Abstract
The roles and the underlying mechanisms of M1-type macrophages in angiogenesis and postmyocardial infarction (MI) cardiac repair have remained unclear. In this study, we investigated the role of M1-like macrophage-derived exosomes in a MI microenvironment. We found that the proinflammatory M1-like-type macrophages released an extensive array of proinflammatory exosomes (M1-Exos) after MI. M1-Exos exerted an anti-angiogenic effect and accelerated MI injury. They also exhibited highly expressed proinflammatory miRNAs, such as miR-155. miR-155 was transferred to endothelial cells (ECs), leading to the inhibition of angiogenesis and cardiac dysfunction by downregulating its novel target genes, including Rac family small GTPase 1 (RAC1), p21 (RAC1)-activated kinase 2 (PAK2), Sirtuin 1 (Sirt1), and protein kinase AMP-activated catalytic subunit alpha 2 (AMPKα2). M1-Exos depressed Sirt1/AMPKα2–endothelial nitric oxide synthase and RAC1–PAK2 signaling pathways by simultaneously targeting the five molecule nodes (genes), reduced the angiogenic ability of ECs, aggravated myocardial injury, and restrained cardiac healing. The elucidation of this mechanism provides novel insights into the functional significance of M1 macrophages and their derived exosomes on angiogenesis and cardiac repair. This mechanism can be used as a novel potential therapeutic approach for the prevention and treatment of MI.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myocardial infarction (MI), one of the main causes of mortality and morbidity worldwide, results in ischemia and irreversible death of cardiomyocytes (CMs). Clinically, the immediate recovery of blood supply (e.g., percutaneous coronary intervention) that maximizes the survival of CMs and preserves cardiac function is the principal approach of MI treatment. However, reperfusion treatment in clinical practice has many problems, including ischemia–reperfusion injury [12] and opportunity loss for patients who miss the optimal timing. Therapeutic angiogenesis has been widely explored in terms of ischemic heart disease treatment [7]. Therapeutic angiogenesis plays a remarkable role in promoting MI repair and preventing adverse ventricular remodeling [7]. Therefore, extensive research is needed to determine the exact influencing factors and underlying mechanisms of angiogenesis after MI.
Immuno-inflammation, especially macrophage modulation, serves as an imperative regulator during cardiac repair and remodeling post-MI [9, 25, 52]. After MI, CM death induces an inflammatory response to degrade and remove cells and extracellular materials. Several hours after MI, monocytes invade the infarcted myocardium and then gradually differentiate to macrophage in a MI microenvironment. Macrophages act as dominant immune cells that control the progression and resolution of inflammation. Macrophages are functionally and phenotypically polarized into two typical subpopulations, namely, classically activated macrophages (M1) and alternatively activated macrophages (M2), which are induced in a special cardiac microenvironment [5, 31, 40, 42, 51]. Though the M1/M2 paradigm is an oversimplified classification, this notion is strongly rooted in the field. This model is often used for the sake of clarity. After MI in mouse, M1 macrophages dominate on the first 5 days (peak on days 3–4) in the first inflammatory stage, and M2 macrophages dictate after day 5 in the second inflammatory phase [5, 40, 43]. M1 and M2 macrophages exhibit distinct functions and subsequently aggravate or resolve inflammation in the infarcted heart tissue. M1 cells create high levels of proinflammatory cytokines (e.g., tumor necrosis factor-alpha [TNF-α], interleukin-1 beta [IL-1β]) and proteinases (e.g., matrix metalloproteinases), displaying phagocytic and proteolytic properties, which aid in clearing cellular debris [5, 31, 40, 42, 51]. By contrast, M2 macrophages produce anti-inflammatory/reparative cytokines (e.g., interleukin-10, vascular endothelial growth factor [VEGF]), mediate the resolution of inflammation, and promote injured tissue repair/remodeling [5, 31, 40, 42, 48, 51].
Although M1 macrophages play a positive role, their prolonged presence extends the proinflammatory state and enhance extracellular matrix degradation and cellular death. This condition results in the expansion of the infarcted area and inadvertent impediment of cardiac repair after MI [9, 20, 33, 46]. The excessive activities of M1 macrophages promote CM death [20], constrain myocardial regeneration [46], propagate fibrosis [14], and suppress neovascularization [49], leading to the high risk of cardiac rupture [21]. Experimental studies have proven that target M1 monocytes/macrophages, which lessen the duration of the proinflammatory phase, can recover functional cardiac output after MI [11]. Promoting the M1/M2 switch or dynamic balance has become an appealing concept of a novel target of MI therapy [3, 48, 51]. Though the entire macrophage population has been defined as a proangiogenic effector because the deletion or pharmaceutical inhibition of macrophages impairs endothelial angiogenesis during MI healing [1, 2], disputes have been raised for the different subsets of macrophages acting as distinct regulator in angiogenesis. Anti-inflammatory M2 macrophages promote angiogenesis rather than proinflammatory M1 macrophages [24]. As a proinflammatory responder and inducer, the antiangiogenic effect of M1 macrophages has been observed in vitro and in vivo [29, 59]. Therefore, the valid influence and exact underlying mechanisms of M1 macrophages in angiogenesis remain unclear and require further investigations.
Exosomes are 30–150 nM cell-derived nanovesicles present in all tissue spaces, body fluids, including blood and urine, and cell culture medium. Exosomes possess specialized functions and play a significant role in intercellular signaling during versatile physiological processes and pathological settings by transferring embodied cargos, such as miRNAs and proteins, into recipient cells. Recently, we revealed that an infarcted heart becomes a harsh microenvironment, which contains detrimental CM-derived exosomes to accelerate transplanted bone marrow mesenchymal stem cell injury [19]. Monocytes/macrophages are important sources of body exosomes [57]. The biomolecular composition of macrophage-released exosomes reflects the particular features of the original cells: strong immune or inflammatory regulatory activity [57]. The role of exosomes or extracellular vesicles derived from M1 phenotype macrophages has been recently elucidated. For instance, human monocyte exosomes provoked by lipopolysaccharides (LPS)/interferon-γ (IFN-γ) [50] or hypertensive conditions [44] induce the activation of nuclear factor-kappa B activation and the expression of intercellular cell adhesion molecule-1 (ICAM-1), chemokine ligand-2, and cytokine interleukin-6 (IL-6), leading to the dysfunction of endothelial cells (ECs). M1-like macrophage release exosomes derived from obese adipose tissue encompass abundant miRNAs, such as miR-155 and miR-146a, causing insulin resistance in mice [58]. Exosomes from human macrophages suppress EC migration by controlling integrin trafficking [32] and participate in cardiac repair/remodeling, including the inhibition of cardiac fibroblast (CF) proliferation and the promotion of inflammation after MI [54].
In this study, we hypothesized that M1-like macrophage-derived exosomes transfer specific miRNA into ECs, exerting a proinflammatory role and influencing angiogenesis and repair after MI. We investigated the underlying mechanisms to improve our understanding on angiogenesis and therapeutic implication for the potential clinical manipulation strategy of MI.
Materials and methods
All animal protocols were in accordance with the Guidelines for the Care and Use of Laboratory Animals published by the US Nation Institutes of Health (8th edition, 2011) and approved by the Animal Care and Use Committee of Guangzhou Medical University (Guangzhou, China; permit number: 2018-071).
Supplemental Methods are available within the article and in the online Data Supplement.
Major resource
The detailed description of all the antibodies, chemicals, commercial kits, cell lines, mouse strains, primers and oligonucleotides, softwares, and algorithms used in this study is listed in the Major Resource Tables in the Online Data Supplement.
Mice
Eight-to-ten-week-old wild type (WT) C57BL/6 J male mice were purchased from Guangdong Medical Laboratory Animal Center (Guangzhou, China). Rab27a−/− and aSMase−/− mice on a C57BL/6 background were purchased from Cyagen Biosciences Inc. (Guangzhou, China). During the experiment, the mice were kept on a 12 h light/12 h dark cycle in pathogen-free conditions with free access to food and water at the institutional animal care facility. They were anesthetized with 2.0% isoflurane during MI model operation. After the study, all the mice were first anesthetized by allowing them to inhale 2.0% isoflurane and subjected to cervical dislocation.
MI model
The mice were anesthetized with 2.0% isoflurane, ventilated with room air, and placed on a mechanical small animal ventilator (ALCBIO, ALC-V8S) under direct vision in accordance with our previous experience [19, 34] and guidance [35, 36] with the mouse models of MI. The third intercostal space was exposed, and the thoracic cavity was penetrated. The pericardium was dissected, and the left anterior descending (LAD) coronary artery was ligated using a 9–0 polyester suture. Successful ligation was marked with the white anterior wall of the left ventricle, weakened myocardial tissue movement around the apex, and an ST elevation on electrocardiograms. Mice in the sham-operated group underwent the same surgical procedure but without the LAD ligation. Gentian violet infusion of vena cava was performed to confirm the consistency in LAD ligation between animals and groups. For the intraperitoneal injection of BrdU, 100 mg/kg BrdU was administered to the mice every 3 days after MI.
