Abstract
Diblock amphiphilic copolymers form aggregates in some solvents. Such aggregates exhibit different morphologies, depending mainly on the polar/apolar block ratios. Aggregation of copolymers with polar block excess leads to micelle-like aggregates, known as polymeric micelles, which can be used as vehicles for drug and gene delivery, water decontamination, and catalysis. Here, we synthesized by RAFT polymerization three different polymers namely [dimethyl 2-(aminoethyl) methacrylate] (PDMAEMA), poly (methyl methacrylate) (PMMA) copolymers, and PDMAEMA-block-PMMA and characterized their aggregates by NMR spectroscopy and pH titrations. We investigated correlations between their chemical structure, aggregation behavior, protonation degree, and chain conformation in the corona. Decreased amine protonation in the copolymers reduced the electrostatic repulsion, and the apparent pKa of the amino groups approached that of isolated amine. These effects increased compactness and sizes of the polymers and their aggregates at higher pH as reflected by the increased NMR line widths.

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Diblock amphiphilic copolymers can form aggregates in some solvents [1]. In water, such aggregates exhibit different morphologies, depending primarily on the polar/apolar block ratios [2, 3]. Aggregation of polymers with polar block excess leads to micelle-like aggregates, known as polymeric micelles [4, 5]. Among many other applications, polymeric micelles can be used as vehicles for drug and gene delivery [6, 7], water decontamination [8], and catalysis [9].
The molar mass of the polymeric micelles and the core size is determined by the apolar block, while the molar mass of the polar block determines the thickness of the corona. Current polymerization techniques allow size and composition control of these polymers and, as a consequence, of their aggregates [10, 11].
PMMA is an apolar polymer studied mainly as a drug delivery system mainly due to its biocompatibility [12]. PMMA as copolymer block in aqueous solutions forms the core of micelles, or other types of aggregates, where relatively hydrophobic molecules can be loaded [13].
PMMA homopolymers present glass transition temperature (Tg) well above room temperature (~ 105 °C for the atactic form). The high Tg of PMMA (apolar block) in polymeric micelles leads to a state where micelles do not disaggregate as the copolymer concentration drops, as the core lacks motional freedom [14]. Dilution-stable micelles can be an advantage in many applications.
Depending on the nature of the repetitive unit(s), polymeric micelles may vary in size and morphology upon changes in temperature [15], pH [16], or light [17], allowing the control of the aggregate properties. Examples are block copolymers synthesized with dimethyl 2-(aminoethyl) methacrylate, DMAEMA, and poly (methyl methacrylate), PMMA. The tertiary amino group of the homopolymer PDMAEMA and many of its copolymers determine that effective charge and hydrophilicity are pH-dependent. These polymers, which usually exhibit a lower critical solution temperature (LCST), interact with water more strongly below LCST, so that micelles are likely to precipitate, or shrink, above LCST [18].
PDMAEMA protonation in the corona of polymeric micelles determines the drug loading capacity, the solubility and stability of the micelles, and the interaction with negatively charged species such as nucleic acids. Many other potential applications of PDMAEMA polymeric micelles are strongly associated with the degree of protonation in the corona. PDMAEMA-based copolymers usually have an LCST that is evident even when aggregated as micelles. Above this temperature, corona dehydration can lead to load release, micelle aggregation/precipitation, or both [19]. As temperature effects are also affected by medium acidity, pH can be used together with the temperature for controlling drug release [20].
Despite the breadth of potential applications, detailed understanding of the relationships between pH and conformational changes of PDMAEMA aggregates is not available. The importance of understanding PDMAEMA protonation at the micelle corona, as well as how these changes affect chain conformation is of both fundamental and technological interest. Here, we have used NMR spectroscopy and pH titration assays to characterize micelles of three different PDMAEMA-block-PMMA (Fig. 1) polymers synthesized by RAFT polymerization. Different conditions were examined to investigate possible correlations between the copolymers chemical structure, aggregation behavior, protonation degree, and chain conformation in the corona.
Material and methods
Materials
Methanol, n-hexane and acetone (analytical grade), and triethylamine, TEA (HPLC grade), were from J. T. Baker. Tetrahydrofurane, THF, deuterated chloroform, CDCl3, deuterated water, D2O, 1-(N-phenyl-amino) naphthalene, NPN, 2-(N-morpholino)ethanesulfonic acid, MES, tris(hydroxymethyl)aminomethane, Tris, and N-cyclohexyl-3-aminopropanesulfonic acid, CAPS, were from Sigma-Aldrich and used without further purification. Methyl methacrylate (MMA) 99% and dimethyl 2-(aminoethyl) methacrylate (DMAEMA) 98%, were from Sigma-Aldrich. The polymerization inhibitor was removed from MMA by extraction with NaOH aqueous solutions. An excess of initiator was added to the copolymerizations in order to consume the inhibitor from DMAEMA.
NaH2PO4, boric acid, HCl, and NaOH were from Merck KGaA, and benzoyl peroxide (BPO) was from Vetec, Brazil. Cumyl dithiobenzoate, CDB, was synthesized as described [21].
