Abstract
It is well known that AM symbiosis provides several ecosystem services leading to plant adaptation in different environmental conditions and positively affects physiological and production features. Although beneficial effects from grapevine and AM fungi interactions have been reported, the impact on growth-defence tradeoff features has still to be elucidated. In this study, the potential benefits of an inoculum formed by two AM fungal species, with or without a monosaccharide addition, were evaluated on young grapevine cuttings grafted onto 1103P and SO4 rootstocks. Inoculated and non-inoculated plants were maintained in potted vineyard substrate under greenhouse conditions for 3 months. Here, agronomic features were combined with biochemical and molecular techniques to assess the influence of the different treatments. Despite the opposite behaviour of the two selected rootstocks, in AM samples, the evaluation of gene expression, agronomic traits and metabolites production revealed an involvement of the whole root microbiome in the growth-defence tradeoff balancing. Noteworthy, we showed that rootstock genotypes and treatments shaped the root-associated microbes, stimulating plant growth and defence pathways. Progresses in this field would open new perspectives, enabling the application of AMF or their inducers to achieve a more sustainable agriculture also in light of the ongoing climate change.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grapevine is one of the most cultivated crops worldwide since its great economic importance resulting from grape and wine production, and commercialization (Chitarra et al. 2017). For this reason, over the years, viticulture industry has selected several cultivars showing different traits (i.e. flavour, yields, colour) influenced by geology, soil-scape and climate features, driving some major wine peculiarities (Priori et al. 2019). These components, and their interactions, concur to define the terroir of a particular environment (Resolution OIV/VITI 333/2010). Besides scion variety features, rootstocks are able to strongly affect scion performances by means of water transport, biochemical and molecular processes, impacting the whole plant functions and its response to biotic/abiotic stress factors (Chitarra et al. 2017). In the last decade, research on scion/rootstock interactions strongly increased, aiming to develop more sustainable practices against pests and ameliorating plant adaptability to the ongoing climate change (Lovisolo et al. 2016; Warschefsky et al. 2016; Zombardo et al. 2020). Key drivers influencing defence features and adaptive traits are thought to be the microbial communities residing in plant tissues. To date, several studies reported evidence about their influence on physiological performances (e.g. production of flavours, hormones, VOCs) in many plants, including grapevine, where residing microbiota contribute to defining the microbial terroir (Gilbert et al. 2014).
According to the Intergovernmental Panel on Climate Change (IPCC 2014), an increase in the global surface temperature is expected over the next years, affecting crop production as a consequence of the predicted occurrence of biotic and abiotic stresses (Mittler and Blumwald 2010). To achieve resilience to stress, numerous efforts have been done over the years, such as the adoption of specific breeding programs and genetic engineering approaches (Cushman and Bohnert 2000). Researchers have been focusing just recently their attention on the exploitation of ‘native’ plant defence mechanisms (e.g. hormone signalling, plant immunity activation) against biotic and abiotic stressful factors (Feys and Parker 2000; Jones and Dangl 2006; Hirayama and Shinozaki 2007). The triggering of these responses can occur using chemical treatments (Balestrini et al. 2018), root-associated microorganisms and RNA interference technologies (Alagna et al. 2020), leading plants in a state of alertness—‘primed state’ or ‘priming’—and enabling them to respond more quickly and robustly in case of the exposure to a stress (Beckers and Conrath 2007).
Among soil beneficial microorganisms, arbuscular mycorrhizal fungi (AMF) establish symbioses with the majority of land plants showing an important role in providing nutrients, particularly phosphate and N, but also water and other elements to the host plant (Jacott et al. 2017; Balestrini and Lumini 2018). Mycorrhizal symbiosis is able to influence plant growth and productivity and enhance the tolerance to biotic and abiotic stresses as demonstrated in many crops (Balestrini and Lumini 2018; Balestrini et al. 2018; Alagna et al. 2020). In addition, AM fungi are able to increase aggregation of soil surrounding roots, improving soil matrix stability and physicochemical characteristics (Uroz et al. 2019). Grapevine roots are naturally colonized by native AM fungi with a great impact on growth, yield, quality and development performances (Deal et al. 1972; Karagiannidis et al. 1995; Linderman and Davis 2001; Trouvelot et al. 2015). Thanks to the application of metagenomics approaches to soil and roots, new insights about the AMF living in symbiosis with grapevine have been discovered (Balestrini et al. 2010; Holland et al. 2014; Balestrini and Lumini 2018).
Rootstocks-mediated adaptation to a specific environment is based on the growth-defence tradeoff-mediated mechanisms (Chitarra et al. 2017). Tradeoff phenomenon was firstly observed in forestry plants-insect interaction studies and is based on the idea that the limited carbon resources produced by photosynthesis are allocated toward growth or defence processes in order to maximize the adaptation strategies and fitness costs in diverse environments (Huot et al. 2014; Chitarra et al. 2017; Züst and Agrawal 2017). Stresses impair plant growth, redirecting energy and carbon sources toward defence, reducing growth and reproduction performances (Bandau et al. 2015; Züst and Agrawal 2017). Recently, it was suggested that through a meta-analysis, the increased plant resistance promoted by Epichloë fungal endophytes does not compromise plant growth, eliminating the tradeoff between growth and defence (Bastías et al. 2021). A role in tradeoff balance has been demonstrated also for AM symbioses, improving nutrient uptake, disease tolerance and abiotic stress resilience (Jacott et al. 2017).
In this study, we aimed to evaluate if AM fungi and rootstocks can concomitantly contribute to fine-tuning growth-defence tradeoff features in grapevine, thus enabling plants to trigger earlier and enhanced defence responses against a potential stressor. The use of specific molecules that can promote the AM fungal colonization has been proposed to improve mycorrhizal inoculum applications under practical field condition (Bedini et al. 2018). In this context, an affordable strategy is the application at low doses of oligosaccharides (i.e. glucose, fructose and xylose) that have a stimulant effect on AM symbiosis colonization (Lucic and Mercy 2014—Patent application EP2982241A1). These compounds, initially called as elicitors, in relation to the impact on plant defence, can promote mycorrhizal performances and, for this reason, the term ‘inducer’ was proposed (Bedini et al. 2018). In this work, the impact of an inoculum formed by two AMF species (Funneliformis mosseae and Rhizophagus irregularis), already reported among the species present in vineyards (Berruti et al. 2017), with or without the addition of a monosaccharide (D-glucose) at low dose (the so called inducer), has been evaluated on young grapevine cuttings cv. Glera grafted onto 1103 Paulsen and SO4 rootstocks, well known to trigger an opposite growth-defence behaviour in the scion. The effect of the several treatments on the root-associated microbiota has been also evaluated, to verify the response mediated by the AM and its recruited mycorrhizosphere.
Materials and methods
Biological materials and experimental setup
Two hundred 1-year-old dormant vines of ‘Glera’ cultivar grafted onto 1103 Paulsen (1103P) and SO4 rootstocks certified as ‘virus free’ were purchased from an Italian vine nursery (Vivai Cooperativi Rauscedo, Italy; http://www.vivairauscedo.com). Vine roots were washed with tap water and cut to about 4 cm before plantation in 2-L pot containers filled with not sterilized substrate mixture of vineyard soil/Sphagnum peat (8:2, v:v) to better simulate the field conditions. The substrate composition was a sandy loam soil (pH 7.8; available P 10.4 mg kg−1; organic matter 1.80%; cation exchange capacity 20.11 mew 100 g−1).
Grapevine cuttings were inoculated with AMF mixed inoculum (INOQ GmbH, Germany, 238.5 million propagule per kg inoculum) at planting time by placing it in the hole and in contact with the roots following the manufacturer’s instructions. Mycorrhizal inoculum, a powder-based mycorrhizal root fragment (Advantage Grade II, 2016—INOQ GmbH), contained 50% Rhizoglomus irregulare (syn. Rhizophagus irregularis; 450 million propagules per kg) and 50% Funneliformis mosseae (27 million propagules per kg). The fungal lines were produced ex vitro, on Zea mays and Plantago lanceolata (sand/vermiculite, v/v). Both AMF inoculum and D-glucose at low dose (i.e. the inducer) were prepared by Louis Mercy (INOQ GmbH; patent EP2982241A1). The containers were prepared according to treatments as follows: (i) 25 plants for each rootstock as uninoculated control plants (C); (ii) 25 plants for each rootstock inoculated with 50 mg/L of AMF mixed inoculum (M); (iii) 25 plants for each rootstock inoculated with 50 mg/L of AMF mixed inoculum + inducer (M + I); (iv) 25 plants for each rootstock amended with 50 mg/L of inducer to stimulate the exploitation of native AMF symbiosis (I). Daily watered grapevine plants were kept under partially climate-controlled greenhouse, under natural light and photoperiod conditions for 3 months.
After 3 months, at the end of the experiment, engraftment, growth index and chlorophyll content were recorded. Leaf and root samples for molecular and biochemical analysis were collected from at least three randomly selected plants and immediately stored at − 80 °C. A part of the root apparatus was used to estimate the level of mycorrhiza formation as described (Balestrini et al. 2017).
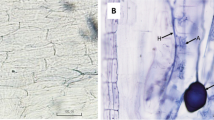
Morphological observations in the colonized fragments of thin roots allowed to identify the presence of the typical structures of the symbiosis, regardless of the thesis. However, the patchy level of colonization, and the quality of the root segments after the staining, made morphological quantification difficult, and therefore, the AMF presence has been assessed by molecular analyses (see below).
