Abstract
Objective
The aim of this study was to explore whether polymorphisms in miR-146a, miR-499 and IRAK1 are associated with susceptibility to inflammatory arthritis.
Methods
Manual searches performed in the MEDLINE and EMBASE databases were used to identify published articles in which the roles of microRNA (miRNA) and IRAK1 polymorphisms in inflammatory arthritis were determined. A meta-analysis was conducted to investigate associations of the miR-146a rs2910164, miR-499 rs3746444, IRAK1 rs3027898 and IRAK1 rs1059703 polymorphisms with susceptibility to inflammatory arthritis.
Results
Nine studies containing 1224 patients and 1841 controls were included in the meta-analysis. The meta-analysis revealed no association between inflammatory arthritis and the rs2910164 C allele of miR-146a (odds ratio, OR = 0.974; 95 % confidence interval, CI = 0.810–1.091; p = 0.650). Stratification by ethnicity or disease type revealed no association between the miR-146a C allele and inflammatory arthritis in European, Middle Eastern or Asian patients with rheumatoid arthritis (RA) or juvenile idiopathic arthritis (JIA). However, the meta-analysis revealed an overall association between RA and the miR-499 rs374644 C (OR = 1.123, 95 % CI = 1.019–2.586, p = 0.041); stratification by ethnicity revealed a particular association in Middle Eastern populations (OR = 1.943, 95 % CI = 1.508–2.504, p = 2.7 × 10–8). The meta-analysis of IRAK1 polymorphisms revealed an association between inflammatory arthritis and the rs3027898 CC genotype (OR = 2.602, 95 % CI = 1.387–4.879, p = 0.003). An analysis using the homozygote contrast showed the same pattern for the rs3027898 CC genotype (OR = 2.472, 95 % CI = 1.300–4.700, p = 0.006). No association between inflammatory arthritis and the rs1059703 polymorphism was found.
Conclusion
This meta-analysis suggests that the miR-499 rs374644 and IRAKI rs3027898 polymorphisms are associated with susceptibility to inflammatory arthritis.
Zusammenfassung
Ziel
Ziel der Studie war die Erforschung möglicher Zusammenhänge zwischen Polymorphismen in den Genen miR-146a, miR-499 und IRAK1 mit der Suszeptibilität für inflammatorische Arthritiden.
Methoden
Manuell wurde in den Datenbanken MEDLINE und EMBASE nach Veröffentlichungen zur Rolle von microRNA(miRNA)- und IRAK1-Polymorphismen bei inflammatorischer Arthritis gesucht. Zur Überprüfung von Assoziationen zwischen miR-146a rs2910164-, miR-499 rs3746444-, IRAK1 rs3027898- und IRAK1 rs1059703-Polymorphismen und der Suszeptibilität für inflammatorische Arthritis wurde eine Metaanalyse durchgeführt.
Ergebnisse
Neun Studien mit 1224 Patienten und 1841 Kontrollen wurden in die Metaanalyse aufgenommen. Die Metaanalyse zeigte keine Assoziation zwischen inflammatorischer Arthritis und dem rs2910164 C-Allel von miR-146a (Odds Ratio, OR,= 0,974; 95 %-Konfidenzintervall, 95%-KI = 0,810–1,091; p = 0,650). Die Stratifizierung nach Ethnizität bzw. Erkrankungstyp ergab keine Assoziation zwischen dem miR-146a C-Allel und inflammatorischer Arthritis bei Patienten aus Europa, dem nahen Osten und Asien mit rheumatoider Arthritis (RA) bzw. juveniler idiopathischer Arthritis (JIA). Doch die Metaanalyse deckte einen Zusammenhang insgesamt auf zwischen RA und miR-499 rs374644 C (OR = 1,123, 95 %-KI = 1,019–2,586, p = 0,041) und die Stratifizierung nach Ethnizität eine besondere Assoziation im Nahostkollektiven (OR = 1,943, 95 %-KI = 1,508–2,504, p = 2,7 × 10–8). Die Metaanalyse von IRAK1 –Polymorphismen zeigte eine Assoziation zwischen inflammatorischer Arthritis und dem rs3027898 CC-Genotyp (OR = 2,602, 95 %-KI = 1,387–4,879, p = 0,003). Eine Analyse unter Verwendung von „homozygote contrast“ ergab das gleiche Muster für den rs3027898 CC-Genotyp (OR = 2,472, 95 %-KI = 1,300–4,700, p = 0,006). Keine Assoziation fand sich zwischen inflammatorischer Arthritis und dem rs1059703-Polymorphismus.