Induction of M0 macrophage and M1 macrophage
Human THP-1 monocytes were plated in RPMI 1640 medium with 10% fetal bovine serum (FBS) and phorbol 12-myristate 13-acetate (PMA) (10 ng/mL). After 48 h of differentiation, the cell supernatant was discarded. Unattached cells were removed with PBS, and a new 1640 medium with 10% exosome-depleted FBS (ViVaCell, #C38010050, China) was added. For mouse RAW264.7 macrophages, the cells were replaced with DMEM (10% exosome-depleted FBS) at 70%–80% confluency. PMA-induced THP-1 or RAW264.7 cells were then stimulated with human or mouse GM-CSF (50 ng/mL) with IFN-γ (20 ng/mL), IFN-γ (20 ng/mL) alone, TNF-α (100 ng/mL), IL-1β (100 ng/mL), or H2O2 (25 μM) for 24 h. Alternatively, PMA-induced THP-1 or RAW264.7 cells were cultured in a new glucose-free 1640 (Gibco, Cat#11879-020) or glucose-free DMEM (Gibco, Cat#11966-025) with 10% exosome-depleted FBS. The cells were exposed to a hypoxic condition created using a hypoxic work incubator with 1% O2, 94% N2, and 5% CO2 at 37 °C for 24 h. After the treatment, macrophages and culture supernatants were collected for analysis. For mouse peritoneal macrophages (PMs) or bone marrow-derived macrophages (BMDMs), the cells were stimulated with 50 ng/mL GM-CSF and 20 ng/mL IFN-γ for 24 h.
Exosome purification
Exosomes were isolated from the heart through differential ultracentrifugation in accordance with previously described methods with modifications [23] and guideline recommendation [4]. In brief, the piece of heart tissues was incubated at 37 °C in a shaking water bath with 275 U/mL collagenase type II solution for 30 min. The tissue was returned to ice and diluted with PBS containing protease and phosphatase inhibitors. The tissue solution was mixed gently with a pipette tip. After brief centrifugation, the sample pellet was prepared for protein concentration assay, whereas the supernatant was filtered through a 70 μm mesh filter (352350, BD Falcon) and then to a 40 μm mesh filter (352340, BD Falcon). The filtrate was then passed through a 100 kD cut-off filter. The samples were sequentially centrifuged at 300×g for 10 min at 4 °C, 2000×g for 10 min at 4 °C, and 10,000×g for 30 min at 4 °C to discard cells, membranes, and debris. The supernatant was filtered through a 0.22 μm filter (Merck Millipore) and centrifuged at 100,000×g for 90 min at 4 °C to pelletize the exosomes. The exosomes were then washed with cold PBS and recentrifuged at the same speed using a SW41 rotor (Beckman Coulter) and resuspended in 100 μL of 0.22 μm filtered cold PBS.
Plasma exosomes were purified through differential ultracentrifugation. In brief, blood from mice was drawn into blood collection tubes containing buffered sodium citrate as anticoagulants and then diluted 1:1 in ice-cold PBS. After centrifugation at 2000×g for 30 min at 4 °C, the plasma was systematically collected without disturbing the lower layers that contained blood cells and platelets and then transferred to new tubes. The plasma was centrifuged at 10,000×g for 45 min at 4 °C to remove cell debris and microvesicles. The supernatant was systematically transferred to new tubes and centrifuged at 120,000×g for 2 h at 4 °C using a 45Ti rotor (Beckman Coulter). The pellets were resuspended in 2 mL of PBS and diluted to a large volume. The suspension was filtered through a 0.22 μm filter to remove contaminating apoptotic bodies, microvesicles, and cell debris and collected in fresh ultracentrifuge tubes at 120,000×g for 70 min at 4 °C. The pellets were dissolved in 50–100 μL of 0.22 μm filtered PBS for downstream analysis.
Exosomes from the culture medium supernatant were isolated through differential ultracentrifugation. In brief, conditioned medium from THP-1, RAW264.7, PMs, or BMDMs with different treatments were centrifuged at 300×g for 10 min at 4 °C to pelletize the cells. The supernatant was centrifuged at 2,000×g for 20 min at 4 °C, transferred to new tubes, and centrifuged at 10,000×g for 40 min at 4 °C. The supernatant was filtered through a 0.22 μm filter to remove cellular debris, apoptotic bodies, and microvesicles. The filtered solution was ultracentrifuged at 120,000×g for 90 min at 4 °C. All pellets were washed with 10 mL of PBS and recentrifuged at the same speed. After the supernatant was removed, the pellets were dissolved in 50–200 μL of 0.22 μm filtered PBS.
Transmission electron microscopy (TEM) imaging
Exosomes were imaged through TEM. In brief, 20 μL of the fresh sample of exosomes that were not subjected to freezing temperatures at any step during processing was loaded on carbon-coated copper electron microscopy grids for 2 min. The samples were then negatively labeled with phosphotungstic acid solution for 5 min. The grids were washed thrice with PBS to remove extra phosphotungstic acid solution and maintained under a semi-dry condition using filter paper. Images were obtained with a Hitachi H7650 TEM (Japan) at 80 kV.
Nanoparticle tracking analysis (NTA)
The size distribution and concentration of exosomes were analyzed using a NanoSight NS300 system equipped with a 405 nm laser. In brief, 10 μL of exosomes with 990 μL of 0.22 μm filtered sterile PBS was pushed slowly using a 1 mL syringe and illuminated with a laser. Their movements under Brownian motion were recorded in 30 s sample videos, which were analyzed with the NTA analytical software (NanoSight, version 3.0). At least three videos were captured for each individual sample. The capture and analysis settings were manually set in accordance with the manufacturer’s instructions.
Uptake of exosomes and treatment of human coronary artery ECs (HCAECs)
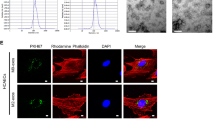
Purified exosomes were labeled with a PKH26 red fluorescent cell linker. Exosomes were resuspended in 1 mL of diluent C and 4 μL of PKH26 ethanolic dye was added to 1 mL of diluent C to prepare a 4 × 10−6 M dye solution. Exosome suspension was mixed with the dye solution for 5 min and then incubated in 1% BSA for 1 min to stop the staining. Labeled exosomes were ultracentrifuged at 100,000×g for 70 min, washed with PBS, ultracentrifuged again, and resuspended in 50 μL of PBS. PKH26-labeled exosomes were incubated with HCAECs at 37 °C for 12 h. ECs were fixed with 4% PFA and washed with PBS. The cytoskeleton was labeled with Phalloidin-iFluor 488 at room temperature for 30 min, whereas the nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI). The uptake of the labeled exosomes by HCAECs was determined through microscopy or confocal microscopy.
For the treatment, HCAECs were cultured in EBM-2 supplemented with 5% exosome-free FBS and endothelial growth media supplement mix under standard cell culture conditions (37 °C and 5% CO2). After the cells were grown to 70–80% confluency, M0-Exos (2 × 109/mL), M1-Exos (2 × 109/mL), or PBS was added. Analysis was conducted after 24 h.
miR-155 translocation analysis
THP-1 cells were transfected with Cy3-miR-155-5p for 48 h and then induced into M1-like macrophages. M1-Exos were ultracentrifuged from the conditioned medium and cocultured (2 × 109/mL) with HCAECs for 12 h. HCAECs were washed with PBS and fixed with 4% PFA, the cytoskeleton of HCAECs was selectively stained with Phalloidin-iFluor 488 at room temperature for 30 min, and the nucleus was stained with DAPI. The internalization of exosomal miR-155-5p was confirmed through confocal microscopy.
Alternatively, M1-Exos were pretreated with 0.5% Triton X-100 for 5 min and then treated with 5 μL of RNase (20 U/μL) for 1 h. Exosomes were washed with PBS and ultracentrifuged at 120,000×g for 90 min at 4 °C to remove residual RNase and Triton X-100. The expression levels of miR-155 in ECs were determined through qPCR.
Bioinformatics analysis
HCAECs were infected with Lentivirus-hsa-miR-155-5p (LV-miR-155) or Lentivirus-negative control (LV-NC) for 72 h. RNA-sequencing was then performed with Illumina HiSeqTM2500 (Gene Denovo Biotechnology Co., China). mRNAs were screened in accordance with the criteria of change ratio ≤ − 2.0 for further bioinformatics analysis. Online softwares, including microRNA.org (www.microrna.org), miRWalk (www.mirwalk.umm.uni-heidelberg.de/), and miRTarBase (www.mirtarbase.mbc.nctu.edu.tw), were used to perform bioinformatics analysis between miR-155-5p and candidate target genes.
Quantification and statistical analysis
The results are shown as mean ± SEM unless otherwise specified. The differences between groups were analyzed using Student’s t test for two groups or by one- or two-way ANOVA followed by Tukey–Kramer post hoc analysis when more than two groups were compared. The survival rate was estimated by the Kaplan–Meier method and compared via a log-rank test. Statistical significance was considered at P < 0.05. All analyses were performed with SPSS version 16.0 (SPSS Inc.) or Excel.