Growth index, engraftment and chlorophyll content
At the end of the experiment, phenological stages were recorded and classified according to Biologische Bundesanstalt, bundessortenamt und CHemische industrie (BBCH) scale (from 00 to 12, from dormancy to 9 or more leaves unfolded, respectively). BBCH scales have been developed for many crops, including grapevine, and it is based on a decimal code system that identifies the growth stage (Lancashire et al. 1991); engraftment % (i.e. rooting %) were visually determined for each plant and treatment. Chlorophyll content was determined using a portable chlorophyll meter SPAD (Konica Minolta 502 Plus). Readings were collected from the second or third leaf from the top on at least three leaves per plant on five randomly selected vines for each experimental condition (Chitarra et al. 2016).
Targeted metabolite analyses
Contents of trans-resveratrol, viniferin and abscisic acid (ABA) were quantified on at least three biological replicates per condition according to the protocol previously described (Pagliarani et al. 2019, 2020; Mannino et al. 2020). Leaves and roots from two randomly selected plants were pooled to form a biological replicate, immediately frozen in liquid nitrogen, freeze-dried and stored at − 80 °C until use. Briefly, about 100 mg of freeze-dried sample (leaf or root) was transferred with 1 mL of methanol:water (1:1 v/v) acidified with 0.1% (v/v) of formic acid in an ultrasonic bath for 1 h. Samples were centrifuged for 2 min at 4 °C and 23.477 g, and the supernatant was analysed by high-performance liquid chromatography (HPLC). Original standards of resveratrol (purity ≥ 99%), viniferin (purity ≥ 95%) and ABA (purity ≥ 98.5%, Sigma-Aldrich) were used for the identification by comparing retention time and UV spectra. The quantification was made by external calibration method. The HPLC apparatus was an Agilent 1220 Infinity LC system (Agilent R, Waldbronn, Germany) model G4290B equipped with gradient pump, auto-sampler and column oven set at 30 °C. A 170 Diode Array Detector (Gilson, Middleton, WI, United States) set at 265 nm (ABA and IAA) and 280 nm (for stilbenes) was used as detector. A Nucleodur C18 analytical column (250 × 4.6 mm i.d., 5 μm, Macherey Nagel) was used. The mobile phases consisted in water acidified with formic acid 0.1% (A) and acetonitrile (B), at a flow rate of 0.500 mL min−1 in gradient mode, 0–6 min, from 10 to 30% of B; 6–16 min, from 30 to 100% of B; and 16–21 min, 100% of B. Twenty microliters was injected for each sample.
Total N, soluble carbohydrate content in leaf and net nitrate uptake in root
The Kjeldahl method was performed according to method 981.10 of the AOAC International (2016), using VELP Scientifica DKL 20 Automatic Kjeldahl Digestion Unit and UDK 159 Automatic Kjeldahl Distillation and Titration System. Approximately 0.2 g of leaf raw material was hydrolysed with 15-mL concentrated sulfuric acid (H2SO4) containing one catalyst tablets (3.47 g K2SO4 + 0.003 Se, VELP Scientifica, Italy) in a heat block (DK Heating Digester, VELP Scientifica, Italy) at 300 °C for 2 h. After cooling, H2O was added to the hydrolysates before neutralization with NaOH (30%) and subsequently distilled in a current of steam. The distillate was collected in 25 mL of H3BO3 (1%) and titrated with HCl 0.1 M. The amount of total N in the raw materials was calculated.
Leaf soluble carbohydrate content was quantified (Chitarra et al. 2018).
At the end of the experiment, white non-lignified roots (0.5–1 g) were collected from four randomly selected plants for each treatment and rootstock. Root samples were washed in 0.5 mmol L−1 CaSO4 for 15 min, and then transferred to a 20-mL aerated uptake solution containing 0.5 mmol L−1 Ca(NO3)2 and 0.5 mmol L-1 CaSO4. Net uptake of NO3− was measured removing samples of uptake solution (aliquot of 200 µL) for its determination every 2 min for 10 min (Tomasi et al. 2015). The aliquots were carefully mixed with 800 µL of salicylic acid (5% w/v in concentrated H2SO4) and incubated for 20 min at room temperature following the addition of 19 mL of 2 mol L−1 NaOH. After cooling, nitrate concentration was measured at the absorbance of 410 nm (Shimadzu UV Visible Spectrophotometer UVmini-1240, Kyoto, Japan) and the net nitrate uptake was expressed as µmol (g FW h−1).
RNA isolation and RT-qPCR
Expression changes of target transcripts were profiled on root and leaf samples (three independent biological replicate for each treatment) by quantitative real-time PCR (RT-qPCR) (Chitarra et al. 2018). Total RNA was isolated from the same lyophilized samples (leaves and roots) used for HPLC–DAD analysis and cDNA synthesis was performed as previously reported (Chitarra et al. 2016). The absence of genomic DNA contamination was checked before cDNA synthesis by qPCR using VvUBI specific primers of grapevine. RT-qPCR reactions were carried out in a final volume of 15 μL containing 7.5 μL of Rotor-Gene™ SYBR® Green Master Mix (Qiagen), 1 μL of 3 μM specific primers and 1:10 of diluted cDNA. Reactions were run in the Rotor Gene apparatus (Qiagen) using the following program: 10-min preincubation at 95 °C, followed by 40 cycles of 15 s at 95 °C and 30 s at 60 °C. Each amplification was followed by melting curve analysis (60–94 °C) with a heating rate of 0.5 °C every 15 s. All reactions were performed with at least two technical replicates. The comparative threshold cycle method was used to calculate relative expression levels using plant (elongation factors, actin and ubiquitin, VvEF and VvUBI for root and VvACT and VvEF for leaf tissue) reference genes, while R. irregularis and F. mosseae elongation factors (RiEF1, FmEF, respectively) were used to normalize the expression of the AMF phosphate transporter (PT) genes. Oligonucleotide sequences are listed in Supplementary Table 1. Gene expression data were calculated as expression ratio (relative quantity, RQ) to Control 1103P plants (C 1103P).
Root DNA isolation and sequencing
Root samples were lyophilized prior to DNA extraction. About 30 to 40 mg of freeze-dried and homogenized material were used to extract total DNA following the manufacturer’s instruction of plant/fungi DNA isolation kit (Norgen Biotech Corp., Thorold, ON, Canada) as previously reported (Nerva et al. 2019). Total DNA was quantified using a NanoDrop One spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and DNA integrity was inspected running the extracted samples on a 1% agarose electrophoretic gel. Before sending DNA to sequencing, a further quantification was performed using a Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
To inhibit plant material amplification, we added a mixture of peptide nucleotide acid (PNA) blockers oligos (Kaneka Eurogentec S.A., Belgium) targeted at plant mitochondrial and chloroplast 16S rRNA genes (mitochondrial and plastidial) and plant 5.8S nuclear rRNA. Mitochondrial sequence was derived from Lundberg et al. (2013) with a 1-bp mismatch, internal transcribed sequence was derived from Cregger et al. (2018). PNA was custom-designed for V. vinifera (VvpPNA: GGCTCAACCCTGGACAG; Vv-ITS-PNA: CGAGGGCACGCCTGCCTGG; Vv-mPNA: GGCAAGTGTTCTTCGGA). Thermal cycler conditions were maintained as suggested by the Illumina protocol as previously reported (Nerva et al. 2019).
Sequences were deposited in NCBI database under the BioProject PRJNA718015, BioSamples SAMN18520793 to SAMN18520808 and SRR14089924 to SRR14089939.
Rhizoplane metaphylogenomic analyses, taxonomic distributions
A first strict quality control on raw data was performed with PrinSeq v0.20.4 (Schmieder and Edwards 2011) and then processed with Qiime2 (Bolyen et al. 2019). A previously reported and specific pipeline was used for fungal analysis: retained reads were used to identify the start and stop sites for the ITS region using the hidden Markov models (HMMs) (Rivers et al. 2018), created for fungi and 17 other groups of eukaryotes, which enable the selection of ITS-containing sequences. Briefly, the software allows to distinguish true sequences from sequencing errors, filtering out reads with errors or reads without ITS sequences. To distinguish true sequences from those containing errors, sequences have been sorted by abundance and then clustered in a greedy fashion at a threshold percentage of identity (97%). Trimmed sequences were analysed with DADA2 (Callahan et al. 2016) and sequence variants were taxonomically classified through the UNITE (Abarenkov et al. 2010) database (we selected the reference database built on a dynamic use of clustering thresholds). For graphic representation, only genera with an average relative abundance higher than the settled threshold (1%) were retained.
A 16S specific pipeline was used for bacteria: quality filtering was performed with DADA2 which is able to perform chimera removal, error-correction and sequence variant calling with reads truncated at 260 bp and displaying a quality score above 20. Feature sequences were summarized and annotated using the RDP classifier (Cole et al. 2014) trained to the full length 16S database retrieved from the curated SILVA database (v132) (Quast et al. 2012).
Statistics
Metagenome analyses were performed using R version 3.6.3 (2020–02-29). Fungal and bacterial data were imported and filtered with phyloseq package (version 1.28.0) (McMurdie and Holmes 2013), keeping only the operational taxonomic units (OTUs) with a relative abundance above 0.01 in at least a single sample. Differential abundance of taxa due to the effects of rootstock-treatment interaction was then tested using DESeq2 (version 1.24.0) (Love et al. 2014) package.
