SNP marker
Immunohistochemistry with LIS1 antibody
To analyze whether the frequently lost LIS1 DNA clone is also down-regulated at the protein level, we performed an immunohistochemical analysis in 40 neurocytomas (including all 20 samples applied for array-CGH). We revealed strong cytoplasmic LIS1 protein reactivity in all tumors examined and all samples showed greater than 70% of immunostained cells. Intercellular fibrillary zones and rosette-like structures were also immunopositive. The intensity of LIS1 staining was similar to that revealed in the human brain tissue applied as a positive control.
Discussion
To identify recurrent copy-number imbalances in 20 central neurocytomas, we used low-density DNA microarray assay with the GenoSensor 300 platform, which allows for the rapid investigation of multiple, cancer-associated abnormalities in chromosomal loci throughout the entire tumor genome [1, 15, 21]. During the current array-CGH screening, we found a set of frequently altered DNA clones that may include candidate genes for tumorigenesis of these infrequent CNS neoplasms. The results of microarray experiments were validated by FISH and QRT-PCR data analyses. Interestingly, neurocytomas, which are traditionally biologically benign neoplasms, contained significantly fewer CNAs per tumor than highly malignant glioblastomas, studied previously with the GenoSensor Array 300 [15].
Notably, some current array-CGH findings such as gains at 2p, 10q, and 18q are consistent with the data obtained previously by conventional CGH analysis [32]. Also, frequent deletions at both arms of chromosome 17 are in line with data obtained by Cerda-Nicolas et al. [4], who observed monosomy 17 in neurocytoma. The histopathological resemblance of neurocytomas with oligodendrogliomas prompted us to analyze 1p/19q status in these tumors. We found that small deletions including single DNA clones on 1p36.3 and 1p34.2 are frequent in neurocytomas, whereas array-CGH oligodendroglioma analyses usually disclosed stretched submegabase deletions at 1p [10, 19, 26]. Thus, our findings are in concordance with data of LOH analysis performed by Tong et al. [31], who found a few interstitial deletions on 1p in six of nine neurocytomas but most of the markers examined were retained. In contrast to oligodendrogliomas, we established deletions of clones from 19q13.3 in a single sample only.
A set of imbalanced DNA clones harbored a few genes that appeared to be promising candidates for the molecular pathogenesis of neurocytomas. At first view, the frequent deletion of LIS1 clone is a very attractive finding because this gene encodes a protein that regulates neuronal migration from the periventricular matrix. Loss of this locus is associated with cortical malformations (lissencephaly and subcortical band heterotopia) [17]. Nevertheless, expression analysis revealed appropriate mRNA levels for this gene accompanied by a strong immunoreactivity of LIS1 protein product in all examined cases. Similarly, other potent candidate genes showed no alterations at the transcriptional level. Moreover, a few genes from deleted DNA clones (SERPINE1, RB1, BRCA1, ZNF439) revealed strong mRNA expression in the majority of tested samples, suggesting dosage overcompensation mediated by the remaining allele. Probably, inactivation of the other closely linked genes included in these imbalanced DNA clones may be functionally significant for neurocytoma development. The results of allelelic discrimination analysis with 14 intragenic SNP markers spanning 17p13.3 locus confirm this suggestion.
Nevertheless, six genes showed strong concordance between imbalanced DNA clones and relative mRNA expression levels, which strongly implicates their relevance in the molecular pathogenesis of central neurocytomas. A DNA clone at 2p24.1, which contains a MYCN proto-oncogene, was found to be gained in 70% of neurocytomas. Correspondingly, QRT-RT-PCR analysis revealed strong overexpression of MYCN mRNA in all samples tested. MYCN amplification/overexpression is frequently observed in neuroblastomas and medulloblastomas [11]. On the other hand, embryological studies showed that MYCN plays a significant role in neurogenesis, because its expression is essential for the rapid expansion of neural progenitor cells during CNS development and inhibition of neuronal differentiation [12]. Thus, MYCN may be a “driver oncogene” in neurocytoma pathogenesis and aid in renewal of the tumor cells population.