Results
M1-like macrophages secrete exosomes under MI microenvironment
On day 3, the exosomes were collected and separated from the heart tissue of the mouse MI model through serial, differential centrifugation, and ultracentrifugation steps (detailed in the “Materials and methods” section). The morphological characteristics of the isolated pellets were directly examined via TEM, and the particles were revealed as cup-shaped membrane-bound vesicles with a diameter of approximately 100 nm (Fig. 1a). To illustrate the purified particles in detail, we performed NTA to measure the size distribution of the vesicles. The diameters of almost all particles ranged between 30 and 150 nm, with an average of 104 ± 11 nm (Fig. 1b). Furthermore, in comparison with tissue lysates by immunoblot analysis, vesicle preparations highly enriched the exosomal marker proteins Alix, CD63, and CD81 but not Calnexin, which is an endoplasmic reticulum marker (Fig. 1c). Therefore, the property analysis demonstrated that the isolated vesicles released from the MI tissue comprised the characteristic features of exosomes.
Secretion of exosomes derived from M1-like macrophages in a MI microenvironment. a TEM images of the isolated exosomes from cardiac tissues in the MI model on day 3 (n = 4). Bar: 100 nm. b Representative images of the NTA of the distribution and average diameter of exosome size (n ≥ 6). c Characterization of the exosomal markers of exosomes derived from heart tissues (n = 3). Proteins (15 μg) of the sample was loaded in each well. d NTA shows the concentration of the isolated exosomes derived from cardiac tissues (left panel) or circulating blood (right panel) on day 3 in the sham or MI model. n = 4–5. ***P < 0.001; **P < 0.01, MI vs. sham. e Heart tissues from MI mice on day 3 were immunostained against cTnI (Red) or CD63 (green) compared with those of the sham control. Note that CD63 staining increases in the MI group. Bar: 50 μm. n = 4. f Rab27a−/− or aSMase−/− inhibited the release of exosomes on either MI or sham treatment in cardiac tissues rather than that of the WT group. Exosomes were isolated from the MI model on day 3. ***P < 0.001, MI vs. sham. ###P < 0.001; ##P < 0.01, knockout vs. WT. g Time course analysis of M1-Exos following MI (n = 4). GAPDH served as external control, indicating that the same amounts of heart tissues were used to isolate exosomes between treatments. Right panel: statistical bar. *P < 0.05; **P < 0.01; ***P < 0.001, indicating treatment vs. sham. #P < 0.05; ##P < 0.01, Day 3 vs. Day 1. h NTA revealed that human THP-1 (left panel) and mouse RAW246.7 (right panel) M1-like macrophages released more exosomes (M1-Exos) because of GM-CSF/IFN-γ, IFN-γ alone, TNF-α, IL-1β, OGD, or H2O2 treatment for 24 h. n = 3–6 biological replicates. ***P < 0.001; **P < 0.01; *P < 0.05, indicating treatment vs. control. i Peritoneal macrophages (PMs) or bone marrow-derived macrophages (BMDMs) M1 released more exosomes (M1-Exos) than that of M0 macrophages (M0-Exos). Cells were treated with GM-CSF (50 ng/mL)/IFN-γ (20 ng/mL) for 24 h. n = 4 biological replicates. *P < 0.05; **P < 0.01, M1 vs. M0
We then observed that the number of exosomes derived from either the cardiac tissue or the circulating blood increased on day 3 post-MI compared with that of the sham control (Fig. 1d). The expression level of CD63 detected through immunohistochemistry in the myocardium was significantly higher than that of the sham control. This result confirmed that exosomes accumulated between cardiac tissues after MI (Fig. 1e). Rab27a is required for the docking of multivesicular bodies to the plasma membrane and the secretion of exosomes [45]. Acid sphingomyelinase (aSMase) mediates the biogenesis of extracellular vesicles [53]. We used Rab27a−/− and aSMase−/− mice to evaluate the influence of post-MI exosomes in the heart. We found that either Rab27a or aSMase deletion diminished the extent of the increased exosomes induced by the MI stimulus compared with that of the WT within the cardiac tissue (Fig. 1f).
Immuno-inflammation plays a vital role in cardiac repair after MI [9, 25]. Considering that M1 macrophages definitively dominate on day 3 post-MI and their activities reach their peak [5, 40], we hypothesized that exosomes derived from M1 macrophages (hereafter abbreviated as M1-Exos) on day 3 exerted crucial functions in cardiac repair/remodeling. Within the isolated exosomes from the time-course of MI hearts, we found that the marker proteins of M1 macrophage, including CD86, CD11c, and inducible nitric oxide synthase (iNOS), peaked on day 3 (Fig. 1g). This pattern was consistent with the dynamic change of exosomal markers, the expressions of CD9 and CD63 (Fig. 1g).
Macrophage polarization is deterministically dictated by the signals from the tissue microenvironment. Following MI, cardiac tissues are filled with early pro-M1 macrophage mediators, including ischemic and hypoxic stress, and massive chemokines and proinflammatory cytokines, including granulocyte–macrophage colony-stimulating factor (GM-CSF), IFN-γ, TNF-α, and IL-1β in vivo [5, 31, 40, 42, 51]. Each of these factors can generate the polarization of M1 macrophages to certain degrees and shape their phenotype and function [31, 37, 42, 63]. First, to ascertain these factors, we observed that GM-CSF, IFN-γ, TNF-α, IL-1β, and oxygen and glucose deprivation ([OGD] for simulating the ischemic and hypoxic status) promoted M1 macrophage markers, including the mRNA levels of TNF-α, IL-1β, IL-6, interleukin-12beta (IL-12β), C–C motif chemokine receptor 7 (CCR7), and C–X–C motif chemokine 10 (CXCL10) (Fig. S1a), and the protein expression levels of CD86 and iNOS (Fig. S1b). These results reflected the differentiation of M1 macrophages. We then used commercial exosome-depleted FBS (Fig. S2) to culture macrophages. The expression levels of exosomal markers, including CD63 and CD81, were identified in M1-Exos in the same way, except for Calnexin (Figs. S3a–S3b). We found that human M1 THP-1 and mouse RAW246.7 macrophages, which were elicited by GM-CSF with IFN-γ; IFN-γ alone; or TNF-α, IL-1β, OGD, or H2O2 treatment, released more exosomes (M1-Exos) (Fig. 1h). Considering that IFN-γ with GM-CSF or LPS is widely used to induce M1 macrophage differentiation in vitro [5, 31, 40, 42, 51], we induced the polarization of M1-like macrophages through IFN-γ plus GM-CSF treatment [42] to mimic the in vivo setting (Figs. S1a–S1b) because no LPS existed during MI. We also confirmed that PM and BMDM M1 cells released more exosomes (M1-Exos) than that of M0 macrophages (M0-Exos), which were induced by GM-CSF/IFN-γ (Fig. 1i). Collectively, these data demonstrate that the proinflammatory factors, such as GM-CSF, IFN-γ, TNF-α, IL-1β, and hypoxia and ischemia, promote the significant secretion of M1-Exos within the first inflammatory phase in a MI microenvironment.
M1-Exos exacerbate MI injury and inhibit cardiac angiogenesis
We hypothesized that M1-Exos played a significant role in cardiac repair after MI. To test this hypothesis, we delivered M0-Exos or M1-Exos stemmed from mouse RAW246.7 macrophages through intramyocardial injection in the MI model. Under the consistency in LAD ligation between animals and groups (Fig. S4), the survival rate of the M1-Exos treatment was lower than that of M0-Exos treatment or MI alone (Fig. 2a). The M0-Exos treatment showed no clear change in the cardiac function compared with that of MI alone 28 days after infarction. By contrast, M1-Exos injection further declined the ejection fraction (EF) and fractional shortening (FS) values compared with that of the M0-Exos treatment or MI alone, accelerating the inhibition of left ventricular contractility and cardiac function (Fig. 2b). M1-Exos expanded the infarct size based on the triphenyltetrazolium chloride (TTC) staining assay (Fig. 2c). Angiogenesis was enhanced during post-MI healing to support cardiac repair under normal conditions. However, the handling of M1-Exos inhibited the expression levels of alpha-smooth muscle actin (α-SMA) and CD31 through immunohistochemistry analysis compared with that of MI alone or M0-Exos treatment both on days 14 and 28 (Fig. 2d). BrdU/CD31 double staining obtained similar observation (Fig. S5). Furthermore, M1-Exos, not M0-Exos, inhibited the VEGFA-VEGF receptor 2 (VEGFR2) singling (Fig. 2e) in the heart. These data suggest that M1-Exos impeded angiogenesis in vivo after MI. Therefore, M1-Exos aggravate cardiac injury and inhibit heart repair after MI.