For phenotypic, biochemical and RT-qPCR data, when ANOVA indicated that for either Rootstock (R, 1103P and SO4), Inducer (I, NI) and Myc inoculum (M, Myc and NMyc) factors or their interaction was significant, mean separation was performed according to Tukey’s HSD test at a probability level of P ≤ 0.05. ANOVA and Tukey’s HSD test were also used to analyse the treatment effects for each rootstock individually. The standard deviation (SD) or error (SE) of all means was calculated.
Results
Growth, primary metabolism and N uptake and accumulation
The impact of an AM inoculum, an inducer and a combination of both was evaluated on growth parameters (both rooting % and growth stages coded by BBCH scale) in two grapevine rootstock genotypes (R, 1103P and SO4). Four conditions for each genotype were considered: C, not inoculated plants; I, plants treated with the inducer (I); M, AM-inoculated plants; M + I, AM-inoculated plants + inducer.
Results showed a similar impact of the three treatments on the cutting growth parameters (Fig. 1, Table S2), independently from the genotype. Particularly, in SO4 genotype, both the rooting % and the BBCH values were higher in treated plants with respect to the control (Fig. 1a, b). Chlorophyll Content Index (CCI) has been evaluated at the end of the experiment, showing no strong differences among the genotypes and treatments (Fig. 1c), although it was significantly influenced by root colonization (M), the inducer (I) and the M × I interaction in both rootstock genotypes.
Growth-related traits and metabolites. a Growth index according to BBCH scale recorded for each treatment at the end of the experiment (n = 25). Upper picture showed an overview of the cuttings’ development in response to the treatments at the end of the experiment. b Rooting % of cuttings at the end of the experiment (n = 25). c Chlorophyll Content Index (CCI) measured at the end of the experiment (n = 25). d Quantification of soluble carbohydrates contents in leaves at the end of the experiment (n = 4). All data are expressed as mean ± SD. ns, *, ** and ***: non-significant or significant at P ≤ 0.05, P ≤ 0.01 and P ≤ 0.001, respectively. Different lowercase letters above the bars indicate significant differences according to Tukey HSD test (P ≤ 0.05), considering R × I × M interaction. Analysis of variance on the single variables is reported in Table S2. Different uppercase letters above the bars indicate significant differences according to Tukey HSD test (P ≤ 0.05) considering the two rootstocks independently. C, control plants; I, inducer-treated plants; M, AMF mixed inoculum-treated plants; M + I AMF mixed inoculum + inducer-treated plants for 1103P and SO4 selected rootstocks
Treatments generally led to slightly lower values of carbohydrates content in leaves with the exception of M, and only R and I factors significantly influenced this measurement (Fig. 1d). In detail, for each rootstock, I and M + I plants showed significant lower levels of carbohydrates (Fig. 1d).
Net nitrate uptake (NNU) was evaluated (Fig. 2a Table S2), showing that it was significantly affected by M factors and the interaction M × I with lower values in treated samples for both genotypes, particularly in M SO4 plants with respect to C SO4 ones (Fig. 2a).
Net nitrate uptake in roots and total N in leaves. a In vivo Net nitrate uptake. b Total N in leaves (g kg−1 DW). All data are expressed as mean ± SD (n = 3). ns, *, ** and ***: non-significant or significant at P ≤ 0.05, P ≤ 0.01 and P ≤ 0.001, respectively. Different lowercase letters above the bars indicate significant differences according to Tukey HSD test (P ≤ 0.05), considering R × I × M interaction. Analysis of variance on the single variables is reported in Table S2. Different uppercase letters above the bars indicate significant differences according to Tukey HSD test (P ≤ 0.05) considering the two rootstocks independently. C, control plants; I, inducer-treated plants; M, AMF mixed inoculum-treated plants; M + I AMF mixed inoculum + inducer-treated plants for 1103P and SO4 selected rootstocks
As for the CCI, only slight differences in total N content in leaves were evident among genotypes and treatments, although it was significantly affected by the M factor and the M × I interactions (Fig. 2b).
ABA content and the expression of ABA-related genes
To complete the physiological characterization of the two genotypes in response to treatments, the concentration of ABA was quantified in roots and leaves (Fig. 3, Table S2). ABA levels showed a complex scenario in roots where all treatments led to higher ABA levels with respect to the control with the greater significant increase recorded in M SO4. Statistical analyses showed that factors influencing its level were R and M, alone or in the interactions with I (R × I, M × I, R × M × I) (Fig. 3a). ABA content in leaves was under the detection limit among the treatments (data not shown).
Expression changes of ABA-related genes and metabolite quantification in both root and leaf tissues. a ABA content in roots. b VvNCED3 in leaf. c VvABA8OH1 in leaf. d VvNCED3 in root. e VvBG1 in root. f VvGT in leaf. g VvBG1 in leaf. h VvGT in root. i VvABA8OH1 in root. All data are expressed as mean ± SD (n = 3). ns, *, ** and ***: non-significant or significant at P ≤ 0.05, P ≤ 0.01 and P ≤ 0.001, respectively. Different lowercase letters above the bars indicate significant differences according to Tukey HSD test (P ≤ 0.05), considering R × I × M interaction. Analysis of variance on the single variables is reported in Table S2. Different uppercase letters above the bars indicate significant differences according to Tukey HSD test (P ≤ 0.05) considering the two rootstocks independently. C, control plants; I, inducer-treated plants; M, AMF mixed inoculum-treated plants; M + I AMF mixed inoculum + inducer-treated plants for 1103P and SO4 selected rootstocks
To better understand the role of ABA in our system, the expression of ABA-related genes was analysed in both leaves and roots. Relative expressions of (i) a gene encoding for a 9-cis-epoxycarotenoid dioxygenase potentially involved in ABA biosynthesis (VvNCED3, VIT_19s0093g00550 previously reported as VvNCED1); (ii) a gene coding for an enzyme involved in conversion of ABA to 8′-hydroxy ABA (VvABA8OH1); (iii) a β-glucosidase (BG) involved in free ABA biosynthesis via hydrolysis of ABA glucose ester to release the ABA active form (VvBG1; Jia et al. 2016); and (iv) a gene encoding an ABA glucosyltransferase (VvGT; Sun et al. 2010) were evaluated in leaves and roots. In leaves, VvNCED3 expression was not affected by rootstock genotype whereas M samples showed significantly higher expression levels with respect to the other samples (Fig. 3b). No significant difference was detected for VvABA8OH1 expression in leaves although 1103P generally showed higher values with respect to SO4 (Fig. 3c). By contrast, VvNCED3 expression in roots was influenced by R, M and I factors as well as by R × I interaction, and values for each rootstock genotype were lower in all treatments when compared to C plants (Fig. 3d). Similar to that observed in leaves, M + I treatment led to the significant lowest VvNCED3 transcripts level in root samples (Fig. 3d). Two pathways promote free ABA accumulation: (1) NCED-mediated de novo synthesis (Qin and Zeevaart 1999) and (2) BG-mediated hydroxylation (Lee et al. 2006). Looking at VvBG1 gene, its expression was significantly influenced by R and I in leaves, while the presence of the AMF was not significantly relevant. In roots, all the factors and interactions significantly affected VvBG1 expression level, with the highest level in C SO4 samples (Fig. 3e, g). Finally, VvGT showed a trend similar to VvBG1 in leaves where its expression was significantly influenced by R, I and I × M with the exception of SO4 samples where its expression was significantly higher only in M SO4 with respect to C SO4 (Fig. 3f). Conversely, in roots, VvGT transcript levels were significantly lower in all the conditions with respect to the C 1103P plants (Fig. 3h).
Although VvABA8OH1, coding for an enzyme involved in ABA conversion, was not significantly regulated among genotypes and treatments in leaves, it results to be affected by all the considered factors and interactions in roots (Fig. 3i) where it appeared significantly upregulated in M 1103P, M SO4 and M + I SO4 plants with respect to their C (Fig. 3i). It is worth noting the low expression in I root samples, suggesting that the inducer may affect ABA catabolism independently from the genotype and the presence of the AM inoculum.
Defence
Stilbenes are the main defence-related metabolites synthesized in grapevine. In this study trans-resveratrol and viniferin levels were measured in leaves among the several conditions tested (Fig. 4, Table S2). Particularly, resveratrol was only affected by the M × I interaction, showing in parallel significantly higher levels in I and M plants, independently from genotype, with respect to M + I and C plants (Fig. 4a). Viniferin, which was not detectable in C plants, was affected by the M × I interaction and by the I factor alone. I-, M- and M + I-treated plants presented in fact significantly higher values of viniferin than C plants in both rootstocks (Fig. 4b). To correlate biochemical data with molecular responses, expression levels of gene coding for two stilbene synthases (VvSTS1 and VvSTS48) were assessed. Results showed that in both rootstocks, VvSTS1 was upregulated mainly in M 1103P, whereas in SO4 plants, an upregulation in both I and M with respect to the other treatments was observed (Fig. 4c). VvSTS48 expression was influenced by all the factors and their interactions, with the highest expression value in leaves of I-treated SO4 plants (Fig. 4d). Looking independently at each rootstock, in 1103P, only I and M induced significant overexpression of VvSTS48, while in SO4 plants, all the treatments showed enhanced gene expression compared to their controls (Fig. 4d).