The GPC standard for the determination of the weight average mass of poly(methyl methacrylate), PMMA, was poly(methyl methacrylate) standard ReadyCal set, Mp 500-2,700,000, from Sigma Aldrich.
Methods
Synthesis of the copolymers
The copolymers were synthesized by RAFT, as reported elsewhere [22]. Briefly, the two-step procedure for each copolymer was as follows: in the first step, in batch, the PMMA block was synthesized using cumyl dithiobenzoate (CDB) as chain transfer agent, benzoyl peroxide (BPO) as initiator and methyl methacrylate. The CDB/BPO ratio was above 3 in all cases, and the reaction temperature was from 70 to 80 °C. Typically, homopolymerization time was 8 h, with roughly 50% conversion. The polymer was purified by precipitation in methanol (three times, after solubilization in THF). PMMA blocks were then used as macro chain transfer agents (macroCTA) in a batch copolymerization using DMAEMA as monomer and BPO as an initiator. Copolymerizations lasted 24 h. The copolymers were purified by solubilization in THF and precipitation in hexane (three times).
Gel permeation chromatography
The polymers were characterized in a Shimadzu liquid chromatograph Sil-20A, equipped with an autosampler, Shimadzu Sil-20A, an LC-20AD pump, and a RID-A Shimadzu refractive index detector. A Phenogel column, (PolySep™-SEC GFC-P 4000, LC, 300 × 7.8 mm) from Phenomenex was used. The injected volume of the sample (10 mg/L) varied from 5 to 10 μL, and the column flow rate was 0.6 cm3/min. The mobile phase was 0.2% TEA v/v in THF, and PMMA standards were used to build the calibration curve. Samples were filtered through Millex PTFE membrane (0.45 μm pore size).
Determination of hydrodynamic diameter of copolymers aggregates by dynamic light scattering
The Dhs were obtained using a Zetasizer Nano 317 at 25 °C. Buffers were previously filtered through Millipore Millex LCR (0.22 μm) and the copolymer solutions through Millipore PTFE filters (0.45 μm). Typical count rates were between 200 and 500 thousand. Autocorrelation function (ACF) was acquired for 60 s and averaged for 1 min. Hydrodynamic diameters were obtained from a mass-weighted size distribution analysis and reported as the mean of triplicate measurements ± SD.
Determination of the pKa of the amino groups of the polymers
The polymers were dissolved in methanol (50 mg/mL), and the solution was stirred for 1 h. An aliquot of this solution (0.25 to 2.0 mL) was added to 30 mL of 0.01 M HCl under constant stirring and N2 flux. The final percentage of methanol was less than 10% for all polymers. The polymer masses in the titration experiments were from 6.8 to 46.8 mg. Aliquots of 0.05 M NaOH were added to the polymer solution, maintained at 25 ± 0.1 °C, and the pH was measured after each addition using a Digimed DM-20 pH meter (São Paulo, Brazil) equipped with a Corning semi-micro combination electrode (Corning, USA).
Determination of the n/m ratio of PMMAn-b-PDMAEMAm
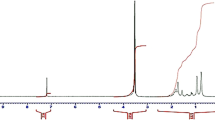
The n/m ratios of PMMAn-b-PDMAEMAm copolymers were determined by 1H-NMR spectra using a Bruker spectrometer operating at 300 MHz (1H frequency). The spectra were obtained with 90° pulses of 8.0 μs and a spectral window of 12 ppm. Signals used for the characterization of the copolymers: c = 3.6 ppm (3 H, O-CH3, PMMA); d = 4.0 ppm (2 H, O-CH2-, PDMAEMA) (Fig. 2). See Results for calculation details.
1H-NMR spectra of the copolymers X, Y, Z, and PDMAEMA. The polymers were renamed according to the number of MMA and DMAEMA determined by NMR and GPC as: X = PMMA50-b-PDMAEMA269 (A), Y = PMMA48-b-PDMAEMA324 (B), Z = PMMA94-b-PDMAEMA88 (C), and PDMAEMA as PDMAEMA265 (D), see Table 2. Polymer concentrations were ca. 10 mg/mL in CDCl3.
1H RMN and NOESY studies of the copolymers
The conformation of the copolymers in water was analyzed by 1H 1D, and 2D NOE NMR experiments carried out at 25 °C on a Bruker Avance III spectrometer operating at 500 MHz (1H frequency). NMR samples consisted of 2 and 10 mg/mL of PMMA48-b-PDMAEMA408 diluted in CDCl3 from a 55 mg/mL stock solution in 99.8% (v/v) methanol-d4. NMR samples in aqueous solution were prepared in 0.01 M sodium phosphate buffer, pH 6.2, 7.0, and 8.2, or 0.01 M borate buffer pH 9.0 containing 2 mg/mL of the copolymer and 10% (v/v) of D2O. 2D NOE experiments were recorded as matrices of 2048 × 700 complex points with a mixing time of 120 ms. Suppression of the water signal was achieved by pre-saturation during the inter-scan delay. All 2D NMR spectra were processed using NMRPipe and analyzed with CCPN Nmr Analysis [23].