Inhibition of cardiac function and post-MI repair by M1-Exos. a Intramyocardial delivery of mouse M1-Exos decreased the survival rate of mice. b M1-Exos reduced the EF and FS values more than M0-Exos treatment on day 28 after MI. Echocardiography images (left panel), EF value (middle panel), and FS value (right panel) are shown. c TTC assay showed that M1-Exos aggravated infarct size compared with that of M0-Exos treatment or MI alone. d M1-Exos repressed the staining of α-SMA and CD31 in MI heart tissues on day 14 and 28. M1-Exos were derived from mouse RAW246.7 M1 macrophages. Bar: 50 μm. n = 4. ***P < 0.001; **P < 0.01, MI alone or M0-Exos vs. the sham group. ###P < 0.001; ##P < 0.01; #P < 0.05, M1-Exos vs. M0-Exos or MI alone. e M1-Exos inhibited the expression levels of VEGF-A and VEGFR2 (n = 3)
M1-Exos prevent the angiogenic capability of ECs
Exosomes, intercellular communication mediators, transmit cargos and influence recipient cells. To assess the direct effect of M1-Exos on ECs, we cocultured human THP-1-derived M1-Exos with HCAECs. We observed that PHK-26-labeled M1-Exos could be absorbed by ECs (Fig. 3a). We then found that M1-Exos downregulated the phosphorylation levels of Akt and endothelial nitric oxide synthase (eNOS) compared with that of M0-Exos (Fig. 3b), reflecting the inhibition of Akt–eNOS signaling pathway. M1-Exos enhanced the secretion of soluble ICAM-1 (sICAM-1) and soluble vascular cell adhesion molecule-1 (sVCMA-1) (Fig. 3c), corroborating the EC injury.
Inhibition of EC function and angiogenic ability by M1-Exos. a PHK26-labeled M1-Exos were taken up by HCAECs. PKH26-labeled exosomes were incubated with ECs at 37 °C for 12 h. Exosome-depleted PHK-26 solution serving as the negative control (NC) at the lower panel. Bar: left, 50 μm; right, 10 μm. n = 3 biological replicates. b M1-Exos decreased the expression levels of p-Akt and p-eNOS. n ≥ 4. c M1-Exos increased the release of sVCAM-1 and sICAM-1 assayed through ELISA. n = 3 biological replicates. d–h CCK-8 detection, EdU staining, Transwell and scratch/wound assay, and tube formation analysis demonstrated that M1-Exos impeded cell viability (d), proliferation (e), migration (f and g), and tube formation ability (h) of ECs. M1-Exos were derived from human THP-1 M1 macrophages. HCAECs were treated with exosomes for 24 h. Bar: 400 μm in (e); 100 μm in (f); and 500 μm in (g) and (h). n = 3–7 biological replicates. ***P < 0.001; **P < 0.01; *P < 0.05, M1-Exos vs. M0-Exos or PBS treatment
Furthermore, M1-Exos inhibited the viability of ECs in the CCK-8 assay (Fig. 3d). ECs treated with M1-Exos had a less EdU staining intensity, indicating a decrease in cell proliferation (Fig. 3e). Using a Transwell (Fig. 3f) or scratch/wound migration assay (Fig. 3g), we found that M1-Exos were associated with the attenuated migration of ECs into the well or wound areas compared with that of M0-Exos or PBS control. M1-Exos stimuli decreased the tube formation ability on ECs (Fig. 3h). In summary, these data indicate that M1-Exos impair the normal EC function and reduce the endothelial angiogenesis capability.
M1-Exos transfer miR-155 into ECs
Exosomes transfer heterogeneous components, such as miRNAs and proteins, to initiate or suppress diverse signaling pathways in recipient cells. miR-155 has been proven as a particular marker and mediator miRNA for M1 macrophage differentiation [18, 22, 39]. miR-155 is one of the high abundant miRNA in M1-Exos [54, 58]. We found that the miR-155 level in the heart was upregulated on day 3 following MI (Fig. 4a), and this result was similar to a previous finding [54]. This increase is mainly attributed to macrophages rather than CMs or CFs [54]. Macrophage-derived exosomes contain enriched miR-155 after MI [54]. We confirmed that exosomal miR-155 was enhanced during cardiac tissues on day 3 post-MI (Fig. 4a). Furthermore, under either GM-CSF plus IFN-γ stimuli, TNF-α, IL-1β, or OGD treatment, both cellular miR-155 and exosomal miR-155 levels were amplified in human THP-1 or mouse RAW246.7 macrophages (Fig. 4b). We also confirmed that the cellular miR-155 and exosomal miR-155 levels in mouse PM or BMDM M1 macrophages increased (Fig. 4c). M1-Exos could be taken up by the ECs through a co-culture assay, as shown in Fig. 3a. Then, we isolated the exosomes from the conditioned medium of THP-1 M1 macrophages that were transfected with a Cy3-miR-155 mimic and added them to the cultured HCAECs. The appearance of a red fluorescent Cy3 dye in the ECs demonstrated that M1-like macrophages could secrete exosomal miR-155, which was then transported into target ECs (Fig. 4d). Subsequently, we observed that M1-Exos treatment increased the miR-155 level in ECs compared with that of M0-Exos or PBS control (Fig. 4e). Moreover, treatment with Triton X-100 with RNase, which destroyed the membrane structure of the exosomes and digested RNAs, eliminated this increase (Fig. 4e). This phenomenon indicated that the enlarged miR-155 in ECs was relocated from M1-Exos. The data demonstrate that M1-Exos translocate miRNAs, especially miR-155, into ECs.
Transmission of miR-155 from M1-Exos into ECs. a miR-155 level was enriched in cardiac tissues or exosomes derived from MI heart on day 3. n = 4–5. **P < 0.01, MI vs. sham. b Cellular (left panel) and exosomal (right panel) miR-155 levels enhanced in THP-1 and RAW264.7 M1 macrophages under GM-CSF/IFN-γ, TNF-α, IL-1β, or OGD stimulus for 24 h. n = 3–7 biological replicates. ***P < 0.001; **P < 0.01; *P < 0.05, indicated treatment vs. control. c Cellular (left panel) and exosomal (right panel) miR-155 levels increased in mouse M1 PMs and BMDMs. Cells were treated with GM-CSF (50 ng/mL)/IFN-γ (20 ng/mL) for 24 h. n = 3 biological replicates. ***P < 0.001; **P < 0.01, M1 vs. M0. d Translocation of exosomal Cy3-miR-155 from M1-like macrophages to ECs (n = 3 biological replicates). Confocal imaging of HCAECs. Bar: 10 μm. e Triton X-100/RNase treatment prevented the transfer of miR-155 from M1-Exos into HCAECs. n = 3 biological replicates. ***P < 0.001, M1-Exos vs. M0-Exos or PBS. ###P < 0.001, Triton X-100/RNase treatment vs. M1-Exos alone
miR-155 reduces angiogenesis
miR-155, a classic multifunctional miRNA, is one of the most important inflammatory regulators. Increasing evidence has indicated that miR-155 plays a proinflammatory role in M1-like macrophages as mentioned previously. We also confirmed that miR-155 promoted the M2/M1 macrophage switch (Fig. S6). Here, we aimed to determine the direct function of miR-155 on ECs and angiogenesis. We found that hsa-miR-155 mimic inhibited the Akt–eNOS signaling pathway, whereas anti-miR-155 (miR-155 inhibitor) improved this pathway (Fig. 5a). miR-155 mimic promoted the secretion of sICAM-1 and sVCMA-1, although anti-miR-155 did not decrease the baseline of the secretion of these adhesion molecules (Fig. 5b). This result reflected the detrimental role of miR-155 on ECs. Moreover, miR-155 mimic restrained the cell viability, proliferation, migration, and tube formation on ECs, whereas anti-miR-155 slightly exerted the opposite effects (Fig. 5c–f and Fig. S7); this observation was consistent with a previous study on miR-155 knockout mice [47]. Furthermore, the intramyocardial delivery of lentivirus-mmu-miR-155-5p (LV-miR-155) (Fig. S8) inhibited the expression levels of α-SMA and CD31 (Fig. 5g), whereas lentivirus-anti-mmu-miR-155-5p (LV-anti-miR-155) enhanced the protein levels of α-SMA and CD31 (Fig. 5g). LV-miR-155 or LV-anti-miR-155 decreased or increased the VEGFA-VEGFR2 level after MI, respectively (Fig. 5h). Overall, miR-155 inhibits angiogenesis.
Effect of miR-155 on angiogenesis. a miR-155 decreased the phosphorylation level of Akt and eNOS on HCAECs (n > 6 biological replicates). ECs were transfected with the indicated duplex for 48 h. b ELISA revealed that miR-155 increased the secretion of sICAM-1 and sVCMA-1. n = 3 biological replicates. *P < 0.05, miR-155 vs. NC (negative control). c–f, miR-155 lessened the cell viability (c), proliferation (d), migration (e), and tube formation (f). n = 3–7 biological replicates. ***P < 0.001; **P < 0.01; *P < 0.05, miR-155 vs. NC. ###P < 0.001; ##P < 0.01; #P < 0.05, anti-miR-155 vs. anti-NC. g Effect of miR-155 on α-SMA or CD31 staining in MI heart tissues. Bar: 50 μm. n = 3. **P < 0.01; *P < 0.05, miR-155 vs. NC. ##P < 0.01, anti-miR-155 vs. NC. h miR-155 reduced the VEGF-A and VEGFR2 expression levels (n = 3)
miR-155 aggravates post-MI cardiac injury
Next, we evaluated the effect of miR-155 on cardiac function. The intramyocardial delivery of LV-miR-155 reduced the survival rate (Fig. 6a), decreased the EF and FS values (Fig. 6b), and expanded the infarct size (Fig. 6c) in the mouse MI model after 28 days compared with those of the NC control. By contrast, LV-anti-miR-155 improved the cardiac function and diminished the cardiac injury (Fig. 6). Overall, miR-155 accelerates MI injury, and this effect is similar to the outcome of M1-Exos.