Fazit
Die Metaanalyse verweist auf einen Zusammenhang zwischen den miR-499 rs374644- und IRAKI rs3027898-Polymorphismen und der Suszeptibilität für inflammatorische Arthritis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Inflammatory arthritis comprises a group of relatively common complex diseases that are known to be caused by interactions between genetic and environmental factors. Although the etiologies of these diseases have not been determined, genetic studies have established that susceptibility to them has a genetic component [1].
MicroRNAs (miRNAs) are noncoding RNA molecules approximately 22 nucleotides in length that participate in transcriptional and translational regulation [2]. They act by binding to the 3′ untranslated region of target mRNA, leading to either degradation of mRNA or repression of protein translation [3]. Therefore, miRNAs play a role in cell proliferation, differentiation and apoptosis [4]. miR-146a is involved in modulating the expression of inflammatory cytokines and the miR-146a polymorphism rs2910164 has been associated with several diseases, such as cancer and autoimmune diseases [5, 6]. The miR-146a rs2910164 polymorphism involves a G→C nucleotide substitution, which leads to a change from a G:U pair to a C:U mismatch in the structure of the miR-146a precursor. The CC genotype results in a lower expression level of mature miR-146a and promotes proliferation of cancer cells [7]. In addition, the minor C allele of the miR-146a polymorphism causes mispairing within the miR-146a hairpin and decreases the expression of its mature form, leading to diverse functional alterations [8]. Single nucleotide polymorphisms (SNPs) in miRNA sequences may alter miRNA expression. miR-146a regulates TNF-α via TRAF-6/IRAK1. IRAK1 is a target of miR-146a and plays an important role in the activation of NF-kB [9]. The most commonly studied polymorphisms of the IRAK1 gene in inflammatory arthritis are the rs3027898 (in the 3′ untranslated region) and rs1059703 (exon 12) polymorphisms. The miR-499 rs4746444 polymorphism is located in the mature miRNA region and may affect target mRNA binding and the pre-miRNA maturation process. The latter polymorphism has been associated with susceptibility to rheumatoid arthritis (RA) [10].
miRNAs play a role in cell proliferation, differentiation and apoptosis
Some studies have shown that the polymorphisms in miR-146a, miR-499 and IRAK1 are associated with inflammatory arthritis, but other reports have found no such associations [11, 12, 13, 14, 15, 16, 17, 18]. These disparities are probably caused by small sample sizes, low statistical power and/or clinical heterogeneity. Therefore, to overcome the limitations of individual studies, resolve inconsistencies and reduce the likelihood that random errors are responsible for false-positive or false-negative associations, we conducted a meta-analysis [19, 20, 21]. The aim of the present study was to determine whether the miR-146a, miR-499 and IRAK1 polymorphisms are associated with susceptibility to inflammatory arthritis.
Methods
Identification of eligible studies and data extraction
We performed searches for studies that examined associations between the miR-146a, miR-499 and IRAK1 polymorphisms and inflammatory arthritis. The MEDLINE and EMBASE citation databases were used to identify published articles in which the miR-146a, miR-499 and/or IRAK1 polymorphisms were analyzed in patients with inflammatory arthritis. Combinations of keywords such as ‘microRNA,’ ‘miR-146a,’ ‘miR-499,’ ‘IRAK1,’ ‘polymorphism,’ ‘arthritis’ and the names of individual diseases were entered as Medical Subject Headings (MeSH) and text words. References in the identified studies were also investigated to identify additional studies not indexed by MEDLINE and EMBASE. No restrictions were placed on language, race, ethnicity or geographic area. Studies were included if they: (1) were published before June 2014, (2) contained original data and (3) provided sufficient genotypic data to calculate odds ratios (ORs). The following were excluded: (1) studies containing overlapping data, (2) studies in which numbers of null and wild-type genotypes/alleles could not be ascertained and (3) studies in which family members had been studied (for example, a transmission disequilibrium test) because these analyses are based on linkage considerations. Data on methods and results were extracted from original studies by two independent reviewers. Discrepancy between the reviewers was resolved by consensus or a third reviewer. The following information was obtained for each study: author, year of publication, ethnicity of the study population, demographics and numbers of cases and controls. Frequencies of alleles were calculated from genotype distributions.