Determination of the critical aggregation concentration of the polymers
An aliquot (4 μL) of a methanolic solution of NPN 1 × 10−3 M was added to 2 mL of buffer, final concentration was 2 × 10−6 M. Increasing volumes of a concentrated solution of the polymers, dissolved in the adequate buffer, were added with stirring, and the fluorescence spectra were registered (352 to 600 nm) using λexc = 353 nm (25 °C). The fluorescence emission at 418 nm was used to calculate the CAC from the intersection of the plot of I/Io vs. log (polymer). The fluorescence, at each copolymer concentration, was corrected by the dilution of the sample.
Elemental analysis of the copolymers was performed at the Analytical Instrumentation Center of the Chemistry Institute of the University of São Paulo.
Results
The macro chain transfer agents (macroCTA) of PMMA generating each PMMA block of the copolymers, and the PMMA-b-PDMAEMAs, were analyzed by GPC (see Methods). The values of Mw, Mn, and PDI of PMMA macroCTA, determined by GPC using the standard curves prepared with PMMA are in Table 1.
Due to the lack of appropriate standards for the PMMA-b-PDMAEMA copolymers, we considered the total molar mass of the copolymer as given by the sum of the respective PMMA and PDMAEMA blocks molar masses. For each copolymer, the PMMA block molar mass was measured by GPC while the PDMAEMA block molar mass was calculated from the number of DMAEMA units obtained by NMR. These materials presented, as expected, different Mw versus hydrodynamic ratio relationships compared to (homo) PMMA, making direct GPC analysis inaccurate, particularly to determine the composition.
MMA/DMAEMA ratios were determined using 1H-NMR (Fig. 2) as mentioned above. Chemical shift assignments were taken from the literature [24, 25]. The NMR spectrum of PDMAEMA displayed the expected peaks for the side chain methyl and methylene protons (Fig. 2D, peaks labeled a, b, and d) and two sets of peaks for the main chain methyl protons (e). Peaks due to the main chain methylene protons were difficult to observe due to exchange broadening (Fig. 2D). The presence of PMMA gave rise to a new peak at 3.55 ppm (c) due to the PMMA methyl protons (Fig. 2A–C) and to increased peak intensities of the main chain methylene protons (f) that could arise from both PMMA and PDMAEMA blocks. Increasing proportions of PMMA led to greater NMR signal intensities from the PMMA block. Interestingly the NMR signals due to the main chain methylene peaks (f), which are broad in the NMR spectrum of PDMAEMA alone, become sharper after addition of MMA probably due to better solubility of the copolymer in CDCl3.
PDMAEMA and PMMA ratios in the different copolymers were obtained using eqs. 1 and 2.
The ratios of the relative areas under the 1H NMR peak corresponding to the side chain methylene group of the ester of PDMAEMA (d), and the methyl group of the ester of MMA (c) (Fig. 2) are represented in Eq. 1, where
Ad refers to the area under the peak of PDMAEMA methylene group, while Ac is the area under the peak due to the PMMA methyl group. The coefficients 2 and 3 normalize the areas for the number of hydrogens from the methylene and methyl groups, respectively; n is the number of units of DMAEMA and m of PMMA.
The total polymer mass is given by the relative composition of the two monomers as described in Eq. 2:
where (PDMAEMA) and (PMMA) are the monomer molecular masses, i.e., 157.9 and 100, respectively. Results are in Table 2.
Elemental analysis
The elemental composition of all polymers was determined, and the theoretical values of C, H, and N calculated using the molar mass determined by GPC and NMR. The experimental values obtained by elemental analysis were in good agreement with the proposed copolymer structure (Table 3).
Critical aggregation concentration of the copolymers
The CACs of the copolymers were determined at pH 6.0, 8.5, and 10.0 using 1-(N-phenylamino) naphthalene (NPN) as a probe. NPN fluorescence increases when solubilized in a nonpolar medium such as the copolymer aggregate core [26]. The NPN concentration used here (2 × 10−6 M) was ca. hundred times lower than that of the polymer, unlikely to affect the measured values of CACs (see Supplementary Material, Figs. 1–4). The CACs of PMMA50-b-PDMAEMA269, PMMA48-b-PDMAEMA324, and PDMAEMA265 decreased with pH while that of PMMA94-b-PDMAEMA88 was pH-independent (Table 4). For a given polymer, the differences in CAC between pH 8.5 and 10.0 were small because above pH 8.5 polymers are essentially deprotonated (see below). For PDMAEMA265, the same aggregation occurred (not shown) even at pH 6, in which the amino groups start dissociating (see below). A more explicit relationship between the copolymers CAC and hydrophobic/hydrophilic ratios could be obtained using copolymers with structures of greater ratio differences than those studied here [22, 27]. Xiao and co-workers (2012), working with similar materials, found comparable values, although they indicated that CAC passes through a minimum when the PDMAEMA block increases and the PMMA block is kept constant (55 repeating units of PMMA per chain) [28]. The values of CACs reported here for materials with similar molar mass and composition are comparable to published data (Table 4) [1, 28].