Aggravation of post-MI cardiac injury by miR-155. a Lentivirus-miR-155 (LV-miR-155) repressed the survival rate. *P < 0.05, miR-155 vs. NC. b–c, LV-NC, LV-miR-155, and LV-anti-miR-155 were delivered through intramyocardial injection in the MI model. After 28 days, miR-155 decreased the EF and FS values (b) and increased the infarct size (c), whereas anti-miR-155 exerted the opposite effects. ***P < 0.001; **P < 0.01; *P < 0.05, MI vs. sham. ##P < 0.01; #P < 0.05, miR-155 or anti-miR-155 vs. NC
Rac family small GTPase 1 (RAC1), p21 (RAC1) activated kinase 2 (PAK2), Sirtuin 1 (Sirt1), and protein kinase AMP-activated catalytic subunit alpha 2 (AMPKα2) are the novel target genes of miR-155
To clarify the target genes of miR-155 on EC, we performed mRNA sequencing (Supplemental Table 1) combined with bioinformatics analysis and screened dozens of potential targets of miR-155. Among the predicted targets, we noticed that RAC1, PAK2, Sirt1, and AMPKα2 might be new candidate genes of miR-155 (Fig. 7a) aside from the well-known targets, such as eNOS. Consistent with the sequencing data and the prediction that RAC1 3′-UTR possessed a miR-155 binding site (Fig. 7a), we found that miR-155 mimic strikingly downregulated the luciferase activity of the WT rather than the mutant RAC1 3′-UTR (Figs. 7a, b). Similarly, the results of the dual luciferase reporter assays confirmed that miR-155 targeted the 3′-UTR of PAK2 and Sirt1 (Figs. 7a, b). Noticeably, the 3′-UTR of AMPKα2 enclosed two indispensable miR-155 binding sites (Figs. 7a, b), and each mutation responded to the effect of miR-155 (Fig. 7b).
Novel target genes for miR-155: RAC1, PAK2, Sirt1, and AMPKα2. a Bioinformatics analysis suggested that RAC1, PAK2, Sirt1, and AMPKα2 are the new candidate target genes of miR-155. b Dual luciferase reporter assays showed that the 3′-UTR of RAC1, PAK2, Sirt1, and AMPKα2 contained the miR-155 binding sites. 20 nM of miR-155 mimic or NC duplex and 150 ng of pmirGLO plasmid that contained either WT or mutant 3′-UTR of target genes were cotransfected into HEK293T cells for 36 h. n = 3–6 biological replicates. ***P < 0.001; **P < 0.01; *P < 0.05, WT vs. Mut (mutation). #P < 0.05, Mut1 or Mut2 vs. Mut1/Mut2. c miR-155 repressed the mRNA levels of RAC1, PAK2, Sirt1, AMPKα2, and eNOS. n = 3–5 biological replicates. ***P < 0.001; **P < 0.01, miR-155 vs. NC. d miR-155 inhibited the protein expression of RAC1, PAK2, Sirt1, AMPKα2, and eNOS and the RAC1–PAK1/2 and Sirt1/AMPK–eNOS pathways (n ≥ 4 biological replicates)
To further investigate whether miR-155 could downregulate these target genes in ECs, we transfected the NC or miR-155 mimic into HCAECs and evaluated the mRNA levels. We observed that miR-155 mimic inhibited the mRNA expression levels of RAC1, PAK2, Sirt1, AMPKα2, and eNOS (Fig. 7c). The immunoblot assay revealed that miR-155 overexpression reduced the protein levels of RAC1, PAK2, Sirt1, AMPKα2, and eNOS, whereas anti-miR-155 treatment upregulated the expression levels of these proteins in ECs (Fig. 7d). Interestingly, these five targets are exactly the five molecule nodes in RAC1–PAK2 and Sirt1/AMPKα2–eNOS pathways. Therefore, we examined whether miR-155 repressed these two signaling pathways. In addition to the total protein expressions, their phosphorylation levels (p-PAK1/2, p-AMPK, and p-eNOS) and activity of downstream enzymes (p-LIMK1/2 and p-ACC) were decreased by miR-155 mimic and increased by anti-miR-155 (Fig. 7d).
Sirt1/AMPK-eNOS signaling is a vital pathway that maintains endothelial function and mediates angiogenesis [28]. RAC1–PAK1/2 also plays a key proangiogenic role [26, 41]. Consequently, these data indicated that miR-155 prevents these two pathways by simultaneously targeting RAC1, PAK2, Sirt1, AMPKα2, and eNOS, leading to the downregulation of EC function and angiogenesis.
Anti-miR-155 partially rescues the influence of M1-Exos
We next evaluated the relative contribution of miR-155 within the effect of M1-Exos on ECs. We found that LV-anti-miR-155 (lentivirus-anti-hsa-miR-155-5p) moderately increased the protein levels of RAC1, PAK2, Sirt1, AMPKα2, and eNOS, which were inhibited by M1-Exos (Fig. 8a). Accordingly, LV-anti-miR-155 comparatively negated the inhibitory effect of M1-Exos on cell viability (Fig. 8b), proliferation (Fig. 8c), migration (Fig. 8d), and tube formation (Fig. 8e) in ECs. These results demonstrate that miR-155 inhibition partially rescues the effects of M1-Exos on ECs, implying that other biomolecules (e.g., miRNAs or proteins), except miR-155, play a part in the outcome of M1-Exos.
Alleviation of the effect of M1-Exos by anti-miR-155. a Lentivirus-mediated overexpression of anti-miR-155 (LV-anti-hsa-miR-155-5p) partially restored the expression levels of RAC1, PAK2, Sirt1, AMPKα2, and eNOS on HCAECs under M1-Exos treatment. ECs were infected with indicated lentivirus for 72 h. b Anti-miR-155 partially rescued the cell viability, which decreased by M1-Exos injury. c Anti-miR-155 promoted EC proliferation. Bar, 400 μm. d Anti-miR-155 enhanced EC migration compared with NC. Bar, 500 μm. e Anti-miR-155 alleviated the inhibition role of M1-Exos on tube formation ability in ECs. Bar, 500 μm. Each experiment in this figure was replicated at least thrice. ***P < 0.001; **P < 0.01, M1-Exos vs. M0-Exos. ###P < 0.001, anti-miR-155 vs. NC
Discussion
Direct and indirect effects of proinflammatory factors on angiogenesis in a MI microenvironment
Recently, the coronary circulation including endothelial dysfunction has become a new important frontier in cardioprotection research [12, 15, 16]. As previously mentioned, the influence of M1 macrophages on angiogenesis remains unclear. Both the antiangiogenic and proangiogenic effects have been documented. Their influence might be related to the factors that M1 macrophages react to and release, such as GM-CSF, TNF-α, IL-1β, and IFN-γ, and ischemia and hypoxia, which involve complex and even bidirectional regulative mechanisms of angiogenesis. During the first phase of inflammation in the MI microenvironment, proangiogenic inducers, including hypoxia, ischemia, GM-CSF, IL-1β, and IL-6, emerge to heal injured tissues [6]. By contrast, IFN-γ acts as a well-known antiangiogenic mediator. Interestingly, TNF-α is generally believed to be proangiogenic and antiangiogenic in different contexts [6].
Another reason might be the indirect role of these microenvironmental factors, including the function of diverse tissue cell-secreted extracellular vesicles or exosomes. Here, we indicated that either GM-CSF/IFN-γ, IFN-γ alone, TNF-α, IL-1β, or OGD promoted the release of M1-Exos (Figs. 1, 9). Moreover, we highlighted that M1-Exos impaired EC function, exerted an anti-angiogenic role, and hindered cardiac repair (Figs. 2, 3, and 9).
Graphic illustration of the underlying mechanism. Under the MI microenvironment, the proinflammatory M1-like macrophages release proinflammatory M1-Exos. M1-Exos transfer miR-155 into ECs, which simultaneously target the whole five node genes in RAC1–PAK2 and Sirt1/AMPKα2–eNOS signaling pathways, leading to the inhibition of angiogenesis and cardiac repair
Effect of M1-Exos on angiogenesis, CMs, and CFs during post-MI response
Here, we clearly showed that M1-Exos not only injured ECs but also inhibited their cell viability, proliferation, migration, and tube formation in vitro (Fig. 3). Furthermore, M1-Exos decreased post-MI angiogenesis and impeded cardiac repair in vivo (Fig. 2). Lee et al. [32] established that human macrophage-derived exosomes suppress the migration of ECs by controlling integrin trafficking. They demonstrated that exosomes released from human macrophages promotes the degradation of integrin β1 through internalization and ubiquitination [32]. We also confirmed that both M0-Exos and M1-Exos decreased the protein level of integrin β1 in HCAECs (Fig. S9a).