Evaluation of publication bias
Funnel plots are often used to detect publication bias. However, due to the limitations of funnel plotting—which requires a range of studies of varying sizes involving subjective judgments—publication bias was evaluated using Egger’s linear regression test [22], which measures funnel plot asymmetry using a natural logarithm scale of ORs.
Evaluations of statistical associations
A χ-square test was used to determine whether the observed genotype frequencies conformed to Hardy–Weinberg (HW) expectations (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). Meta-analyses were performed using: (1) allelic contrast, (2) homozygote contrast, (3) recessive and (4) dominant models. Subgroup analyses were performed according to ethnicity, disease type and Hardy-Weinberg equilibrium (HWE) status to evaluate ethnic- and disease-specific effects. Point estimates of risks, ORs and 95 % confidence intervals (CIs) were estimated for each study. Cochran’s Q statistic was used to assess intra- and interstudy variations and heterogeneities. This heterogeneity test assesses the null hypothesis that all studies evaluated the same effect. I2 values were used to quantify the effect of heterogeneity. I2 values range between 0 and 100 % and represent the proportion of interstudy variability attributable to heterogeneity rather than chance [23]. I2 values of 25, 50 and 75 % were nominally defined as low, moderate and high estimates, respectively. The fixed effects model assumes that a genetic factor has the same effect on disease susceptibility across all studies investigated and that observed variations between studies are caused by chance alone. The random effects model assumes that different studies show substantial diversity and assesses both intrastudy sampling error and interstudy variance. When study groups are homogeneous, the two models are similar; but where this is not the case, the random effects model usually provides wider CIs than the fixed effects model. Furthermore, the random effects model is used in the presence of significant interstudy heterogeneity [24]. Statistical manipulations were undertaken using the Comprehensive Meta-Analysis computer program (Biostat, Englewood, NJ, USA).
Results
Studies included in the meta-analysis
Electronic and manual searching identified 115 reports and 10 were selected for full-text review based on title and abstract details. Two reports were excluded (one had no control data; the other was a review) and eight reports thus met the inclusion criteria [11, 12, 13, 14, 15, 16, 17, 18]. One of these reports contained data on two different groups [17] and we analyzed these reports independently. Therefore, a total of nine separate studies were considered in the meta-analysis, which contained, in total, 1224 patients and 1841 controls. Three studies were based on a European population, two on Middle Eastern populations, two on East Asian populations, one on a South Asian population and one on a South American population (Fig. 1, Tab. 1). An ethnicity-specific meta-analysis was conducted on European, Middle Eastern and East Asian populations. These studies included patients with RA (n = 5), juvenile inflammatory arthritis (JIA; n = 2), psoriatic arthritis (PsA; n = 1) and ankylosing spondylitis (AS; n = 1). A disease-specific meta-analysis was performed on RA and JIA. All the studies except for one (which showed only allelic data of the polymorphisms) provided genotypic data of the polymorphisms. Eight studies examined the rs2910164 (miR-146a) polymorphism, three the rs3746444 (miR-499) polymorphism, four the rs3027898 (IRAK1) polymorphism and three studies examined the rs1059703 (IRAK1) polymorphism. A meta-analysis was thus performed on the rs2910164, rs3746444, rs3027898 and rs1059703 polymorphisms. Selected characteristics of these studies related to the association between miR-146a, miR-499 and IRAK1 polymorphisms and inflammatory arthritis diseases are summarized in Tab. 1.