Potentiometric titration of the copolymers
The pKa of the N,N-dimethylaminoethylamine, DMAEMA, in water, is 8.3 at 25 °C [29]. The titration curves of PDMAEMA and the copolymers, obtained at concentrations above their CACs, showed two endpoints: the first one corresponding to the titration of excess HCl and the second corresponding to the complete dissociation of the ammonium groups (Fig. 3). The dissociation of the ammonium groups started between pH 5.5–6.0, and complete dissociation was observed between pH 7.5–8.5, a range of ca. three pHs units. The apparent pKa (pKap) of the polymer dimethylammonium groups was significantly lower than the pKa of the monomer. The average pKap of each polymer, calculated from the pH value between the two inflection points in Fig. 3, is in Table 5.
Potentiometric titration of the copolymers. Titrations of the polymers were done in 30 mL of HCl. The mass of polymer was dissolved directly in the HCl solution. For each titration, the mass of the polymer and the concentration of HCl and NaOH are given. (A) PMMA48-b-PDMAEMA324 = 46.8 mg in HCl 0.0106 M, titrated with NaOH 0.083 M; (B) PMMA94-b-PDMAEMA88 = 15.9 mg in 0.012 M HCl and NaOH 0.1013 M; (C) PMMA50-b-PDMAEMA269 = 6.8 mg in HCl 0.012 and NaOH 0.065 M; (D) PDMAEMA265 = 39.5 mg in 0.0109 M HCl and NaOH 0.0792 M
The low pH range of dissociation of the amino groups in the polymer, compared with that of the monomer, may be explained by the electrostatic interactions influencing the dissociation of the protonated amino groups. When many charged groups are close to each other as in a charged cationic micelle (the expected morphology of the aggregates), a local anion condensation at the surface of the micelle is likely to occur.
Size determination of the copolymers aggregates in aqueous solutions by dynamic light scattering
The effect of pH on the hydrodynamic diameter (Dh) of the aggregates was determined using dynamic light scattering (DLS) (Fig. 4). Simple calculations (using the values of Dhs, the molecular weights, and a rough estimate of their density) showed that the aggregates contained approximately 800 monomers and, based on the packing parameter, they are, most probably, spherical [30]. The Dhs for PMMA50-b-PDMAEMA269 and PDMAEMA265 (35–40 nm) were similar at pH 6.0, and higher pH deprotonation of the ammonium groups led to a decrease in Dh for both polymers (Fig. 4). The presence of the small portion of the hydrophobic chain of PMMA in PMMA50-b-PDMAEMA269 was not enough to decrease the highly hydrophilic characteristic of this copolymer since PMMA comprised only ca. 14%.
Effect of the pH on the hydrodynamic diameter, Dh, of the polymer aggregates: (□) [PMMA50-b-PDMAEMA269] = 1.03 mg/mL M; (▲) [PMMA48-b-PDMAEMA324] = 0.99 mg/mL; (∆) [PMMA94-b-PDMAEMA88] = 0.99 mg/mL; (●) [PDMAEMA]265 = 0.9 mg/mL. The buffers used were: CAPS 0.01 M, pH 6.0; tris/HCl 0.01 M, pH 8.0, and borate 0.01 M, pH 10
Similarly, increasing the pH led to a Dh decrease for PMMA94-b-PDMAEMA88 and PMMA50-b-PDMAEMA269 (Fig. 4). With these two aggregates, the decrease in the hydrodynamic diameter can be attributed to the formation of more compact aggregates due to the decrease in electrostatic repulsion in the corona. For PMMA48-b-PDMAEMA324 increasing the pH from 6 to 10 the Dh increased only ca. 18%, reaching a value like that of PMMA94-b-PDMAEMA88. This indicates the delicate balance of forces between the neutral, but still polar, the amine group of PDMAEMA, and the hydrophobic character of PMMA.
The hydrodynamic diameters of PDMAEMAX-b-PMMA55 (X ranging from 11 to 337) micelles at pH = 7 are close to those reported here [28] (Xiao et al. 2012). Micelle diameters increase with pH (pH = 4.0; 7.0, and 9.0). The diameters have an inverse relationship with the PDMAEMA block: the larger the block, the smaller the aggregate. All their data can be rationalized by the packing parameter theory [31]. From our data, the packing parameter theory fails to explain the behavior of polymeric micelles with different PMMA cores such as the PMMA94-b-PDMAEMA88 aggregates presented here. In this case, the shrinking of the corona by protonation can exceed the change on the packing parameter.
Based on these results, it seems that there is a PDMAEMA/PMMA threshold ratio above which the aggregation number does not change (decrease). In this case, the increase on the molar mass of the PDMAEMA block leads to the enlargement of the aggregate, although the packing parameter change points to a smaller aggregate size. In a spherical aggregate, this proposal seems quite reasonable, since as the PDMAEMA chains are distant from the core, the mean distance between chains gets more substantial, and the weaker repulsion is not enough to induce a smaller aggregation number (number of chains per aggregate and size of the core do not change above the threshold ratio). We suggest that the aggregates of PMMA50-b-PDMAEMA269 e PMMA48-b-PDMAEMA324 are above the threshold ratio. At pH = 10, the collapse of the extended PDMAEMA corona of PMMA48-b-PDMAEMA324 changed the packing parameter to a more cylinder like, so the Dh increased at this pH.