Although we illustrated that M1-Exos reduced the cardiac function and expanded the infarct size, we did not investigate the direct influence of M1-Exos on CMs. Wang et al. [54] showed that macrophage exosomes delay the proliferation of CFs and promote their inflammation after MI. In view of their proinflammation properties, the role of M1-Exos on the apoptosis and regeneration of CMs during healing should be investigated in future studies. Furthermore, either the Sirt1/AMPK–eNOS pathway or the RAC1–PAK1/2 pathway is identified as a crucial survival mediator, key metabolism, and function regulating signaling in cardiac physiology. Therefore, M1-Exos may play a regulatory role on CMs.
Inhibition of miR-155 appears to be a potential therapeutic target in cardiac repair after MI
miR-155, a typical multifunctional miRNA, is located within chromosome 21 and transcribed from a B cell integration cluster in humans. Hundreds of experimentally validated target genes for hsa-miR-155-5p are found on the collection of databases, such as miRTarBase (https://mirtarbase.mbc.nctu.edu.tw/php/search.php?org=hsa&opt=mirna_id&kw=miR-155). As a pivotal factor that regulates inflammation, miR-155 controls multiple pathophysiological pathways in immunity, inflammation, tumor biology, and cardiovascular disease [8, 25]. Macrophage- or fibroblast-expressed miR-155 plays a major role in the pathological status of the heart. Silencing miR-155 attenuates cardiac injury and dysfunction in viral carditis by promoting M2 macrophage polarization [60]. miR-155 conditional knockout in fibroblasts reduces infarct size, ameliorates collagen deposition, and cardiac remodeling, and improves cardiac function because it targets tumor protein p53-inducible nuclear protein 1 [13]. Furthermore, miR-155 is enhanced in exosomes derived from macrophages after MI (Fig. 1 and [54]). Macrophage exosomes can transfer miR-155 into CFs and attenuate their proliferation by decreasing the expression of the Son of Sevenless 1 and promote inflammation by downregulating the expression of suppressor of cytokine signaling 1 [54]. Given that the myocardial intramuscular injection of LV-miR-155 aggravates MI size and inhibits cardiac function and that LV-anti-miR-155 treatment exhibits the opposite effect (Fig. 6), we believe that miR-155 influences the cardiac repair/remodeling process.
Despite the crucial role of miR-155 in vascular remodeling post-MI, the contradictory results of miR-155 (either promoting or preventing angiogenesis) have previously raised a debate. Using plasmid transfection, Liu et al. [38] found that miR-155 attenuates angiotensin II-impaired migration and capillary tubule formation abilities on human umbilical vein ECs (HUVECs). With lentiviral infection or synthesized oligonucleotide transfection, Kong et al. [27] reported that miR-155 promotes tumor angiogenesis. Conversely, Zhu et al. [64] observed that the overexpression of miR-155 oligonucleotides reduces the migration of HUVECs in response to angiotensin II. Weber et al. [55] verified that miR-155 decreases the migration and proliferation of HUVECs. miR-155 deficiency in ECs is validated as the stimulatory effect on proliferation and angiogenic tube formation in vitro and in vivo [47]. Smooth muscle cells release exosomes and transfer miR-155 to induce endothelial injury and promote atherosclerosis [61]. The distinction in these observations may be due to special basic conditions, such as angiotensin II pretreatment. In the present study, we applied various delivery methods, including lentivirus infection, and synthesized oligonucleotide transfection of mimic (miR-155) or inhibitor (anti-miR-155) on different types of ECs, such as HCAECs, HUVECs, and human cardiac microvascular ECs, to determine the exact role of miR-155 on angiogenesis. We verified that miR-155 prevented the migration, proliferation, and tube formation of ECs and depressed their relative pathways, such as RAC1–PAK1/2 and eNOS signaling in vitro (Figs. 5, 7, and partial data not shown). Furthermore, miR-155 impaired cardiac function and delayed heart repair in vivo (Fig. 6). These outcomes coincide with previous studies on miR-155 [10, 17, 47].
miRNA constantly produces effects by regulating downstream genes. We screened the potential target genes of miR-155 through gene sequencing combined with bioinformatics analysis. Among the candidates, RAC1, PAK2, Sirt1, and AMPKα2 were validated to be the novel targets (Fig. 7). Intriguingly, combined with the well-characterized target eNOS, the five nodes (enzyme/protein molecules) in RAC1–PAK2 and Sirt1/AMPKα2–eNOS signaling were the simultaneous targets of miR-155. Sirt1 and AMPK respectively deacetylates or phosphorylates and then activates eNOS. Sirt1/AMPK–eNOS is a well-studied and key pathway in EC function maintenance and angiogenesis [28]. RAC1–PAK1/2 signaling is essential for lumen formation in vascular morphogenesis and mediates vascular homeostasis [26, 41]. Therefore, miR-155 can decline the migration, proliferation, and tube formation of ECs and depress angiogenesis by downregulating the RAC1–PAK1/2 and Sirt1/AMPKα–eNOS pathways. In the current study, we also confirmed that miR-155 inhibited the VEGF–VEGFR2 signaling (Fig. 5h) and the expression of integrin β1 (Fig. S9b).
Opposing functions of M2-Exos and M1-Exos
A fine-tuned balance between M1 and M2 macrophages states is necessary for myocardial repair. After MI, the timely resolution of inflammation by the effect of M2 macrophages is required for favorable cardiac repair and inhibition of adverse remodeling. M2 macrophages are anti-inflammatory and pro-reparative, producing proangiogenic factors (e.g., IL-10, VEGF, and TGF-β) to encourage angiogenesis and tissue healing. Given that M2 macrophages always perform contrary functions to M1, the function of M2-Exos (exosomes derived from M2 macrophages) should be further investigated. Indeed, M2-Exos display an opposite influence to M1-Exos in tumor biology. That is, the former stimulates tumor growth [30, 62], whereas the latter act as inhibitors. Furthermore, the miR-155 expression is lower in M2 macrophages than in M1 [22, 39]. We also observed that M2 macrophage cytokines, such as VEGF, decreased the expression levels of cellular and exosomal miR-155 on macrophages (Jing Chen and Shiming Liu, unpublished data). M2-Exos might have modulated the signaling of different biomolecules from M1-Exos. Elucidating the expression profiles of miRNAs or proteins within M1-Exos and comparing them with those of M2-Exos using systemic biology techniques (e.g., miRNA sequencing or proteomics) or single-cell approaches [56], help reveal specific regulatory molecules and the precise underlying mechanisms of the M1/M2 paradigm.