Expression changes of stilbenes-related genes and metabolites quantification in leaf tissues. a trans-resveratrol quantification. b Viniferin quantification. c VvSTS1 gene expression changes. d VvSTS48 gene expression changes. All data are expressed as mean ± SD (n = 3). ns, *, ** and ***: non-significant or significant at P ≤ 0.05, P ≤ 0.01 and P ≤ 0.001, respectively. Different lowercase letters above the bars indicate significant differences according to Tukey HSD test (P ≤ 0.05), considering R × I × M interaction. Analysis of variance on the single variables is reported in Table S2. Different uppercase letters above the bars indicate significant differences according to Tukey HSD test (P ≤ 0.05) considering the two rootstocks independently. C, control plants; I, inducer-treated plants; M, AMF mixed inoculum-treated plants; M + I AMF mixed inoculum + inducer-treated plants for 1103P and SO4 selected rootstocks
RT-qPCR was also applied to detect the expression levels of several target genes as markers of diverse defence response pathways (Fig. S1, Table S2). Two genes were studied both in leaves and roots (a sugar transporter, VvSPT13, and a class III chitinases, VvChitIII), and three genes only in leaves (a callose synthase, VvCAS2; a lipoxygenase VvLOX; and the Enhanced Disease Susceptibility 1, VvEDS1) (Fig. S1a–g). Expression of all the considered genes was influenced by I factor, while influence by M was more variable, suggesting a different impact of the treatments on plant metabolism. Among these genes, VvSTP13, encoding a sugar transporter, in leaves of both rootstocks was significantly upregulated in all treatments with respect to their C plants (Fig. S1a), while in root, only M-treated plants showed significantly higher expression values (Fig. S1). VvChitIII showed a different pattern in leaves and roots. In leaves, VvChitIII transcript was significantly induced in M- and M + I-treated plants (Fig. S1c), while in roots, an upregulation was observed only in M-treated ones (Fig. S1d). VvCAS2, coding for a callose synthase (Santi et al. 2013), showed a downregulation in all the treatments, while VvLOX gene, encoding a lipoxygenase involved in the jasmonic acid biosynthesis, was upregulated in all the treatments: among them, the lowest value was observed in M SO4 plants (similar to the C 1103P leaves), suggesting a different response to symbioses in the two genotypes (Fig. S1 e–f). VvEDS1, selected as marker of systemic acquired responses (SAR) mediated by salicylic acid (SA), was influenced by I and M, showing an upregulation trend in I-treated leaves. Conversely, this gene was downregulated in M-treated plants (Fig. S1g).
Rhizoplane metaphylogenomic analyses
Bacterial community was analysed at both order and genus level: the number of retained sequences after chimera removal and taxonomical assignment was always above 35,000 (detailed results of sequencing are reported in Table S3). Shannon index diversity indicated that the only significant difference was observed for the I SO4 samples which show higher index values (Table S4). No significant differences were observed among samples comparing the Shannon index on the fungal community (Table S5). Similar to Shannon index, non-metric multidimensional scaling (NMDS) based on Bray–Curtis dissimilarity matrixes showed that the bacterial community (Fig. 5a) is more affected by treatments than the fungal one (Fig. S2).
Distinct root-associated bacteria community composition among treatments. NMDS algorithm based on Bray–Curtis distances matrixes was used to reduce into a bi-dimensional scaling data obtained for bacteria community (a). Relative abundance of bacterial genera (b) among treatments. Only genera representing at least the 1% over the total number of classified amplicons were retained (n = 3). C, control plants; I, inducer-treated plants; M, AMF mixed inoculum-treated plants; M + I AMF mixed inoculum + inducer-treated plants for 1103P and SO4 selected rootstocks
The bacteria community composition for each sample type at order and genus levels is reported in Table S6. Statistical results of pairwise comparisons among genera are reported in Table S7. To simplify, results are described for the orders and genera that represent at least the 1% of the bacterial community (Fig. 5b). Comparison of the bacterial community between the two rootstocks (1103P vs SO4) revealed that 1103P has a significant higher relative abundance of Pseudomonas species, whereas SO4 has a significant higher relative abundance of Bacillus species. Among the bacterial genera, which display significant differences among the treatments, M 1103P vines stimulated the presence of Bacillus species but repressed the interaction with Pseudomonas ones. In parallel, when comparing treatments on SO4 rootstock, a positive interaction between the mycorrhizal inoculation and the Pseudomonas abundance was observed, whereas the inducer treatment showed a negative impact on Flavobacterium abundance.
The fungal community composition for each sample type at order and genus levels is reported in Table S6. Statistics of the pairwise comparisons among genera are reported in Table S8. Results for the fungal orders and genera that represent at least the 1% of the fungal community are reported in Fig. S3. Focusing on AMF, results confirm the presence of Rhizophagus and Funneliformis in inoculated plants. However, AMF were detected also in the I-treated plants (Fig. 6a). Despite the presence of AMF associated to these roots, gene expression analysis on fungal PT genes showed the presence of RiPT and FmPT transcripts only in M-inoculated plants. Surprisingly, absent or low expression levels were detected in I-treated plants (Fig. 6b, c; Table S2). Indeed, fungal PT genes were expressed in a different way in the two genotypes, suggesting a different symbiosis efficiency of the two rootstocks. This finding was further confirmed by a plant PT gene (VvPT1-3), which expression level was mainly affected by R and M factors, and by ‘R × I’ interaction. It was upregulated in 1103P roots, independently by treatment, with respect to C 1103P, and strongly upregulated in M SO4 ones (Fig. 6d, Table S2).
Mycorrhiza genera and expression changes of plant and fungus phosphate transporter (PT) genes as markers of functional symbioses. a Relative abundances of mycorrhiza genera (n = 3). b RiPT. c FmPT. d VvPT1-3. Gene expression data are expressed as mean ± SD (n = 3). ns, *, ** and ***: non-significant or significant at P ≤ 0.05, P ≤ 0.01 and P ≤ 0.001, respectively. Different lowercase letters above the bars indicate significant differences according to Tukey HSD test (P ≤ 0.05), considering R × I × M interaction. Analysis of variance on the single variables is reported in Table S2. Different uppercase letters above the bars indicate significant differences according to Tukey HSD test (P ≤ 0.05) considering the two rootstocks independently. C, control plants; I, inducer-treated plants; M, AMF mixed inoculum-treated plants; M + I AMF mixed inoculum + Inducer-treated plants for 1103P and SO4 selected rootstocks. Insets: microscope images of typical AM symbioses structures in 1103P and SO4 M-colonized roots
Comparing the fungal composition in C, 24 genera with significant differences of relative abundance were observed. Among the analysed genera, Clonostachys displayed a significant negative correlation with all the treatment in both rootstock genotypes. Focusing on significant genera, usually involved in pathogenic interaction, such as Fusarium, Rhizoctonia and Ilyonectria (Fig. S4), the concomitant use of mycorrhizal inocula with the inducer brought to a significant reduction of Ilyonectria in both rootstocks. Conversely, Fusarium abundance was stimulated in all treatments except for the inoculation with AMF in the 1103P rootstock. Finally, Rhizoctonia genus was positively influenced by the inducer, but only in the SO4 rootstock.
Discussion
Treatments and genotypes differently shape the root-associated bacterial and fungal communities
The importance of root-associated microbes was extensively demonstrated in several crops including grapevine, with the potential to exploit biocontrol strategies that rely on the beneficial traits of plant growth-promoting microorganisms (PGPBs) naturally associated with plants (Verbon and Liberman 2016; Marasco et al. 2018; Yu et al. 2019). Among them, AMF and their impacts on diverse plant species, including economically important crops, have been largely studied highlighting the importance of this relationship that can positively affect both growth and defence traits (Jacott et al. 2017). However, despite these advantages, grapevine breeders normally focus their attention more on phenotypic or metabolic peculiarities rather than on the improvement of the interactions with root-associated microbes (Marín et al. 2021).
Grapevine roots are commonly colonized by different AMF taxa depending on the considered environment, season and soil management making them relevant in defining the ‘microbial terroir’ of a specific grape cultivar (Massa et al. 2020). Svenningsen et al. (2018) reported that AMF ecosystem services might be suppressed by some bacterial groups belonging to Acidobacteria, Actinobacteria, Firmicutes, Chitinophagaceae and Proteobacteria. Our results showed an inverse correlation between the presence of some of these bacteria (i.e. Acidobacteria, genus Vicinamibacter and Actinobacteria genus Gaiella) and AMF ‘functionality’, although ITS sequencing showed a similar level in terms of AMF abundance. It is also necessary to consider that ITS was used in the present work as universal fungal marker (Schoch et al. 2012; Lindahl et al. 2013) to better define the overall fungal population despite ribosomal large subunit (LSU) region consistently shows greater utility for taxonomic resolution for AMF (Xue et al. 2019a). Despite the latter approach can give better results, it has rarely been used in environmental studies of AMF because of sequencing and bioinformatics challenges (Delavaux et al. 2021). Similarly, for a better description of the AMF population, it was recently reported that the use of AMF specific primers, coupled to nested PCR, can greatly help in better defining the AMF population (Suzuki et al. 2020).