Analysis of 1H 1D and 2D-NOE at pH 6.2, 7.0, 8.2, and 10.0 of PMMA48-b-PDMAEMA324
Monomer structure in the aggregates, as well as interactions between the MMA and DMAEMA groups as a function of the pH, was investigated by NMR. We recorded the 1D 1H NMR spectra of PMMA48-b-PDMAEMA324 in CDCl3 solutions containing two polymer concentrations, 2 mg/mL−1 and 10 mg/mL−1. At high polymer concentration, the NMR peaks of the methylene groups of the copolymer main-chain split in two broader peaks slightly downfield shifted (Fig. 5f). We excluded viscosity differences as a possible explanation for these spectral changes since, despite the differences in solute concentration, any change in viscosity would essentially affect the line widths. These concentration-dependent changes of the main chain methylene signals are likely to be explained by polymer aggregation in deuterated chloroform at a concentration of 10 mg/mL−1. The spectrum of PMMA48-b-PDMAEMA324 at 10 mg/mL (Fig. 5) was similar to that of PMMA94-b-PDMAEMA88 (Fig. 2C). In the last case, we note the existence of two peaks (which can be ascribed to two different CH2 f-protons) also suggesting the formation of copolymer aggregates due to the high copolymer concentration. The CAC for PMMA94-b-PDMAEMA88 in water was ca. 0.005 mg/mL at all pH (Table 5). It could also be inferred that the block polymer chains in the core structure have substantially restricted mobility compared to the block in bulk chloroform, and this fact would justify the lower intensity of the 1H signals of the –CH2– groups in the aggregates. Additionally, it is likely that the less-shielded signal is associated with the more hydrophilic block. In summary, these observations are consistent with the proposed aggregation of the PMMA48-b-PDMAEMA324 at higher concentrations in chloroform.
The 1D 1H NMR spectra of the copolymers in water were strongly affected by pH due to the changes in the ionization state of the DMAEMA ammonium groups. 1H-NMR spectra of PMMA50-b-PDMAEMA269, PMMA48-b-PDMAEMA324, and PMMA94-b-PDMAEMA88 were obtained in 0.01 M phosphate buffer, pH 6.2 and 8.2 and in borate buffer, 0.01 M, pH 10.0 at the same concentration of 2.8 mg/mL (Fig. 6). Consistent with their proximity to DMAEMA amine nitrogen, the DMAEMA amine methyl group resonances (Fig. 6A) shifted approximately 0.2 ppm upfield, and their intensity decreased, as the pH was increased from 6.2 up to 10. Similar trends were observed for the resonances of the side chain methylene protons (Fig. 6B), two bonds away from the DMAEMA amine group. These resonances were shifted upfield (from 2.93 and 2.88 to 2.62 ppm) and lost intensity (Fig. 6). The pH effects were not restricted to protons a few bonds away from DMAEMA amine group. They were also detected at the main chain methylene groups (f Fig. 6) where resonances become barely detectable at higher pH.
The nuclear Overhauser effect (NOE)—To get insights on conformational changes associated with the ionization of the copolymers, we obtained a series of 2D NOE spectra of aqueous solutions of PMMA48-b-PDMAEMA324 (Fig. 7). At low pH, the NOEs reflect mainly short-range correlations between the methylene protons of the DMAEMA main chain and side chain (peaks e and d, respectively) and all other protons in the polymeric chain as schematically summarized in Fig. 8 and observed in Fig. 7. Additional correlations between the neighboring MMA methyl protons and the main chain methylene protons (peaks f and e, respectively) were also observed. In contrast, the MMA side chain methyl group (peak c) did not show any NOE correlation, suggesting that this methyl group is positioned far away from the rest of the chain and may experience greater conformational freedom. All NOE correlations become progressively stronger as the pH increased from 6.2 to 10.0. This effect is apparent for the NOEs between the methylene (peak e) and the methyl protons (peak f) and the DMAEMA side chain (peaks a and b) (Figs. 7 and 8) despite detectable broadening of the main chain resonances (Fig. 6). The stronger NOEs could be explained by the increased compactness of the polymers at higher pH. Such a greater degree of compactness reflects on reduced flexibility at the DMAEMA side chains (a and b) whose signals display increased line widths at higher pH. Besides, the aggregates increase in size at higher pH, which also affects the NOE intensities. In combination, the pH effects on size and degree of compactness of the aggregates make the interpretation of the NOEs in terms of local structural changes a difficult task.
Scheme of the NOEs observed between protons of the copolymer at pH 6.2 and pH 10.0. The blue dashed lines indicate the NOE interactions at pH 6.2. The solid blue lines indicate the NOEs that were observed at pH 6.2 but became more evident at pH 10. The pink lines indicate the NOEs that are observable only at pH 10
Discussion
It is attractive to postulate that for a polymer where the hydrophobic/hydrophilic ratio approaches one, aggregation is controlled by the hydrophobic component rendering the pH effects on interfacial repulsion less critical. For a given polymer, the differences in CAC between pH 8.5 and 10.0 were small because above pH 8.5 polymers are essentially deprotonated (see below). For PDMAEMA265, aggregation occurred (not shown) even at pH 6, in which the amino groups start dissociating (see below). A more explicit relationship between the copolymers CAC and hydrophobic/hydrophilic ratios could be obtained using copolymers with structures with different ratios than those studied here [22, 27].