References
Aurora AB, Porrello ER, Tan W, Mahmoud AI, Hill JA, Bassel-Duby R, Sadek HA, Olson EN (2014) Macrophages are required for neonatal heart regeneration. J Clin Invest 124:1382–1392. https://doi.org/10.1172/JCI72181
Ben-Mordechai T, Holbova R, Landa-Rouben N, Harel-Adar T, Feinberg MS, Abd Elrahman I, Blum G, Epstein FH, Silman Z, Cohen S, Leor J (2013) Macrophage subpopulations are essential for infarct repair with and without stem cell therapy. J Am Coll Cardiol 62:1890–1901. https://doi.org/10.1016/j.jacc.2013.07.057
Ben-Mordechai T, Palevski D, Glucksam-Galnoy Y, Elron-Gross I, Margalit R, Leor J (2015) Targeting macrophage subsets for infarct repair. J Cardiovasc Pharmacol Ther 20:36–51. https://doi.org/10.1177/1074248414534916
Botker HE, Hausenloy D, Andreadou I, Antonucci S, Boengler K, Davidson SM, Deshwal S, Devaux Y, Di Lisa F, Di Sante M, Efentakis P, Femmino S, Garcia-Dorado D, Giricz Z, Ibanez B, Iliodromitis E, Kaludercic N, Kleinbongard P, Neuhauser M, Ovize M, Pagliaro P, Rahbek-Schmidt M, Ruiz-Meana M, Schluter KD, Schulz R, Skyschally A, Wilder C, Yellon DM, Ferdinandy P, Heusch G (2018) Practical guidelines for rigor and reproducibility in preclinical and clinical studies on cardioprotection. Basic Res Cardiol 113:39. https://doi.org/10.1007/s00395-018-0696-8
Cheng Y, Rong J (2018) Macrophage polarization as a therapeutic target in myocardial infarction. Curr Drug Targets 19:651–662. https://doi.org/10.2174/1389450118666171031115025
Corliss BA, Azimi MS, Munson JM, Peirce SM, Murfee WL (2016) Macrophages: an inflammatory link between angiogenesis and lymphangiogenesis. Microcirculation 23:95–121. https://doi.org/10.1111/micc.12259
Deveza L, Choi J, Yang F (2012) Therapeutic angiogenesis for treating cardiovascular diseases. Theranostics 2:801–814. https://doi.org/10.7150/thno.4419
Eisenhardt SU, Weiss JB, Smolka C, Maxeiner J, Pankratz F, Bemtgen X, Kustermann M, Thiele JR, Schmidt Y, Bjoern Stark G, Moser M, Bode C, Grundmann S (2015) MicroRNA-155 aggravates ischemia-reperfusion injury by modulation of inflammatory cell recruitment and the respiratory oxidative burst. Basic Res Cardiol 110:32. https://doi.org/10.1007/s00395-015-0490-9
Frangogiannis NG (2015) Inflammation in cardiac injury, repair and regeneration. Curr Opin Cardiol 30:240–245. https://doi.org/10.1097/HCO.0000000000000158
Guo J, Liu HB, Sun C, Yan XQ, Hu J, Yu J, Yuan Y, Du ZM (2019) MicroRNA-155 promotes myocardial infarction-induced apoptosis by targeting RNA-binding protein QKI. Oxid Med Cell Longev 2019:4579806. https://doi.org/10.1155/2019/4579806
Harel-Adar T, Ben Mordechai T, Amsalem Y, Feinberg MS, Leor J, Cohen S (2011) Modulation of cardiac macrophages by phosphatidylserine-presenting liposomes improves infarct repair. Proc Natl Acad Sci USA 108:1827–1832. https://doi.org/10.1073/pnas.1015623108
Hausenloy DJ, Chilian W, Crea F, Davidson SM, Ferdinandy P, Garcia-Dorado D, van Royen N, Schulz R, Heusch G (2019) The coronary circulation in acute myocardial ischaemia/reperfusion injury: a target for cardioprotection. Cardiovasc Res 115:1143–1155. https://doi.org/10.1093/cvr/cvy286
He W, Huang H, Xie Q, Wang Z, Fan Y, Kong B, Huang D, Xiao Y (2016) MiR-155 knockout in fibroblasts improves cardiac remodeling by targeting tumor protein p53-inducible nuclear protein 1. J Cardiovasc Pharmacol Ther 21:423–435. https://doi.org/10.1177/1074248415616188
Hesketh M, Sahin KB, West ZE, Murray RZ (2017) Macrophage phenotypes regulate scar formation and chronic wound healing. Int J Mol Sci. https://doi.org/10.3390/ijms18071545
Heusch G (2016) The coronary circulation as a target of cardioprotection. Circ Res 118:1643–1658. https://doi.org/10.1161/CIRCRESAHA.116.308640
Heusch G (2019) Coronary microvascular obstruction: the new frontier in cardioprotection. Basic Res Cardiol 114:45. https://doi.org/10.1007/s00395-019-0756-8
Hu J, Huang CX, Rao PP, Cao GQ, Zhang Y, Zhou JP, Zhu LY, Liu MX, Zhang GG (2019) MicroRNA-155 inhibition attenuates endoplasmic reticulum stress-induced cardiomyocyte apoptosis following myocardial infarction via reducing macrophage inflammation. Eur J Pharmacol 857:172449. https://doi.org/10.1016/j.ejphar.2019.172449
Hu J, Huang CX, Rao PP, Zhou JP, Wang X, Tang L, Liu MX, Zhang GG (2019) Inhibition of microRNA-155 attenuates sympathetic neural remodeling following myocardial infarction via reducing M1 macrophage polarization and inflammatory responses in mice. Eur J Pharmacol 851:122–132. https://doi.org/10.1016/j.ejphar.2019.02.001
Hu M, Guo G, Huang Q, Cheng C, Xu R, Li A, Liu N, Liu S (2018) The harsh microenvironment in infarcted heart accelerates transplanted bone marrow mesenchymal stem cells injury: the role of injured cardiomyocytes-derived exosomes. Cell Death Dis 9:357. https://doi.org/10.1038/s41419-018-0392-5
Hu Y, Zhang H, Lu Y, Bai H, Xu Y, Zhu X, Zhou R, Ben J, Xu Y, Chen Q (2011) Class A scavenger receptor attenuates myocardial infarction-induced cardiomyocyte necrosis through suppressing M1 macrophage subset polarization. Basic Res Cardiol 106:1311–1328. https://doi.org/10.1007/s00395-011-0204-x
Ishikawa S, Noma T, Fu HY, Matsuzaki T, Ishizawa M, Ishikawa K, Murakami K, Nishimoto N, Nishiyama A, Minamino T (2017) Apoptosis inhibitor of macrophage depletion decreased M1 macrophage accumulation and the incidence of cardiac rupture after myocardial infarction in mice. PLoS ONE 12:e0187894. https://doi.org/10.1371/journal.pone.0187894
Jablonski KA, Gaudet AD, Amici SA, Popovich PG, Guerau-de-Arellano M (2016) Control of the inflammatory macrophage transcriptional signature by miR-155. PLoS ONE 11:e0159724. https://doi.org/10.1371/journal.pone.0159724
Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, Coffey RJ (2019) Reassessment of exosome composition. Cell 177(428–445):e418. https://doi.org/10.1016/j.cell.2019.02.029
Jetten N, Verbruggen S, Gijbels MJ, Post MJ, De Winther MP, Donners MM (2014) Anti-inflammatory M2, but not pro-inflammatory M1 macrophages promote angiogenesis in vivo. Angiogenesis 17:109–118. https://doi.org/10.1007/s10456-013-9381-6
Jung M, Dodsworth M, Thum T (2018) Inflammatory cells and their non-coding RNAs as targets for treating myocardial infarction. Basic Res Cardiol 114:4. https://doi.org/10.1007/s00395-018-0712-z
Koh W, Mahan RD, Davis GE (2008) Cdc42- and Rac1-mediated endothelial lumen formation requires Pak2, Pak4 and Par3, and PKC-dependent signaling. J Cell Sci 121:989–1001. https://doi.org/10.1242/jcs.020693
Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, Lancaster JM, Coppola D, Sellers TA, Djeu JY, Cheng JQ (2014) Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene 33:679–689. https://doi.org/10.1038/onc.2012.636
Kroller-Schon S, Jansen T, Tran TLP, Kvandova M, Kalinovic S, Oelze M, Keaney JF Jr, Foretz M, Viollet B, Daiber A, Kossmann S, Lagrange J, Frenis K, Wenzel P, Munzel T, Schulz E (2019) Endothelial alpha1AMPK modulates angiotensin II-mediated vascular inflammation and dysfunction. Basic Res Cardiol 114:8. https://doi.org/10.1007/s00395-019-0717-2
Lambert JM, Lopez EF, Lindsey ML (2008) Macrophage roles following myocardial infarction. Int J Cardiol 130:147–158. https://doi.org/10.1016/j.ijcard.2008.04.059
Lan J, Sun L, Xu F, Liu L, Hu F, Song D, Hou Z, Wu W, Luo X, Wang J, Yuan X, Hu J, Wang G (2019) M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res 79:146–158. https://doi.org/10.1158/0008-5472.CAN-18-0014
Lawrence T, Natoli G (2011) Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol 11:750–761. https://doi.org/10.1038/nri3088
Lee HD, Kim YH, Kim DS (2014) Exosomes derived from human macrophages suppress endothelial cell migration by controlling integrin trafficking. Eur J Immunol 44:1156–1169. https://doi.org/10.1002/eji.201343660
Leuschner F, Dutta P, Gorbatov R, Novobrantseva TI, Donahoe JS, Courties G, Lee KM, Kim JI, Markmann JF, Marinelli B, Panizzi P, Lee WW, Iwamoto Y, Milstein S, Epstein-Barash H, Cantley W, Wong J, Cortez-Retamozo V, Newton A, Love K, Libby P, Pittet MJ, Swirski FK, Koteliansky V, Langer R, Weissleder R, Anderson DG, Nahrendorf M (2011) Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat Biotechnol 29:1005–1010. https://doi.org/10.1038/nbt.1989
Li J, Li SH, Wu J, Weisel RD, Yao A, Stanford WL, Liu SM, Li RK (2018) Young bone marrow Sca-1 cells rejuvenate the aged heart by promoting epithelial-to-mesenchymal transition. Theranostics 8:1766–1781. https://doi.org/10.7150/thno.22788