Additionally, results obtained from the microbiome analysis confirm that the response of microbial communities to the different treatments are genotype dependent (Marasco et al. 2018). This is particularly clear for the bacterial community, where the addition of the mycorrhizal inoculum promoted the Pseudomonas genus in 1103P and the Bacillus genus in SO4. It is important to remind that both these genera were largely investigated in grapevine because of their ability to protect vine plants against several fungal pathogens. Pseudomonas genus was studied for its ability to impair Botrytis, Neofusiccocum, Ilyonectria, Aspergillus, Phaeomoniella and Phaeoacremonium genera, which are all well-known grape fungal pathogens (Andreolli et al. 2019; Niem et al. 2020). On the other hand, Bacillus species were studied for their ability to reduce the impact of black foot disease (mainly due to infection by Cylindrocarpon and Ilyonectria species) and downy mildew on grapes (Zhang et al. 2017; Russi et al. 2020). These studies well fit with our data where we observed the lower Ilyonectria abundance in M + I 1103P and concomitantly the higher abundance of Bacillus species. Looking at the fungi, all the treatments promoted the presence of different AMF species, suggesting the recruitment of native AM fungal communities by the I-treated roots, independently from the rootstock genotypes. In detail, it is worth noting a higher diversity in AMF colonization in I 1103P with respect to I SO4 plants, independently from the presence of the AMF inoculum, confirming a diverse recruitment pattern for the two genotypes. Interestingly, Clonostachys genus negatively correlated with all the treatments. This genus was extensively studied for its promising exploitation as biological control agents against soil and root pathogens (Nygren et al. 2018; Sun et al. 2020). Considering that in all treatments the Rhizophagus genus was more abundant than in C, we can confirm that a mutual exclusion between Clonostachys and Rhizophagus genera is present. Although a full explanation for this reciprocally inhibitory interaction is still missing, the complex microbial community modulation mediated by the AM fungi could impair the ability of Clonostachys to endophytically colonize the host plant (Ravnskov et al. 2006; Akyol et al. 2018; Xue et al. 2019b). These findings, in accordance with the increase in defence-related metabolites and the expression data on defence-related genes, well fit with the concept of mycorrhizal-induced resistance (MIR) (Cameron et al. 2013) as a cumulative effect of direct and indirect (i.e. mediated by mycorrhizosphere-associated microorganisms) defence responses. Recently, Emmett et al. (2021) also demonstrated that a conserved community is associated to AMF extraradical hyphae, suggesting an influence on the plant-fungal symbiosis.
AM fungi and root-associated microbes balance rootstocks growth traits showing a different pattern of functional symbioses
The impact of the different treatments on two different rootstock genotypes was evaluated. The selected rootstocks (i.e. 1103P and SO4) were well characterized at both agronomic and molecular level (Chitarra et al. 2017), showing opposite growth and defence attitudes. Among rootstock features, fine root development and density, imparting vigour to the scion, varied considerably with an impact on water and nutrient uptake as well as on the interaction with soil microorganisms. AM colonization showed that SO4 consistently presented higher levels of root colonization, together with Kober 5BB and Ruggieri 140, with respect to the others (Chitarra et al. 2017). This is in agreement with previous works (Bavaresco and Fogher 1996; Bavaresco et al. 2000), who showed a variation in the range of AM-colonized grape rootstocks among genotypes, which could be considered the main factor driving AM recruitment. However, functional symbiosis was strongly influenced also by scion requirements, soil fertility and soil pH (Bavaresco and Fogher 1996; Bavaresco et al. 2000). Here, both rooting and growth parameters, and partially the CCI, clearly showed a compensation effect in the less vigorous SO4 with respect to 1103P, reaching similar values in all the treatments. A role could be attributed to AMF particularly in SO4. To attest this hypothesis, considering that high-affinity PTs in AM have been characterized and it has clearly been demonstrated that plants possess a symbiotic Pi uptake pathway (Berruti et al. 2016), AM fungal PT genes (RiPT and FmPT) have been tested showing a highly expression in M SO4 for both, and also in M + I SO4 for FmPT. Similarly, the plant gene VvPT1-3, homolog of mycorrhiza-inducible inorganic phosphate transporters such as LePT4 and OsPT11 (Balestrini et al. 2017), was significantly upregulated in M SO4. The positive effects exerted by AM symbiosis in growth and physiological features were largely documented in several plants (e.g. Chitarra et al. 2016; Balestrini et al. 2020). Surprisingly, although the ITS sequencing showed a certain abundance of AM genera in both I and M + I, the inducer seemed to lower the expression of plant and fungal genes generally involved in symbiosis functioning. This should be related to the presence of bacteria reported to diminish AMF functionality (Svenningsen et al. 2018). As well, an impact of the inducer on the number of fine roots, which are those colonized by AMF, cannot be excluded also considering that IAA was not detectable in I samples. Looking at the whole microbial community, in addition to a selection based on the rootstock genotype, it is worth noting that I treatment (particularly I SO4) was able to significantly increase diversity of the microbiota (Table S4). Samples treated with the inducer showed higher bacterial diversity hosting many groups of PGPBs such as Burkholderiaceae that might be linked to potassium (K) and phosphorous (P) solubilization and availability (Gu et al. 2020); Pseudomonas and Bacillus spp. were able to produce siderophores, auxin and cytokinins and characterized as phosphate-solubilizing bacteria (Saad et al. 2020; Subrahmanyam et al. 2020) (Table S7). These findings could explain the bacteria-mediated growth effects in I treatments particularly for the SO4 genotype. By contrast, the whole fungal diversity was not significantly affected among the treatments.
Nitrogen (N) is an essential element for all grapevine processes and N transporters were found among the genes upregulated by both a single AMF and a mixed bacterial-fungal inoculum through transcriptomics in grapevine roots (Balestrini et al. 2017). However, although AMF may positively influence plant N compound uptake and transport (Balestrini et al. 2020), negative, neutral or positive AMF effects on N nutrition have been reported (Bücking and Kafle 2015). Due to the fact that several nitrate transporters were found to be regulated by an AMF inoculum (Balestrini et al. 2017), the attention was mainly focused on nitrate uptake. Lower values of nitrate uptake with respect to controls were observed among all treatments, independently from the considered genotypes. Furthermore, any relevant effect on N accumulation in leaves was observed, suggesting that a positive correlation between N content and growth is not relevant in our system or likely due to a biomass dilution effect since the higher growth index recorded particularly in SO4-treated plants. AMF have been reported to show NH4+ preference to be assimilated in extraradical mycelium and translocated to plant roots after completion of the GS-GOGAT cycle (Balestrini et al. 2020). In this respect, to the plants side, the lower NNU observed in M-inoculated plants suggest a role of AMF in regulating root N uptake strategies helping plants in acquiring N.
The plant hormone ABA is a chemical signal involved in the plant response to various abiotic environmental factors, but it can also play a role in interactions with phytopathogens by modulating tissue colonization depending on microorganism type, site and time of infection (Ton et al. 2009). An impact of ABA on AMF colonization has been also reported at diverse colonization stages (Bedini et al. 2018). A role for ABA in the mechanisms by which AM symbiosis influences stomata conductance under drought stress was also suggested (Chitarra et al. 2016). Here, ABA levels were affected by both the genotype and the AMF inoculum. A significant effect of the M treatment was found on the expression of a key gene involved in the ABA synthesis in leaves (VvNCED3), showing a positive correlation with the ABA levels in roots. Our result is in accordance with the fact that ABA produced in leaves is then translocated in roots where it might act as a signal to promote root growth (McAdam et al. 2016). AMF presence led to higher ABA content in M SO4 roots, despite the fact that generally SO4 rootstock was reported to have a low endogenous content (Chitarra et al. 2017), suggesting a potential enhanced tolerance to abiotic stresses in M SO4. As already reported by Ferrero et al. (2018), the relationship between biosynthetic and catabolic processes may be complex and diverse in the different plant organs. Our results showed a different expression pattern of most of the considered genes involved in ABA synthesis and catabolism in leaves and roots. A gene coding for an ABA 8'-hydroxylase (VvABA8OH1), belonging to the CYP707A gene family and with a primary role in ABA catabolism, showed an opposite trend in M and I root apparatus, in agreement with the ABA root accumulation. Overall, obtained data are in accordance with that reported by Martín-Rodríguez et al. (2016) showing that both ABA biosynthesis and catabolism are finely tuned in AM-colonized roots. Although with the activation of different mechanisms depending on the treatment, an impact on ABA homeostasis can be suggested particularly in SO4 genotype.
AM symbiosis triggers defence-related transcripts and metabolites more in 1103P than in SO4 rootstock
Plants finely tune the immune system to control both pathogen infection and beneficial microorganism accommodation. Soil bacteria and fungi play a double role in promoting growth and defence response, helping in maintaining the homeostasis in the whole microbial communities associated to the roots through the Induced Systemic Resistance (ISR) pathways (Liu et al. 2020). In grapevine, stilbenes are phytoalexins with proved antifungal activities (Chalal et al. 2014). Here, resveratrol content was higher in I and M leaves with respect to untreated controls, while viniferin, that is highly toxic for grape foliar pathogens such as downy and powdery mildew (Chitarra et al. 2017), has a similar trend in all the treatments while it was not detected in C plants. These patterns clearly highlight a stimulating effect mediated by root-associated microbes (native or inoculated), with differences that might be related to the diverse microbiome composition. Among the genes involved in stilbene synthesis, VvSTS48, coding for a stilbene synthase reported as induced by downy mildew infection, showed the highest expression value in I SO4 plants, suggesting a different modulation among treatments and genotypes.