The Mw dependence of the Dh of the aggregates is unusual since the hydrodynamic diameter of the aggregates decreases with increasing Mw of the monomer (Fig. 4). However, we note that the polymer with the lowest Mw is that with the highest proportion of PMMA, rendering it more hydrophobic. Relatively small polymers, with high hydrophobicity, associate to form larger aggregates. It has been noted that “the micelle size seems to decrease with increasing alkyl group size” because larger aggregation numbers were calculated for smaller hydrophobic chains [32, 33].
For PMMA48-b-PDMAEMA324 and PMMA94-b-PDMAEMA88, the hydrophobic moiety corresponds to ~ 25 and 50% of the copolymers, respectively. This higher hydrophobicity (compared with the two polymers described above) led to an increase in the Dh of the aggregates, reaching values of 70 and 115 nm, respectively, at pH 6.0. The Dh values, at all pH, were higher for PMMA94-b-PDMAEMA88 than for PMMA48-b-PDMAEMA324 due to the higher content of PMMA and a smaller amount of PDMAEMA.
At pH = 6, the copolymers that have shorter PMMA blocks (leading the smaller nonpolar cores in the micelles) form aggregates with Dh ranging from around 40 up to 70 nm. For comparison, homo PDMAEMA aggregates display Dh = 35 nm. The difference between the Dh of PMMA50-b-PDMAEMA269 (~ 40 nm) and PMMA48-b-PDMAEMA324 (~ 70 nm) aggregates is probably due to the thicker PDMAEMA corona in the second one. At the same pH, PMMA94-b-PDMAEMA88 aggregates show a diameter of 110 nm. The low PDMAEMA/PMMA of this material indicates a form factor that resembles a cylinder (instead of a truncated cone), more likely to lead to larger micelles or even vesicles or lamellae [30].
As pH increases and PDMAEMA protonation drops, PMMA48-b-PDMAEMA324 micelles increase in size. This material has a higher PDMAEMA/PMMA ratio. Deprotonation causes the PDMAEMA corona to shrink, so that the form factor also changes in the direction truncated cone → cylinder, and, once again according to Israelachvili (ref), the aggregate size tends to increase to better accommodate each copolymer chain. PMMA94-b-PDMAEMA88, on the other hand, with a cone shape at pH = 6, shows a decrease on its micelle size, particularly at pH = 10 probably due to the shrink of the PDMAEMA corona caused by deprotonation [30]. From pH = 6 to pH = 10, the diameter of such aggregates come from roughly 115 to 85 nm.
PMMA94-b-PDMAEMA88 aggregates show a very slight pH dependence. Having an average PDMAEMA/PMMA ratio, it seems that the aggregates are subject to a balance of effects: as protonation falls, the form factor shifts to a cone, but the corona shrinks decreasing the overall diameter. The second effect still prevails, and the aggregate diameter decreases from 40 to 35 nm in a very similar way as PDMAEMA aggregates.
Protonation of PDMAEMA polymers in water is more complicated than DMAEMA itself or other monoamines [34]. Multiple amine groups in the side chain lead to complex group interactions such that, at the limit, the apparent pKas (pKap) may be different for each ammonium group. Closely spaced positive charges in a polymer [35] or in a micelle [36] can determine counterion condensation and, as H+ or OH− may bind to ionic micelles, local pH can differ from bulk pH by several pH units. Therefore, even when intrinsic pKas are unchanged in the polymer or aggregate, the measured pKap may be distinct to that of the monomer or undissociated species because of differences in the local pH. As a result, a broad buffered pH region was observed on titration of amine-containing polymers and, due to the positive charge effects, the mean pKap was shifted to lower pH compared to protonated tertiary amines [29, 37]. This is also true if PDMAEMA copolymers self-aggregate to micelles. As protonation/deprotonation ratios affect a variety of micellar properties, understanding and controlling degrees of protonation can lead to polymeric aggregates with a variety of different and exciting properties.
The pKas of the monomer and the polymer in PDMAEMA-b-PS micelles become identical at high ionic strengths, with obvious consequences on the zeta potential of the aggregates [38]. This effect is attributed to a weaker correlation between the acid/basic centers caused by ionic screening [38]. A marked contraction on the hydrophilic corona occurs as the ionic strength and pH increase, providing means to modify the micellar shape by controlling the degree of protonation of corona groups.
There seems to be no general rule for describing the effect of pH on the Dh of the polymer aggregates. This indicates the delicate force balance between the neutral, but still polar, the amine group of PDMAEMA, and hydrophobic character of PMMA.
The low pH range of dissociation of the amino groups in the polymer, compared with that of the monomer, may be explained by the electrostatic interactions influencing the dissociation of the protonated amino groups nearby. When a large number of charged groups are close to each other, such as in a charged cationic micelle (the expected morphology of the aggregates), a local anion condensation, at the surface of the micelle, is likely to occur. In the case of a positively charged surface, such as in the studied copolymers, the OH− concentration [OH−], at the surface particle is larger than that in bulk solution. The same effect is observed in polymers [39]. This high surface [OH−] facilitates the dissociation of protonated amines leading to an apparent decrease of pKap of the amino groups.