Lindsey ML, Bolli R, Canty JM Jr, Du XJ, Frangogiannis NG, Frantz S, Gourdie RG, Holmes JW, Jones SP, Kloner RA, Lefer DJ, Liao R, Murphy E, Ping P, Przyklenk K, Recchia FA, Schwartz Longacre L, Ripplinger CM, Van Eyk JE, Heusch G (2018) Guidelines for experimental models of myocardial ischemia and infarction. Am J Physiol Heart Circ Physiol 314:H812–H838. https://doi.org/10.1152/ajpheart.00335.2017
Lindsey ML, Kassiri Z, Virag JAI, de Castro Bras LE, Scherrer-Crosbie M (2018) Guidelines for measuring cardiac physiology in mice. Am J Physiol Heart Circ Physiol 314:H733–H752. https://doi.org/10.1152/ajpheart.00339.2017
Lindsey ML, Saucerman JJ, DeLeon-Pennell KY (2016) Knowledge gaps to understanding cardiac macrophage polarization following myocardial infarction. Biochim Biophys Acta 1862:2288–2292. https://doi.org/10.1016/j.bbadis.2016.05.013
Liu T, Shen D, Xing S, Chen J, Yu Z, Wang J, Wu B, Chi H, Zhao H, Liang Z, Chen C (2013) Attenuation of exogenous angiotensin II stress-induced damage and apoptosis in human vascular endothelial cells via microRNA-155 expression. Int J Mol Med 31:188–196. https://doi.org/10.3892/ijmm.2012.1182
Lu L, McCurdy S, Huang S, Zhu X, Peplowska K, Tiirikainen M, Boisvert WA, Garmire LX (2016) Time series miRNA-mRNA integrated analysis reveals critical miRNAs and targets in macrophage polarization. Sci Rep 6:37446. https://doi.org/10.1038/srep37446
Ma Y, Mouton AJ, Lindsey ML (2018) Cardiac macrophage biology in the steady-state heart, the aging heart, and following myocardial infarction. Transl Res 191:15–28. https://doi.org/10.1016/j.trsl.2017.10.001
Marinkovic G, Heemskerk N, van Buul JD, de Waard V (2015) The ins and outs of small GTPase Rac1 in the vasculature. J Pharmacol Exp Ther 354:91–102. https://doi.org/10.1124/jpet.115.223610
Martinez FO, Gordon S (2014) The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 6:13. https://doi.org/10.12703/P6-13
Mouton AJ, DeLeon-Pennell KY, Rivera Gonzalez OJ, Flynn ER, Freeman TC, Saucerman JJ, Garrett MR, Ma Y, Harmancey R, Lindsey ML (2018) Mapping macrophage polarization over the myocardial infarction time continuum. Basic Res Cardiol 113:26. https://doi.org/10.1007/s00395-018-0686-x
Osada-Oka M, Shiota M, Izumi Y, Nishiyama M, Tanaka M, Yamaguchi T, Sakurai E, Miura K, Iwao H (2017) Macrophage-derived exosomes induce inflammatory factors in endothelial cells under hypertensive conditions. Hypertens Res 40:353–360. https://doi.org/10.1038/hr.2016.163
Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C (2010) Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 12:19–30. https://doi.org/10.1038/ncb2000(sup pp 11–13)
Panizzi P, Swirski FK, Figueiredo JL, Waterman P, Sosnovik DE, Aikawa E, Libby P, Pittet M, Weissleder R, Nahrendorf M (2010) Impaired infarct healing in atherosclerotic mice with Ly-6C(hi) monocytosis. J Am Coll Cardiol 55:1629–1638. https://doi.org/10.1016/j.jacc.2009.08.089
Pankratz F, Bemtgen X, Zeiser R, Leonhardt F, Kreuzaler S, Hilgendorf I, Smolka C, Helbing T, Hoefer I, Esser JS, Kustermann M, Moser M, Bode C, Grundmann S (2015) MicroRNA-155 exerts cell-specific antiangiogenic but proarteriogenic effects during adaptive neovascularization. Circulation 131:1575–1589. https://doi.org/10.1161/CIRCULATIONAHA.114.014579
Podaru MN, Fields L, Kainuma S, Ichihara Y, Hussain M, Ito T, Kobayashi K, Mathur A, D'Acquisto F, Lewis-McDougall F, Suzuki K (2019) Reparative macrophage transplantation for myocardial repair: a refinement of bone marrow mononuclear cell-based therapy. Basic Res Cardiol 114:34. https://doi.org/10.1007/s00395-019-0742-1
Richards J, Gabunia K, Kelemen SE, Kako F, Choi ET, Autieri MV (2015) Interleukin-19 increases angiogenesis in ischemic hind limbs by direct effects on both endothelial cells and macrophage polarization. J Mol Cell Cardiol 79:21–31. https://doi.org/10.1016/j.yjmcc.2014.11.002
Tang N, Sun B, Gupta A, Rempel H, Pulliam L (2016) Monocyte exosomes induce adhesion molecules and cytokines via activation of NF-kappaB in endothelial cells. FASEB J 30:3097–3106. https://doi.org/10.1096/fj.201600368RR
ter Horst EN, Hakimzadeh N, van der Laan AM, Krijnen PA, Niessen HW, Piek JJ (2015) Modulators of macrophage polarization influence healing of the infarcted myocardium. Int J Mol Sci 16:29583–29591. https://doi.org/10.3390/ijms161226187
van der Vorst EPC, Weber C (2019) Novel features of monocytes and macrophages in cardiovascular biology and disease. Arterioscler Thromb Vasc Biol 39:e30–e37. https://doi.org/10.1161/ATVBAHA.118.312002
Verderio C, Gabrielli M, Giussani P (2018) Role of sphingolipids in the biogenesis and biological activity of extracellular vesicles. J Lipid Res 59:1325–1340. https://doi.org/10.1194/jlr.R083915
Wang C, Zhang C, Liu L, Axx X, Chen B, Li Y, Du J (2017) Macrophage-derived mir-155-containing exosomes suppress fibroblast proliferation and promote fibroblast inflammation during cardiac injury. Mol Ther 25:192–204. https://doi.org/10.1016/j.ymthe.2016.09.001
Weber M, Kim S, Patterson N, Rooney K, Searles CD (2014) MiRNA-155 targets myosin light chain kinase and modulates actin cytoskeleton organization in endothelial cells. Am J Physiol Heart Circ Physiol 306:H1192–1203. https://doi.org/10.1152/ajpheart.00521.2013
Williams JW, Giannarelli C, Rahman A, Randolph GJ, Kovacic JC (2018) Macrophage biology, classification, and phenotype in cardiovascular disease: JACC macrophage in CVD series (Part 1). J Am Coll Cardiol 72:2166–2180. https://doi.org/10.1016/j.jacc.2018.08.2148
Wu R, Gao W, Yao K, Ge J (2019) Roles of exosomes derived from immune cells in cardiovascular diseases. Front Immunol 10:648. https://doi.org/10.3389/fimmu.2019.00648
Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, Ofrecio JM, Wollam J, Hernandez-Carretero A, Fu W, Li P, Olefsky JM (2017) Adipose tissue macrophage-derived exosomal mirnas can modulate in vivo and in vitro insulin sensitivity. Cell 171(372–384):e312. https://doi.org/10.1016/j.cell.2017.08.035
Yuan A, Hsiao YJ, Chen HY, Chen HW, Ho CC, Chen YY, Liu YC, Hong TH, Yu SL, Chen JJ, Yang PC (2015) Opposite effects of M1 and M2 macrophage subtypes on lung cancer progression. Sci Rep 5:14273. https://doi.org/10.1038/srep14273
Zhang Y, Zhang M, Li X, Tang Z, Wang X, Zhong M, Suo Q, Zhang Y, Lv K (2016) Silencing microRNA-155 attenuates cardiac injury and dysfunction in viral myocarditis via promotion of M2 phenotype polarization of macrophages. Sci Rep 6:22613. https://doi.org/10.1038/srep22613
Zheng B, Yin WN, Suzuki T, Zhang XH, Zhang Y, Song LL, Jin LS, Zhan H, Zhang H, Li JS, Wen JK (2017) Exosome-mediated miR-155 transfer from smooth muscle cells to endothelial cells induces endothelial injury and promotes atherosclerosis. Mol Ther 25:1279–1294. https://doi.org/10.1016/j.ymthe.2017.03.031
Zheng P, Luo Q, Wang W, Li J, Wang T, Wang P, Chen L, Zhang P, Chen H, Liu Y, Dong P, Xie G, Ma Y, Jiang L, Yuan X, Shen L (2018) Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional apolipoprotein E. Cell Death Dis 9:434. https://doi.org/10.1038/s41419-018-0465-5
Zhou J, Bai W, Liu Q, Cui J, Zhang W (2018) Intermittent hypoxia enhances THP-1 monocyte adhesion and chemotaxis and promotes M1 macrophage polarization via RAGE. Biomed Res Int 2018:1650456. https://doi.org/10.1155/2018/1650456
Zhu N, Zhang D, Chen S, Liu X, Lin L, Huang X, Guo Z, Liu J, Wang Y, Yuan W, Qin Y (2011) Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis 215:286–293. https://doi.org/10.1016/j.atherosclerosis.2010.12.024
Acknowledgements
We thank Letpub Inc. and Minghui Tan (Jinan University) for the help in Graphic illustration image (Fig. 9), as well as other members in the Guangzhou Institute of Cardiovascular Disease for helpful discussions and technical assistance.
Funding
This study was supported by research grants from National Natural Science Foundation of China (No. 81570259, 81873474, 81600350), Special Innovation Projects of Universities in Guangdong Province (No. 2018KTSCX189), Program of Construction of High Level Universities for Guangzhou Medical University (to Shi.L. and to Shao.L.), The Key Medical Disciplines and Specialties Program of Guangzhou (2017–2019).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, S., Chen, J., Shi, J. et al. M1-like macrophage-derived exosomes suppress angiogenesis and exacerbate cardiac dysfunction in a myocardial infarction microenvironment. Basic Res Cardiol 115, 22 (2020). https://doi.org/10.1007/s00395-020-0781-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00395-020-0781-7