Carbohydrate metabolism is also involved in plant defence responses against foliar pathogens (Sanmartín et al. 2020). In tomato, AM symbiosis was reported to be involved in Botrytis cinerea resistance through the mycorrhiza-induced resistance (MIR) mediated by callose accumulation. A tomato callose synthase gene (PMR4) was in fact upregulated by mycorrhization mainly upon biotic infection (Sanmartín et al. 2020). In the present study, attention has been focused on the homolog grape gene VvCAS2. Conversely to that previously observed, VvCAS2 showed a downregulation trend in all the treatments with respect to control plants. These findings suggest a primary role in microbe-mediated stimulating of defence responses against biotic factors in grape. Since a correlation between MIR and sugar signalling pathway was reported (Sanmartín et al. 2020), the expression of a grapevine sugar transporter gene (VvSTP13), homolog to the Arabidopsis STP13, involved in intracellular glucose uptake and in B. cinerea resistance, was followed in leaves and roots. Although total soluble carbohydrates were not affected by treatments in leaves, VvSTP13 expression showed an upregulation trend in all the treatments, particularly in both I sample and M 1103P leaves, suggesting an effect of AMF inoculum in the susceptible genotype. Looking at the roots, VvSTP13 upregulation trend was observed mainly in mycorrhizal roots, in agreement with the fact that expression of genes from the STP family was revealed in arbuscule-containing cells of Medicago truncatula (Hennion et al. 2019). The same trend observed for VvSTP13 was also found for a gene coding for a class III chitinase (VvChitIII). Class III chitinases have been already reported to be markers of functional symbioses (Balestrini et al. 2017), being localized in arbuscule-containing cells (Hogekamp et al. 2011). Finally, the expression of two target genes (VvLOX and VvEDS1), respectively involved in ISR mediated by jasmonate and SAR mediated by salicylic acid, although differently modulated by the inducer and AM fungi, confirmed the role of the whole microbiome on the plant immunity system in the scion of both rootstock genotypes (Cameron et al. 2013).
Conclusion
Overall, our results allowed to provide new insights into growth-defence tradeoff responses in a model fruit crop (Fig. 7). Although molecular mechanisms at the basis of plant priming are still matter of debate, several hypotheses have been proposed. In this study, a finely tune regulation of growth and defence traits has been highlighted considering three main influencing factors, i.e. the plant genotype, an AM inoculum and an oligosaccharide described as involved in AMF colonization induction. The attention has been focused on two rootstocks characterized by opposite tradeoffs. Growth traits have been improved mainly in the low vigour genotype (SO4) by all the treatments probably through the activation of diverse pathways by the root-associated microbes. It is worth noting that all the treatments shaped the microbial communities associated to the roots in both the genotypes. Looking at the defence response, a positive impact on immunity system has been revealed both by the AMF inoculum and the oligosaccharide, although with the activation of different pathways. Results suggest that AM symbiosis triggers a mycorrhiza-induced resistance (MIR) also in a model woody plant such as grapevine.
Overview of phenotypic, biochemical and molecular changes induced by the treatments. Green arrows indicate responses in 1103 Paulsen (1103P) rootstock whereas orange ones are referred to SO4 genotype. Upward arrows indicate an increase whereas downward arrows represent a decrease in content of metabolites or gene relative expression or relative abundance of microbial taxa with respect to control (C) plants. NNU, net nitrate uptake; ABA, abscisic acid
Data availability
Sequences were deposited in NCBI database under the BioProject PRJNA718015, BioSamples SAMN18520793 to SAMN18520808 and SRR14089924 to SRR14089939.
References
Abarenkov K, Henrik Nilsson R, Larsson K, Alexander IJ, Eberhardt U, Erland S, Hoiland K, Kjoller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedresoo L, Ursing BM, Vralstad T, Liimatainen K, Peintner U, Koljalg U (2010) The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol 186:281–285. https://doi.org/10.1111/j.1469-8137.2009.03160.x
Akyol TY, Niwa R, Hirakawa H, Maruyama H, Sato T, Suzuki T, Fukunaga A, Sato T, Yoshida S, Tawaraya K, Saito M, Ezawa T, Sato S (2018) Impact of introduction of arbuscular mycorrhizal fungi on the root microbial community in agricultural fields. Microbes Environ 34:23–32. https://doi.org/10.1264/jsme2.ME18109
Alagna F, Balestrini R, Chitarra W, Marsico AD, Nerva L (2020) Getting ready with the priming: Innovative weapons against biotic and abiotic crop enemies in a global changing scenario. In: Hossain MA, Liu F, Burritt DJ, Fujita M, Huang B (eds) Priming-Mediated Stress and Cross-Stress Tolerance in Crop Plants. Academic Press, Elsevier, London, pp 35–56
Andreolli M, Zapparoli G, Angelini E, Lucchetta G, Lampis S, Vallini G (2019) Pseudomonas protegens MP12: a plant growth-promoting endophytic bacterium with broad-spectrum antifungal activity against grapevine phytopathogens. Microbiol Res 219:123–131. https://doi.org/10.1016/j.micres.2018.11.003
Balestrini R, Brunetti C, Chitarra W, Nerva L (2020) Photosynthetic traits and nitrogen uptake in crops: which is the role of arbuscular mycorrhizal fungi? Plants 9:1105. https://doi.org/10.3390/plants9091105
Balestrini R, Chitarra W, Antoniou C, Ruocco M, Fotopoulos V (2018) Improvement of plant performance under water deficit with the employment of biological and chemical priming agents. J Agric Sci 156:680–688. https://doi.org/10.1017/S0021859618000126
Balestrini R, Lumini E (2018) Focus on mycorrhizal symbioses. Appl Soil Ecol 123:299–304
Balestrini R, Magurno F, Walker C, Lumini E, Bianciotto V (2010) Cohorts of arbuscular mycorrhizal fungi (AMF) in Vitis vinifera, a typical Mediterranean fruit crop. Environmental Microbiology Reports 2:594–604. https://doi.org/10.1111/j.1758-2229.2010.00160.x
Balestrini R, Salvioli A, Dal Molin A, Novero M, Gabelli G, Paparelli E, Marroni F, Bonfante P (2017) Impact of an arbuscular mycorrhizal fungus versus a mixed microbial inoculum on the transcriptome reprogramming of grapevine roots. Mycorrhiza 27:417–430. https://doi.org/10.1007/s00572-016-0754-8
Bandau F, Decker VHG, Gundale MJ, Albrectsen BR (2015) Genotypic tannin levels in Populus tremula impact the way nitrogen enrichment affects growth and allocation responses for some traits and not for others. PLoS ONE 10:e0140971. https://doi.org/10.1371/journal.pone.0140971
Bastías DA, Gianoli E, Gundel PE (2021) Fungal endophytes can eliminate the plant growth-defence trade-off. New Phytol 230:2105–2113. https://doi.org/10.1111/nph.17335
Bavaresco L, Cantù E, Trevisan M (2000) Chlorosis occurrence, natural arbuscular-mycorrhizal infection and stilbene root concentration of ungrafted grapevine rootstocks growing on calcareous soil. J Plant Nutr 23:1685–1697. https://doi.org/10.1080/01904160009382133
Bavaresco L, Fogher C (1996) Lime-induced chlorosis of grapevine as affected by rootstock and root infection with arbuscular mycorrhiza and Pseudomonas fluorescens. Vitis 35:119–123. https://doi.org/10.5073/vitis.1996.35.119-123
Beckers GJ, Conrath U (2007) Priming for stress resistance: from the lab to the field. Curr Op Plant Biol 10:425–431. https://doi.org/10.1016/j.pbi.2007.06.002
Bedini A, Mercy L, Schneider C, Franken P, Lucic-Mercy E (2018) Unraveling the initial plant hormone signaling, metabolic mechanisms and plant defense triggering the endomycorrhizal symbiosis behavior. Front Plant Sci 9:1800. https://doi.org/10.3389/fpls.2018.01800
Berruti A, Desirò A, Visentin S, Zecca O, Bonfante P (2017) ITS fungal barcoding primers versus 18S AMF-specific primers reveal similar AMF-based diversity patterns in roots and soils of three mountain vineyards. Env Microbiol Rep 9:658–667. https://doi.org/10.1111/1758-2229.12574
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Mauricio Caraballo-Rodríguez A, Chase J, Cope EK, Da Silva R, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Bin Kang K, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Turan Naimey A, Navas-Molina JA, Felix Nothias L, Orchanian SB, Pearson T, Peoples SL, Petras D, Lai Preuss M, Pruesse E, Buur Rasmussen L, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Jin Song S, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Zech XuZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotech 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Bücking H, Kafle A (2015) Role of arbuscular mycorrhizal fungi in the nitrogen uptake of plants: current knowledge and research gaps. Agronomy 5:587–612. https://doi.org/10.3390/agronomy5040587