Meta-analysis of the miR-146a and miR-499 polymorphisms in inflammatory arthritis
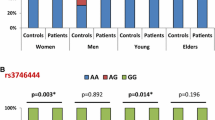
A summary of the meta-analysis findings concerning the associations between the miR-146a rs2910164 polymorphism and inflammatory arthritis is provided in Tab. 2. The meta-analysis revealed no association between inflammatory arthritis and the rs2910164 C allele (OR = 0.974, 95 % CI = 0.810–1.091, p = 0.650; Fig. 2, Tab. 2). Stratification by ethnicity indicated no association between the miR-146a C allele and inflammatory arthritis in European, Middle Eastern or Asian populations (Tab. 2). The disease-specific meta-analysis revealed no association between the miR-146a C allele and RA or JIA (Tab. 2). Furthermore, using the homozygote contrast, no association was found between the miR-146a polymorphism and inflammatory arthritis (Tab. 2). The meta-analysis revealed an association between RA and the miR-499 rs374644 C allele overall (OR = 1.123, 95 % CI = 1.019–2.586, p = 0.041; Fig. 2, Tab. 3). Stratification by ethnicity revealed an association between and the rs374644 C allele and RA in Middle Eastern populations (OR = 1.943, 95 % CI = 1.508–2.504, p = 2.7 × 10–8; Tab. 3). Analysis using the recessive model and homozygote contrast showed the same pattern for the rs374644 C allele (Tab. 3).
Meta-analysis of the rs3027898 and rs1059703 IRAK1 polymorphisms and inflammatory arthritis
Meta-analysis revealed an association between inflammatory arthritis and the rs3027898 CC genotype of IRAK1 (OR = 2.602, 95 % CI = 1.387–4.879, p = 0.003; Fig. 3, Tab. 4). Analysis using the homozygote contrast showed the same pattern for the rs3027898 CC genotype (OR = 2.472, 95 % CI = 1.300–4.700, p = 0.006; Tab. 4). However, an association between inflammatory arthritis and the rs1059703 polymorphism was not found by meta-analysis using allele contrast, the recessive or dominant models, or homozygote contrast (Tab. 4).
Heterogeneity and publication bias
Interstudy heterogeneity was found regarding the relationship between the miR-146a and IRAK1 polymorphisms; however, there was no heterogeneity within each ethnic group. The distribution of miR-146a polymorphism genotypes within the control groups were consistent with HW expectations, except for in two studies on the rs2910164 [13, 17] and one study on the rs374644 polymorphism [12], which suggests there may have been bias in terms of control selection or genotyping errors. When the studies in which controls were not in equilibrium were removed from the analyses, there was no association between the rs2910164 polymorphism and inflammatory arthritis in Middle Eastern populations under homozygote contrast (Tab. 2). However, one study on rs3746444 in which the controls were not out of HWE showed a significant association in Middle Eastern populations under the allele contrast and dominant models (after excluding a study in which controls were not in HWE; Tab. 3). It was difficult to correlate the funnel plot, which is usually used to detect publication bias, because the number of studies included in the analysis was relatively small. Egger’s regression test showed no evidence of publication bias (Egger’s regression test p-values > 0.1).
Discussion
Genetic factors are thought to contribute to inflammatory arthritis and this has encouraged researchers to search for the genes responsible for these diseases. By regulating the activity of immunoregulatory cells and decreasing immune responses, miRNAs play important roles in the pathogenesis of inflammatory diseases [4]. Therefore,
polymorphisms in miRNAs or their target genes could affect immune responses and lead to autoimmune and inflammatory diseases.
Many genes have been studied in this context and studying miRNAs and their target genes will help us to understand the genetic basis of inflammatory arthritis [25].
The present study addresses the association between polymorphisms in miR-146a, miR-499 and IRAK1 and susceptibility to inflammatory arthritis. This meta-analysis revealed no association between inflammatory arthritis and the miR-146a rs2910164 polymorphism. Stratification by ethnicity or disease type revealed no association between the rs2910164 polymorphism and inflammatory arthritis in European, Middle Eastern or Asian populations, or between this polymorphism and RA or JIA. There was also no association found between inflammatory arthritis and the IRAK1 rs1059703 polymorphism. However, meta-analysis revealed an association between RA and the miR-499 rs374644 polymorphism. In addition, the meta-analysis revealed an association between inflammatory arthritis and the IRAK1 rs3027898 polymorphism.