As the pH increases, the number of protonated amines decreases and the distance between the residual positive amines increases. The more considerable distance between the amine groups decreases the intensity of the electrostatic repulsion, thus increasing the apparent pKa of the amino groups which remain protonated. The pKa of these groups then approach that of amine monomers, free of electrostatic interactions [40].
Conclusion
We characterized diblock copolymers containing monomers of PMMA and DMAEMA by gel permeation chromatography and 1H-NMR, determining the molar mass and the ratio between the monomer blocks. In aqueous solution, these polymers form aggregates at a particular concentration (CAC) depend on the ratio between the PMMA and DMAEMA blocks and the number of monomers in each block. Titration of the amines of the copolymers showed a dissociation continuum within a very broad range of pH (about 2 or 3 pH units) confirming that amine dissociation occurs at pH above 5.5 until about pH 8.5. Polymer aggregation was pH dependent due to the protonated tertiary amines of the PMAEMA block which are responsible for the repulsion between the monomers. Thus, the aggregation of the copolymers increases with the deprotonation of the amines. The NMR results show that the pH affects the state of aggregation of these copolymers in water. At pH 6.2 and 7, the interaction between the copolymer monomers is smaller due to the electrostatic repulsion of charged amino groups. At pH > 8.2, as almost all the amino groups are in the neutral form, the copolymer monomers tend to be closer in the aggregate.
References
Riess G (2003) Micellization of block copolymers. Prog Polym Sci 28:1107–1170
Blanazs A, Armes SP, Ryan AJ (2009) Self-assembled block copolymer aggregates: from micelles to vesicles and their biological applications. Macromol Rapid Commun 30:267–277
Nagarajan R (2015) “Non-equilibrium” block copolymer micelles with glassy cores: a predictive approach based on a theory of equilibrium micelles. J Colloid Interface Sci 449:416–427
Smart T, Lomas H, Massignani M, Flores-Merino MV, Perez LR, Battaglia G (2008) Block copolymer nanostructures. Nano Today 3:38–46
Ling P, Xu W, Zhang T (2013) Polymeric micelles, a promising drug delivery system to enhance bioavailability of poorly water-soluble drugs. J Drug Deliv:1–15
Beija M, Salvayre R, Lauth-de Viguerie N, Marty JD (2012) Colloidal systems for drug delivery: from design to therapy. Trends Biotechnol 30:485–496
Guo X, Huang L (2012) Recent advances in nonviral vectors for gene delivery. Acc Chem Res 45:971–979
Huang LQ, Yuan SJ, Lv L, Tan GQ, Liang B, Pehkonen SO (2013) Poly(methacrylic acid)-grafted chitosan microspheres via surface-initiated ATRP for enhanced removal of cd(II) ions from aqueous solution. J Colloid Interface Sci 405:171–182
Ge Z, Xie D, Chen D, Jiang X, Zhang Y, Liu H, Liu S (2007) Stimuli-responsive double hydrophilic block copolymer micelles with switchable catalytic activity. Macromolecules 40:3538–3546
Braunecker WA, Matyjaszewski K (2008) Controlled/living radical polymerization: features, developments, and perspectives (vol 32, pg 93, 2007). Prog Polym Sci 33:165–165
Feng HB, Lu XY, Wang WY, Kang NG, Mays JW (2017) Block copolymers: synthesis, self-assembly, and applications. Polymers 9(10):494–525
Bettencourt A, Almeida AJ (2012) Poly(methyl methacrylate) particulate carriers in drug delivery. J Microencapsul 29:353–367
Lu Y, Park K (2013) Polymeric micelles and alternative nanonized delivery vehicles for poorly soluble drugs. Int J Pharm 453:198–214
Nicolai T, Colombani O, Chassenieux C (2010) Dynamic polymeric micelles versus frozen nanoparticles formed by block copolymers. Soft Matter 6:3111–3118
Agut W, Brûlet A, Schatz C, Taton D, Lecommandoux S (2010) pH and temperature responsive polymeric micelles and polymersomes by self-assembly of poly[2-(dimethylamino)ethyl methacrylate]-b-poly(glutamic acid) double hydrophilic block copolymers. Langmuir 26:10546–10554
Dai S, Ravi P, Tam KC (2008) pH-responsive polymers: synthesis, properties, and applications. Soft Matter 4:435–449
Jochum FD, Theato P (2013) Temperature- and light-responsive smart polymer materials. Chem Soc Rev 42:7468–7483
de Souza JCP, Naves AF, Florenzano FH (2012) Specific thermoresponsiveness of PMMA-block-PDMAEMA to selected ions and other factors in aqueous solution. Colloid Polym Sci 290:1285–1291
Huang Y, Yong P, Chen Y, Gao Y, Xu W, Lv Y, Yang L, Reis RL, Pirraco RP, Chen J (2017) Micellization and gelatinization in aqueous media of pH- and thermo-responsive amphiphilic ABC (PMMA82-b-PDMAEMA150-b-PNIPAM65) triblock copolymer synthesized by consecutive RAFT polymerization. RSC Adv 7:28711–28722