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Cameron DD, Neal AL, van Wees SC, Ton J (2013) Mycorrhiza-induced resistance: more than the sum of its parts? Trends Plant Sci 18:539–545. https://doi.org/10.1016/j.tplants.2013.06.004
Chalal M, Klinguer A, Echairi A, Meunier P, Vervandier-Fasseur D, Adrian M (2014) Antimicrobial activity of resveratrol analogues. Molecules 19:7679–7688. https://doi.org/10.3390/molecules19067679
Chitarra W, Cuozzo D, Ferrandino A, Secchi F, Palmano S, Perrone I, Boccacci P, Pagliarani C, Gribaudo I, Mannini F, Gambino G (2018) Dissecting interplays between Vitis vinifera L. and grapevine virus B (GVB) under field conditions. Mol Plant Path 19:2651–2666. https://doi.org/10.1111/mpp.12735
Chitarra W, Pagliarani C, Maserti B, Lumini E, Siciliano I, Cascone P, Schubert A, Gambino G, Balestrini R, Guerrieri E (2016) Insights on the impact of arbuscular mycorrhizal symbiosis on tomato tolerance to water stress. Plant Physiol 171:1009–1023. https://doi.org/10.1104/pp.16.00307
Chitarra W, Perrone I, Avanzato CG, Minio A, Boccacci P, Santini D, Gilardi G, Siciliano I, Gullino ML, Delledonne M, Mannini F, Gambino G (2017) Grapevine grafting: scion transcript profiling and defense-related metabolites induced by rootstocks. Front Plant Sci 8:654. https://doi.org/10.3389/fpls.2017.00654
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM (2014) Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Ac Res 42:D633–D642. https://doi.org/10.1093/nar/gkt1244
Cregger MA, Veach AM, Yang ZK, Crouch MJ, Vilgalys R, Tuskan GA, Schadt CW (2018) The Populus holobiont: dissecting the effects of plant niches and genotype on the microbiome. Microbiome 6:31. https://doi.org/10.1186/s40168-018-0413-8
Cushman JC, Bohnert HJ (2000) Genomic approaches to plant stress tolerance. Curr Op Plant Biol 3:117–124. https://doi.org/10.1016/S1369-5266(99)00052-7
Deal D, Boothroyd C, Mai W (1972) Replanting of vineyards and its relationship to vesicular-arbuscular mycorrhiza. Phytopathology 62(172):175
Delavaux CS, Sturmer SL, Wagner MR, Schütte U, Morton JB, Beer JD (2021) Utility of large subunit for environmental sequencing of arbuscular mycorrhizal fungi: a new reference database and pipeline. New Phytol 229:3048–3052. https://doi.org/10.1111/nph.17080
Emmett BD, Lévesque-Tremblay V, Harrison MJ (2021) Conserved and reproducible bacterial communities associate with extraradical hyphae of arbuscular mycorrhizal fungi. ISME J 15:2276–2288. https://doi.org/10.1038/s41396-021-00920-2
Ferrero M, Pagliarani C, Novák O, Ferrandino A, Cardinale F, Visentin I, Schubert A (2018) Exogenous strigolactone interacts with abscisic acid-mediated accumulation of anthocyanins in grapevine berries. J Exp Bot 69:2391–2401. https://doi.org/10.1093/jxb/ery033
Feys BJ, Parker JE (2000) Interplay of signaling pathways in plant disease resistance. Trends Genet 16:449–455. https://doi.org/10.1016/S0168-9525(00)02107-7
Gilbert JA, van der Lelie D, Zarraonaindia I (2014) Microbial terroir for wine grapes. Proc Nat Acad Sci USA 111:5–6. https://doi.org/10.1073/pnas.1320471110
Gu Y, Dong K, Geisen S et al (2020) The effect of microbial inoculant origin on the rhizosphere bacterial community composition and plant growth-promotion. Plant Soil 452:105–117. https://doi.org/10.1007/s11104-020-04545-w
Hennion N, Durand M, Vriet C, Doidy J, Maurousset L, Lemoine R, Pourtau N (2019) Sugars en route to the roots. Transport, metabolism and storage within plant roots and towards microorganisms of the rhizosphere. Physiol Plant 165:44–57. https://doi.org/10.1111/ppl.12751
Hirayama T, Shinozaki K (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12:343–351. https://doi.org/10.1016/j.tplants.2007.06.013
Hogekamp C, Arndt D, Pereira PA, Becker JD, Hohnjec N, Küster H (2011) Laser microdissection unravels cell-type-specific transcription in arbuscular mycorrhizal roots, including CAAT-box transcription factor gene expression correlating with fungal contact and spread. Plant Physiol 157:2023–2043. https://doi.org/10.1104/pp.111.186635
Holland TC, Bowen P, Bogdanoff C, Hart MM (2014) How distinct are arbuscular mycorrhizal fungal communities associating with grapevines? Biol Fertil Soils 50:667–674. https://doi.org/10.1007/s00374-013-0887-2
Huot B, Yao J, Montgomery BL, He SY (2014) Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant 7:1267–1287. https://doi.org/10.1093/mp/ssu049
Jacott CN, Murray JD, Ridout CJ (2017) Trade-offs in arbuscular mycorrhizal symbiosis: disease resistance, growth responses and perspectives for crop breeding. Agronomy 7:75. https://doi.org/10.3390/agronomy7040075
Jia H, Wang C, Zhang C, Haider MS, Zhao P, Liu Z, Shangguan L, Pervaiz T, Fang J (2016) Functional analysis of VvBG1 during fruit development and ripening of grape. J Plant Growth Reg 35:987–999. https://doi.org/10.1007/s00344-016-9597-y
Jones JD, Dangl JL (2006) The Plant Immune System Nature 444:323–329. https://doi.org/10.1038/nature05286
Karagiannidis N, Nikolaou N, Mattheou A (1995) Wirkung dreier VA-Mykorrhizapilze auf Ertrag und Nährstoffaufnahme von drei Unterlagen. Vitis 34:85–89
Lancashire PD, Bleiholder H, Van Den Boom T, Langelüddeke P, Stauss R, Weber E, Witzenberger A (1991) A uniform decimal code for growth stages of crops and weeds. Ann App Biol 119:561–601. https://doi.org/10.1111/j.1744-7348.1991.tb04895.x
Lee KH, Piao HL, Kim H-Y, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee I, Hwang I (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126:1109–1120
Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R, Kõljalg U, Pennanen T, Rosendahl S, Stenlid J, Kauserud H (2013) Fungal community analysis by high-throughput sequencing of amplified markers–a user’s guide. New Phytol 199:288–299. https://doi.org/10.1111/nph.12243
Linderman RG, Davis EA (2001) Comparative response of selected grapevine rootstocks and cultivars to inoculation with different mycorrhizal fungi. Am J Enol Vit 52:8–11
Liu H, Brettell LE, Qiu Z, Singh BK (2020) Microbiome-mediated stress resistance in plants. Trends Plant Sci 25:733–743. https://doi.org/10.1016/j.tplants.2020.03.014
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Lovisolo C, Lavoie-Lamoureux A, Tramontini S, Ferrandino A (2016) Grapevine adaptations to water stress: new perspectives about soil/plant interactions. Theor Exp Plant Physiol 28:53–66. https://doi.org/10.1007/s40626-016-0057-7
Lucic E, Mercy L (2014) A method of mycorrhization of plants and use of saccharides in mycorrhization. European Patent Office EP2982241A1
Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL (2013) Practical innovations for high-throughput amplicon sequencing. Nat Methods 10:999–1002. https://doi.org/10.1038/nmeth.2634
Mannino G, Nerva L, Gritli T, Novero M, Fiorilli V, Bacem M, Bertea CM, Lumini E, Chitarra W, Balestrini R (2020) Effects of different microbial inocula on tomato tolerance to water deficit. Agronomy 10:170. https://doi.org/10.3390/agronomy10020170
Marasco R, Rolli E, Fusi M, Michoud G, Daffonchio D (2018) Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome 6:3. https://doi.org/10.1186/s40168-017-0391-2
Marín D, Armengol J, Carbonell-Bejerano P, Escalona JM, Gramaje D, Hernández-Montes E, Intrigliolo DS, Martínez-Zapater JM, Medrano H, Mirás-Avalos JM, Palomares-Rius JE, Romero-Azorín P, Savé R, Santesteban LG, de Herralde F (2021) Challenges of viticulture adaptation to global change: tackling the issue from the roots. Austral J Grape Wine Res 27:8–25. https://doi.org/10.1111/ajgw.12463
Martín-Rodríguez JA, Huertas R, Ho-Plágaro T, Ocampo JA, Turečková V, Tarkowská D, Ludwig-Müller J, García-Garrido JM (2016) Gibberellin–abscisic acid balances during arbuscular mycorrhiza formation in tomato. Frontiers Plant Sci 7:1273. https://doi.org/10.3389/fpls.2016.01273
Massa N, Bona E, Novello G, Todeschini V, Boatti L, Mignone F, Gamalero E, Lingua G, Berta G, Cesaro P (2020) AMF communities associated to Vitis vinifera in an Italian vineyard subjected to integrated pest management at two different phenological stages. Sci Rep 10:9197. https://doi.org/10.1038/s41598-020-66067-w
McAdam SA, Brodribb TJ, Ross JJ (2016) Shoot-derived abscisic acid promotes root growth. Plant Cell Env 39:652–659. https://doi.org/10.1111/pce.12669
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Mittler R, Blumwald E (2010) Genetic engineering for modern agriculture: challenges and perspectives. Ann Rev Plant Biol 61:443–462. https://doi.org/10.1146/annurev-arplant-042809-112116
Nerva L, Pagliarani C, Pugliese M, Monchiero M, Gonthier S, Gullino ML, Gambino G, Chitarra W (2019) Grapevine phyllosphere community analysis in response to elicitor application against powdery mildew. Microorganisms 7:662. https://doi.org/10.3390/microorganisms7120662