Gain of MYCN was inversely correlated with a BIN1 deletion and this gene was decreased at the transcriptional level in all samples tested. BIN1 (bridging integrator 1) encodes for multiple tissue-specific isoforms of the MYC-integrated adaptor protein implicated in tumor suppression and cell death processes in human malignancies [28]. BIN1 expression was reduced in various human tumors. Moreover, the mRNA level of its MYCN-integrating isoform was found to be down-regulated in neuroblastomas [28]. Therefore, BIN1 appears to be a promising candidate suppressor gene for neurocytoma tumorigenesis, whose structural and/or functional alterations can lead to development of these neoplasms. Distinct mechanisms underlying the remaining BIN1 allele inactivation (haploinsufficiency, point mutations, and promoter hypermethylation) should be elucidated in further studies.
The tumor suppressor gene PTEN plays an important role in neuronal development, because loss of its expression is associated with cerebellar dysplasia and other CNS migrational defects [16]. However, PTEN loss does not inhibit neuronal differentiation [16]. PTEN expression fluctuates in neurogenesis and seems to be up-regulated at or near of birth [16]. On the other hand, PTEN overexpression in the pheochromocytoma cell line PC12, inhibits neuronal differentiation and provokes down-regulation of nerve growth factor receptors [20]. Perhaps, strong PTEN overexpression in neurocytomas (in comparison to the normal human cortex) can inhibit further maturation of tumor cells thereby increasing the proliferating cell population. However, support for such an assertion will require additional analyses.
SNRPN encodes a small ribonuclear protein and deletion of this region is critical for development of Prader–Willi syndrome, which is associated with neurodevelopmental disorders [7]. Mice lacking genes responsible for Prader–Willi syndrome display a widespread deficit in neuronal migration, and the extension, arborization, and fasciculation of axons during of CNS development [24]. Deletions of SNRPN locus were discovered in glioblastomas [15] and gastric carcinomas [29] and methylation of this gene was established in germ-cell tumors [3]. The distinct role of frequent SNRPN inactivation for neurocytoma pathogenesis appears to be unclear, although, hypothetically, its tumor suppression function cannot be fully excluded in these tumors.
HRAS allelic deletion was found to be a common abnormality in various tumors and these findings suggest that a putative tumor suppressor closely linked to the HRAS locus on 11p15.5 is involved in human carcinogenesis. On the other hand, the Ras/MAPK signal transduction pathway regulates synaptic plasticity and survival of CNS neurons [5]. Moreover, HRAS protein is essential for differentiation and neurite outgrowth-promoting activity of human embryonic neural cells [2]. Perhaps, frequent HRAS down-regulation in neurocytomas may result in the inhibition of further differentiation in tumor cells.
OR5BF1 belongs to the human olfactory receptor family, which includes more than 900 members distributed throughout the human genome. These genes encode for an immense variety of membrane-bound G-protein-coupled receptors that can represent specific targets for a variety of human diseases including cancer. OR5BF1 is strongly overexpressed in neurocytomas, and it is thought that it may function as a potent oncogene in these tumors. A detailed investigation of its function in neuronal tumors is warranted.
In summary, neurocytoma tumorigenesis appears to be associated with molecular mechanisms that activate the proliferation of tumor cells and, simultaneously, inhibit further neuronal maturation. Also, recurrent genetic alterations in central neurocytomas share some similarities with those defined in neuroblastomas including deletions on 1p36, gain/overexpression of MYCN, and the transcriptional inactivation of BIN1. Thus, despite the striking histological and biological differences between neurocytoma and neuroblastoma, the current findings hint on a certain resemblance of pathobiological mechanisms responsible for the tumorigenesis of various neural-derived neoplasms. Further global microarray-based analyses of neurocytomas are necessary to identify new candidate genes and chromosomal regions having both pathogenetic and prognostic relevance for these enigmatic brain tumors.