Rodríguez-Hernández J, Chécot F, Gnanou Y, Lecommandoux S (2005) Toward ‘smart’ nano-objects by self-assembly of block copolymers in solution. Prog Polym Sci 30:691–724
Baussard J-F, Habib-Jiwan J-L, Laschewsky A, Mertoglu M, Storsberg J (2004) New chain transfer agents for reversible addition-fragmentation chain transfer (RAFT) polymerization in aqueous solution. Polymer 45:3615–3626
de Souza VV, de Carvalho Noronha ML, Alves Almeida FL, Ribeiro Prado CA, Doriguetto AC, Florenzano FH (2011) CMC of PMMA-block-PDMAEMA measured by NPN fluorescence. Polym Bull 67:875–884
Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas P, Ulrich EL, Markley JL, Ionides J, Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins Struct Funct Bioinf 59:687–696
Shen JN, Ye YF, Zeng GN, Qiu JH (2011) Preparation and characterization of PMMA-b-PDMAEMA/polysulfone composite membranes by RAFT polymerization and their permeation performance of carbon dioxide. Advanced Materials Research 284–286:1717–1723
Zhao Y, Guo K, Wang C, Wang L (2010) Effect of inclusion complexation with Cyclodextrin on the cloud point of poly(2-(dimethylamino)ethyl methacrylate) solution. Langmuir 26:8966–8970
Saitoh T, Matsushima S, Hiraide M (2004) Aerosol-OT–γ-alumina admicelles for the concentration of hydrophobic organic compounds in water. J Chromatogr A 1040:185–191
Chatterjee U, Jewrajka SK, Mandal BM (2005) The amphiphilic block copolymers of 2-(dimethylamino) ethyl methacrylate and methyl methacrylate: synthesis by atom transfer radical polymerization and solution properties. Polymer 46:10699–10708
Xiao G, Hu Z, Zeng G, Wang Y, Huang Y, Hong X, Xia B, Zhang G (2012) Effect of hydrophilic chain length on the aqueous solution behavior of block amphiphilic copolymers PMMA-b-PDMAEMA. J Appl Polym Sci 124:202–208
van de Wetering P ME, Schuurmans-Nieuwenbroek NM, van Steenbergen MJ, Hennink WE (1999) Structure-activity relationships of water-soluble cationic methacrylate/methacrylamide polymers for nonviral gene delivery. Bioconjug Chem 10:589–597
Israelachvili J (2011) Intermolecular and surface forces. Elsevier Inc, Amsterdam
Eastoe J (2005) Surfactant chemistry. Wuhan University Press, Wuhan
Barbieri BW, Strauss UP (1985) Effect of alkyl group size on the cooperativity in conformational transitions of hydrophobic polyacids. Macromolecules 18:411–414
Strauss UP, Barbieri BW (1982) Estimation of the cooperative unit size in conformational transitions of hydrophobic polyacids. Macromolecules 15:1347–1349
Lee H, Son SH, Sharma R, Won Y-Y (2011) A discussion of the pH-dependent protonation behaviors of poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA) and poly(ethylenimine-ran-2-ethyl-2-oxazoline) (P(EI-r-EOz)). J Phys Chem B 115:844–860
Manning GS (1979) Counterion binding in polyelectrolyte theory. Acc Chem Res 12:443–449
Quina F, Chaimovich H (1979) Ion-exchange in micellar solutions .1. Conceptual-framework for ion-exchange in micellar solutions. J Phys Chem 83:1844–1850
Přádný M, Ševčík S (1987) Precursors of hydrophilic polymers, 7. Potentiometric properties and structure of copolymers of 2-dimethylaminoethyl methacrylate. Die Makromol Chem 188:227–238
Laaser JE, Jiang Y, Sprouse D, Reineke TM, Lodge TP (2015) pH- and ionic-strength-induced contraction of polybasic micelles in buffered aqueous solutions. Macromolecules 48:2677–2685
McKernan BA, Manning GS, Romsted LS (2003) Estimating concentrations of condensed counterions around a polyelectrolyte by chemical trapping. Conducting Polymers And Polymer Electrolytes: From Biology To Photovoltaics, pp. 184–199
Borukhov I, Andelman D, Borrega R, Cloitre M, Leibler L, Orland H (2000) Polyelectrolyte titration: theory and experiment. J Phys Chem B 104:11027–11034
Acknowledgments
IMC, HC, and RKS are CNPq research scholars. GKVS acknowledges the Projeto Biocomputacional/CAPES (proc. no. 23038.004630/2014-35) and CNPq (proc. 457733/2014-4).
Funding
FAPESP (grant number 2013/08166-5 and 2016/07490-1). IMC has been supported by the INCT-FCx (Instituto Nacional de Ciência e Tecnologia de Fluidos Complexos (CNPq/CAPES/FINEP/FAPESP) and NAP-FCx (Núcleo de Apoio à Pesquisa de Fluidos Complexos da Universidade de São Paulo).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 76 kb)
Rights and permissions
About this article
Cite this article
Saraiva, G.K.V., de Souza, V.V., de Oliveira, L.C. et al. Characterization of PMMA-b-PDMAEMA aggregates in aqueous solutions. Colloid Polym Sci 297, 557–569 (2019). https://doi.org/10.1007/s00396-019-04482-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00396-019-04482-w