Our meta-analysis indicated associations between the miR-499 rs374644 and the IRAK1 rs3027898 polymorphisms and inflammatory arthritis.
miR-499 targets IL-17 receptor B and IL-6, which both play key roles in the pathogenesis of inflammatory arthritis [26]. IL-17 is a proinflammatory cytokine which induces expression of TNF-α and IL-6 and is overexpressed in the synovium of patients with inflammatory arthritis [26]. Therefore, there is a possibility that the miR-499 rs374644 polymorphism increases susceptibility to inflammatory arthritis by affecting IL-17 expression: rs374644 is located in the mature miRNA region and may affect target mRNA binding, which could alter protein expression. miR499 is not only expressed in immune cells, but also by medullary thymic epithelial cells and could, as such, have an impact on central T cell tolerance and subsequently on development of disease [27]. Fekete et al. [28] showed that IRAK1 plays a crucial role in chronic inflammation and the IRAK1 rs3027898 polymorphism lies in the 3′ untranslated region where miRNAs act; however, the functional significance of the polymorphism is unknown. There is also a possibility that the polymorphisms may be in linkage disequilibrium (LD) with nearby causal variants.
Our meta-analysis failed to find an association between the risk of developing inflammatory arthritis and the miR-146a rs2910164 polymorphism. Jazdzewski et al. [8] showed that the miR-146a rs2910164 polymorphism could reduce mature miR-146a expression and affect target mRNA binding. The GG genotype of the miR-146a polymorphism confers a higher expression level of mature miR-146a. Our finding is not consistent with the functional study. However, it remains unknown whether rs2910164 has a functional significance, because mixed results have been reported. We also cannot rule out the possibility that the lack of association may be due the small number of studies used, their low statistical power, or type II errors.
The present study has some limitations that require consideration. Firstly, heterogeneity and confounding factors may have distorted the analysis. In particular, publication bias could have affected our findings, because studies that produced negative results may not have been published or may have been missed. Secondly, our ethnicity-specific meta-analysis included data from European, Middle Eastern and East Asian patients, and our results are thus only applicable to these ethnic groups. Further studies are required in different ethnic populations. Thirdly, we did not stratify and analyze factors such as gender or clinical and environmental variables due to lack of data and the polymorphisms may be associated with clinical manifestations in addition to disease susceptibility. Fourthly, study and subject numbers in the disease-type subgroup analysis were relatively small and, as such, our analysis may have been underpowered. Therefore, additional studies are warranted to explore the associations between inflammatory arthritis and the miRNA polymorphisms.
Conclusion
This meta-analysis of published studies suggests that the miR-499 rs3746444 and IRAK1 rs3027898 polymorphisms are associated with susceptibility to inflammatory arthritis. However, the miR-146a rs2910164 and IRAK1 rs1059703 polymorphisms were not associated with susceptibility to inflammatory arthritis. Further studies are warranted to clarify the role of miRNA genes in the pathogenesis of inflammatory arthritis in different ethnic groups, since miRNA polymorphisms may play different roles in different ethnic populations.