References
Albrecht B, Hausmann M, Zitzelsberger H, Stein H, Siewert JR, Hopt U, Langer R, Hogfler H, Werner M, Walch A (2004) Array-based comparative genomic hybridization for the detection of DNA sequence copy number changes in Barrett’s adenocarnoma. J Pathol 203:780–788
Borasio GD, Markus A, Heumann R, Chezzi C, Sampietro A, Wittinghofer A, Silani V (1996) Ras 21 protein promotes survival and differentiation of human embryonic neural crest-derived cells. Neuroscience 73:1121–1127
Bussey KJ, Lawce HJ, Himoe E, Shu XO, Heerema NA, Perlman EJ, Olson SB, Magenis RE (2001) SNRPN methylation patterns in the germ cell tumors as a reflection of primordial germ cell development. Genes Chromosome Cancer 32:342–352
Cerda-Nicolas M, Lopez-Gines C, Pedro-Alaya A, Llombart-Bosch A (1993) Central neurocytoma: a cytogenetic case study. Cancer Genet Cytogenet 65:173–174
Fivaz M, Meyer T (2005) Reversible intracellular translocation of KRas but not HRas in hyppocampal neurons regulated by Ca2+/calmodulin. J Cell Biol 170:429–441
Fujisawa H, Marukawa K, Hasegawa N, Tohma Y, Hayashi Y, Uchiyama N, Tachibana O, Yamashita J (2002) Genetic differences between neurocytoma and dysembryoplastic neuroepithelial tumor and oligodendroglial tumors. J Neurosurg 97:1350–1355
Horsthemke B, Buiting K (2006) Imprinting defects on human chromosome 15. Cytogenet Genome Res 113:292–299
Horstmann S, Perry A, Reifenberger G, Giangaspero F, Huang H, Hara A, Masuoka J, Rainov NG, Bergmann M, Heppner FL, Brandner S, Chimelli L, Montagma N, Jackson T, Davis DG, Markesvery WR, Elisson DW, Weller RO, Taddei GL, Conti R, Del Bigio MR, Conzales-Campora R, Radharkrishnan VV, Solymezoglu F, Uro-Coste E, Qian J, Kleihues P, Ohgaki H (2004) Genetic and expression profiles of cerebellar liponeurocytoma. Brain Pathol 14:281–289
Jay V, Edwards V, Hoving E, Rutka J, Becker L, Zielenska M, Teshima I (1999) Central neurocytoma: morphological, flow cytometric, polymerase chain reaction, fluorescence in situ hybridization and karyotypic analyses. J Neurosurg 90:348–354
Kitange S, Misra A, Law M, Passe S, Kollmeyer TM, Maurer M, Ballman K, Feuerstein BG, Jenkins RB (2005) Cromosomal imbalances detected by array comparative genomic hybridization in human oligodendrogliomas and mixed oligoastrocytomas. Genes Chromosomes Cancer 42:68–77
Kleihues P, Cavenee WK (eds) (2000) Tumors of the nervous system. Pathology and genetics: World Health Organization International Classification of Tumours. Lyon, France: WHO/IARC
Knoepfler PS, Cheng PF, Eisenman RN (2002) N-MYC is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev 16:2699–2712
Korshunov A, Neben K, Wrobel G, Tews B, Benner A, Hahn M, Golanov A, Lichter P (2003) Gene expression patterns in ependymoma correlate with tumor location, grade and patient age. Am J Pathol 163:1721–1727
Korshunov A, Sycheva R, Golanov A (2004) Molecular stratification of diagnostically challenging high-grade gliomas composed of small cells. The utility of fluorescence in situ hybridization. Clin Cancer Res 10:7820–7826
Korshunov A, Sycheva R, Golanov A (2006) Genetically distinct and clinically relevant subtypes of glioblastoma defined by array-based comparative genomic hybridization. Acta Neuropathol 111:465–474
Lachyankar MB, Sultana N, Schonhoff CM, Mitra P, Poluha W, Lambert S, Quesenberry PJ, Litofsky NS, Recht LD, Nabi R, Miller S, Ohta S, Neel BG, Ross AH (2000) A role for nuclear PTEN in neuronal differentiation. J Neurosci 20:1404–1413
Leventer RJ (2005) Genotype-phenotype associations in lissencephaly and cortical band heterotopia: key questions answered. J Child Neurol 20:307–312
Mendrzyk F, Radlwimmer B, Joos S, Kokocinski F, Benner A, Stange DE, Neben k, Fiegler H, Carter NP, Reifenberger G, Korshunov A, Lichter P (2005) Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J Clin Oncol 23:8853–8862
Mohapatra G, Betensky RA, Miller ER, Carey B, Gaumont LD, Engler DA, Louis DN (2006) Glioma test array for use with formalin-fixed, paraffin-embedded tissue: array comparative genomic hybridization correlates with loss of heterozygosity and fluorescence in situ hybridization. J Mol Diagn 8:268–276
Musatov S, Roberts J, Brooks AI, Pena J, Betchen S, Pfaff DW, Kaplitt MG (2004) Inhibition of neuronal phenotype by PTEN in PC12 cells. Proc Natl Acad Sci USA 101:3627–3631
Nakahara Y, Shiraishi T, Okamoto H, Mineta T, Oishi T, Sasaki T, Tabuchi K (2004) Detrended fluctuation analysis of genome-wide copy number profiles of glioblastomas using array-based comparative genomic hybridization. Neuro-Oncol 6:281–289
Nigro JM, Takahashi MA, Ginzinger DG, Law M, Passe S, Jenkins RB, Aldape K (2001) Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real-time quantitative polymerase chain reaction assay. Am J Pathol 158:1253–1262
Ohgaki H, Eibl RH, Schwab M, Reichel MB, Mariani L, Gehring M, Petersen I, Holl T, Wiestler OD, Kleihues P (1993) Mutations of the p53 tumor suppressor gene in neoplasms of the human nervous system. Mol Carcinogen 8:74–80
Pagliardini S, Ren J, Wevrick R, Greer JJ (2005) Developmental abnormalities of neuronal structure and function in prenatal mice lacking Prader–Willi syndrome gene necdin. Am J Pathol 167:175–191
Rades D, Schild SE (2006). Treatment recommendations for various subgroups of neurocytomas. J Neurooncol 77:305–309
Rossi MR, Galle D, Laduca J, Matsui S, Conroy J, McQuaid D, Chernivsky D, Eddy R, Chen SH, Barnett GH, Nowak NJ, Cowell JK (2005) Identification of consistent novel submegabase deletions in low-grade oligodendrogliomas using array-based comparative genomic hybridization. Genes Chromosomes Cancer 44:85–96
Schmidt MH, Gottfried ON, von Koch CS, Chang SM, McDermott MW (2004) Central neurocytoma: a review. J Neurooncol 66:377–384
Tajiri T, Liu X, Thompson PM, Tanaka S, Suita S, Zhao H, Maris JM, Prendergast GC, Hogarty MD (2003) Expression of a MYCN-integrating isoform of the tumor suppressor BIN1 is reduced in neuroblastomas with unfavorable biological features. Clin Cancer Res 9:3345–3355
Takada H, Imoto I, Tsuda H, Sonoda I, Ichikura T, Mochizuki H, Okanoue T, Inazawa J (2005) Screening of DNA copy number aberrations in gastric cancer cell lines by array-based comparative genomic hybridization. Cancer Sci 96:100–110
Taruscio D, Danesi R, Montaldi A, Cerasoli S, Cenacchi G, Giangaspero F (1997) Nonrandom gain of chromosome 7 in central neurocytoma: a chromosomal analysis and fluorescence in situ hybridization study. Virchows Arch 430:47–51
Tong CY, Ng HK, Pang JC, Hu J, Hui AB, Poon WS (2000) Central neurocytomas are genetically distinct from oligodendrogliomas and neuroblastomas. Histopathology 37:160–165
Yin XL, Pang JC, Hui AB, Ng HK (2000) Detection of chromosomal imbalances in central neurocytomas by using comparative genomic hybridization. J Neurosurg 93:77–81
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Korshunov, A., Sycheva, R. & Golanov, A. Recurrent cytogenetic aberrations in central neurocytomas and their biological relevance. Acta Neuropathol 113, 303–312 (2007). https://doi.org/10.1007/s00401-006-0168-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-006-0168-3