References
Choi SJ, Rho YH, Ji JD et al (2006) Genome scan meta-analysis of rheumatoid arthritis. Rheumatology (Oxford) 45(2):166–170
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294(5543):853–858
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9(2):102–114
Mishra PJ, Bertino JR (2009) MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics 10(3):399–416
Xu WD, Lu MM, Pan HF, Ye DQ (2012) Association of MicroRNA-146a with autoimmune diseases. Inflammation 35(4):1525–1529
Wang J, Bi J, Liu X et al (2012) Has-miR-146a polymorphism (rs2910164) and cancer risk: a meta-analysis of 19 case-control studies. Mol Biol Rep 39(4):4571–4579
Xu T, Zhu Y, Wei QK et al (2008) A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis 29(11):2126–2131
Jazdzewski K, Murray EL, Franssila K et al (2008) Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A 105(20):7269–7274
Taganov KD, Boldin MP, Chang KJ, Baltimore D (2006) NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 103(33):12481–12486
Yang B, Chen J, Li Y et al (2012) Association of polymorphisms in pre-miRNA with inflammatory biomarkers in rheumatoid arthritis in the Chinese Han population. Hum Immunol 73(1):101–106
Singh S, Rai G, Aggarwal A (2014) Association of microRNA-146a and its target gene IRAK1 polymorphism with enthesitis related arthritis category of juvenile idiopathic arthritis. Rheumatol Int
Hashemi M, Eskandari-Nasab E, Zakeri Z et al (2013) Association of pre-miRNA-146a rs2910164 and premiRNA-499 rs3746444 polymorphisms and susceptibility to rheumatoid arthritis. Mol Med Rep 7(1):287–291
El-Shal AS, Aly NM, Galil SM et al (2013) Association of microRNAs genes polymorphisms with rheumatoid arthritis in Egyptian female patients. Joint Bone Spine 80(6):626–631
Jimenez-Morales S, Gamboa-Becerra R, Baca V et al (2012) MiR-146a polymorphism is associated with asthma but not with systemic lupus erythematosus and juvenile rheumatoid arthritis in Mexican patients. Tissue Antigens 80(4):317–321
Yang B, Zhang JL, Shi YY et al (2011) Association study of single nucleotide polymorphisms in pre-miRNA and rheumatoid arthritis in a Han Chinese population. Mol Biol Rep 38(8):4913–4919
Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA (2010) A polymorphism in the 3’-UTR of interleukin-1 receptor-associated kinase (IRAK1), a target gene of miR-146a, is associated with rheumatoid arthritis susceptibility. Joint Bone Spine 77(5):411–413
Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA (2010) The role of microRNA-146a (miR-146a) and its target IL-1R-associated kinase (IRAK1) in psoriatic arthritis susceptibility. Scand J Immunol 71(5):382–385
Qian L, Wang G, Li X et al (2012) Relationship between the single nucleotide polymorphisims in pre-miR-146a rs2910164 and expression of miR-146a in rheumatoid arthritis. J Neurosci 32:253–257
Lee YH, Rho YH, Choi SJ et al (2007) PADI4 polymorphisms and rheumatoid arthritis susceptibility: a meta-analysis. Rheumatol Int 27(9):827–833
Lee YH, Bae SC, Choi SJ et al (2011) Associations between vitamin D receptor polymorphisms and susceptibility to rheumatoid arthritis and systemic lupus erythematosus: a meta-analysis. Mol Biol Rep 38(6):3643–3651
Lee YH, Rho YH, Choi SJ et al (2006) Association of TNF-alpha −308 G/A polymorphism with responsiveness to TNF-alpha-blockers in rheumatoid arthritis: a meta-analysis. Rheumatol Int 27(2):157–161
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315(7109):629–634
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21(11):1539–1558
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3):177–188
Ceribelli A, Yao B, Dominguez-Gutierrez PR et al (2011) MicroRNAs in systemic rheumatic diseases. Arthritis Res Ther 13(4):229
Hillyer P, Larche MJ, Bowman EP et al (2009) Investigating the role of the interleukin-23/-17 A axis in rheumatoid arthritis. Rheumatology (Oxford) 48(12):1581–1579
Ucar O, Tykocinski LO, Dooley J et al (2013) An evolutionarily conserved mutual interdependence between Aire and microRNAs in promiscuous gene expression. Eur J Immunol 43(7):1769–1778
Fekete T, Szabo A, Beltrame L et al (2012) Constraints for monocyte-derived dendritic cell functions under inflammatory conditions. Eur J Immunol 42(2):458–469
Acknowledgement
This study was supported in part by a grant of the Korea Healthcare technology R&D Project, Ministry for Health and Welfare, Republic of Korea (HI13C2124).
Compliance with ethical guidelines
Conflict of interest. G.G. Song, S.-C. Bae, Y. H. Seo, J.-H. Kim, S. J. Choi, J. D. Ji and Y.Ho. Lee state that there are no conflicts of interest. The accompanying manuscript does not include studies on humans or animals.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Song, G., Bae, SC., Seo, Y. et al. The association between susceptibility to inflammatory arthritis and miR-146a, miR-499 and IRAK1 polymorphisms. Z Rheumatol 74, 637–645 (2015). https://doi.org/10.1007/s00393-014-1493-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00393-014-1493-x