Niem JM, Billones-Baaijens R, Stodart B, Savocchia S (2020) Diversity profiling of grapevine microbial endosphere and antagonistic potential of endophytic pseudomonas against grapevine trunk diseases. Front Microbiol 11:477. https://doi.org/10.3389/fmicb.2020.00477
Nygren K, Dubey M, Zapparata A, Iqbal M, Tzelepis GD, Durling MB, Jensen DF, Karlsson M (2018) The mycoparasitic fungus Clonostachys rosea responds with both common and specific gene expression during interspecific interactions with fungal prey. Evolut Applic 11:931–949. https://doi.org/10.1111/eva.12609
Pagliarani C, Boccacci P, Chitarra W, Cosentino E, Sandri M, Perrone I, Mori A, Cuozzo D, Nerva L, Rossato M, Zuccolotto P, Pezzotti M, Delledonne M, Mannini F, Gribaudo I, Gambino G (2019) Distinct metabolic signals underlie clone by environment interplay in “Nebbiolo” grapes over ripening. Front Plant Sci 10:1575. https://doi.org/10.3389/fpls.2019.01575
Pagliarani C, Moine A, Chitarra W, Meloni GR, Abbà S, Nerva L, Pugliese M, Gullino ML, Gambino G (2020) The molecular priming of defense responses is differently regulated in grapevine genotypes following elicitor application against powdery mildew. Int J Mol Sci 21:6776. https://doi.org/10.3390/ijms21186776
Priori S, Pellegrini S, Perria R, Puccioni S, Storchi P, Valboa G, Costantini EAC (2019) Scale effect of terroir under three contrasting vintages in the Chianti Classico area (Tuscany, Italy). Geoderma 334:99–112. https://doi.org/10.1016/j.geoderma.2018.07.048
Qin X, Zeevaart JA (1999) The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Nat Ac Sci USA 96:15354–15361. https://doi.org/10.1073/pnas.96.26.15354
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner O (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Ac Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Ravnskov S, Jensen B, Knudsen IM, Bødker L, Jensen DF, Karliński L, Larsen J (2006) Soil inoculation with the biocontrol agent Clonostachys rosea and the mycorrhizal fungus Glomus intraradices results in mutual inhibition, plant growth promotion and alteration of soil microbial communities. Soil Biol Bioch 38:3453–3462. https://doi.org/10.1016/j.soilbio.2006.06.003
Rivers AR, Weber KC, Gardner TG, Liu S, Armstrong SD (2018) ITSxpress: software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Research 7:1418. https://doi.org/10.12688/f1000research.15704.1
Russi A, Almança MAK, Grohs DS, Schwambach J (2020) Biocontrol of black foot disease on grapevine rootstocks using Bacillus subtilis strain F62. Trop Plant Pathol 45:103–111. https://doi.org/10.1007/s40858-019-00319-7
Saad MM, Eida AA, Hirt H (2020) Tailoring plant-associated microbial inoculants in agriculture: a roadmap for successful application. J Exp Bot 71:3878–3901. https://doi.org/10.1093/jxb/eraa111
Sanmartín N, Pastor V, Pastor-Fernández J, Flors V, Pozo MJ, Sánchez-Bel P (2020) Role and mechanisms of callose priming in mycorrhiza-induced resistance. J Exp Bot 71:2769–2781. https://doi.org/10.1093/jxb/eraa030
Santi S, De Marco F, Polizzotto R, Grisan S, Musetti R (2013) Recovery from stolbur disease in grapevine involves changes in sugar transport and metabolism. Front Plant Sci 4:171. https://doi.org/10.3389/fpls.2013.00171
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. https://doi.org/10.1093/bioinformatics/btr026
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Nat Ac Sci USA 109:6241-6246https://doi.org/10.1073/pnas.1117018109
Subrahmanyam G, Kumar A, Sandilya SP, Chutia M, Yadav AN (2020) Diversity, plant growth promoting attributes, and agricultural applications of rhizospheric microbes In Yadav A., Singh J., Rastegari A., Yadav N. (Eds) Plant Microbiomes for Sustainable Agriculture. Sustainable Development and Biodiversity, vol 25. Springer, Cham, pp 1–52. https://doi.org/10.1007/978-3-030-38453-1_1
Sun L, Zhang M, Ren J, Qi J, Zhang G, Leng P (2010) Reciprocity between abscisic acid and ethylene at the onset of berry ripening and after harvest. BMC Plant Biol 10:257. https://doi.org/10.1186/1471-2229-10-257
Sun Z-B, Li S-D, Ren Q, Xu JL, Lu X, Sun MH (2020) Biology and applications of Clonostachys rosea. J App Microbiol 129:486–495. https://doi.org/10.1111/jam.14625
Suzuki K, Takahashi K, Harada N (2020) Evaluation of primer pairs for studying arbuscular mycorrhizal fungal community compositions using a MiSeq platform. Biol Fertil Soils 56:853–858. https://doi.org/10.1007/s00374-020-01431-6
Svenningsen NB, Watts-Williams SJ, Joner EJ, Battini F, Efthymiou A, Cruz-Paredes C, Nybroe O, Jakobsen I (2018) Suppression of the activity of arbuscular mycorrhizal fungi by the soil microbiota. ISME J 12:1296–1307. https://doi.org/10.1038/s41396-018-0059-3
Tomasi N, Monte R, Varanini Z, Cesco S, Pinton R (2015) Induction of nitrate uptake in Sauvignon Blanc and Chardonnay grapevines depends on the scion and is affected by the rootstock. Austral J Grape Wine Res 21:331–338. https://doi.org/10.1111/ajgw.12137
Ton J, Flors V, Mauch-Mani B (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci 14:310–317. https://doi.org/10.1016/j.tplants.2009.03.006
Trouvelot S, Bonneau L, Redecker D, van Tuinen D, Adrian M, Wipf D (2015) Arbuscular mycorrhiza symbiosis in viticulture: a review. Agronomy Sust Dev 35:1449–1467. https://doi.org/10.1007/s13593-015-0329-7
Uroz S, Courty PE, Oger P (2019) Plant symbionts are engineers of the plant-associated microbiome. Trends Plant Sci 24:905–916. https://doi.org/10.1016/j.tplants.2019.06.008
Verbon EH, Liberman LM (2016) Beneficial microbes affect endogenous mechanisms controlling root development. Trends Plant Sci 21:218–229. https://doi.org/10.1016/j.tplants.2016.01.013
Warschefsky EJ, Klein LL, Frank MH, Chitwood DH, Londo JP, von Wettberg EJB, Miller AJ (2016) Rootstocks: diversity, domestication, and impacts on shoot phenotypes. Trends Plant Sci 21:418–437. https://doi.org/10.1016/j.tplants.2015.11.008
Xue C, Hao Y, Pu X, Penton CR, Wang Q, Zhao M, Zhang B, Ran W, Huang Q, Shen Q, Tiedje JM (2019a) Effect of LSU and ITS genetic markers and reference databases on analyses of fungal communities. Biol Fertil Soils 55:79–88. https://doi.org/10.1007/s00374-018-1331-4
Xue L, Almario J, Fabiańska I, Saridis G, Bucher M (2019b) Dysfunction in the arbuscular mycorrhizal symbiosis has consistent but small effects on the establishment of the fungal microbiota in Lotus japonicus. New Phytol 224:409–420. https://doi.org/10.1111/nph.15958
Yu K, Pieterse CM, Bakker PA, Berendsen RL (2019) Beneficial microbes going underground of root immunity. Plant Cell Environ 42:2860–2870. https://doi.org/10.1111/pce.13632
Zhang X, Zhou Y, Li Y, Fu X, Wang Q (2017) Screening and characterization of endophytic Bacillus for biocontrol of grapevine downy mildew. Crop Protec 96:173–179. https://doi.org/10.1016/j.cropro.2017.02.018
Zombardo A, Mica E, Puccioni S, Perria R, Valentini P, Mattii GB, Cattivelli L, Storchi P (2020) Berry quality of grapevine under water stress as affected by rootstock–Scion interactions through gene expression regulation. Agronomy 10:680. https://doi.org/10.3390/agronomy10050680
Züst T, Agrawal AA (2017) Trade-offs between plant growth and defense against insect herbivory: an emerging mechanistic synthesis. Ann Rev Plant Biol 68:513–534. https://doi.org/10.1146/annurev-arplant-042916-040856
Acknowledgements
Louis Mercy for the preparation of the inoculum and the monosaccharide (Inducer) used in this study. Figure 7 was created with BioRender.com.
Funding
The authors thanks RAVIT project funded by Villa Sandi S.p.A.; BIOPRIME project funded by Mipaaf; Italy and CNR project Green and Circular Economy, FOE-2019 DBA.AD003.139; REVINE-PRIMA project funded by the European Commission, Italian MUR DM n. 1966/2021 Project ID 20114-2.
Author information
Authors and Affiliations
Contributions
WC, RB and LN designed the experimental system. LN, GQ, GG, LM, NB, LL, RP, MGV, MS, FG, RB and WC conducted the wet lab experiments and performed data elaboration. LN, GQ, RB and WC performed RT-qPCR analyses. LN, GG and WC performed the microbiome data analysis of root endophytes. LN, RB and WC wrote the first draft of the manuscript. All the authors carefully revised the final version.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information

Fig. S1
Gene expression changes of defence-related target genes in both leaf and root. a VvSTP13 in leaf. b VvSTP13 in root. c VvChitIII in leaf. d VvChitIII in root. e VvCAS2 in leaf. f VvLOX in leaf. g VvEDS1 in leaf. h VvHNT1 in leaf. All data are expressed as mean ± SD (n = 3). ns, *, ** and ***: non-significant or significant at P ≤ 0.05, P ≤ 0.01 and P ≤ 0.001, respectively. Different lowercase letters above the bars indicate significant differences according to Tukey HSD test (P ≤ 0.05), considering R × I × M interaction. Analysis of variance on the single variables is reported in Table S2. Different uppercase letters above the bars indicate significant differences according to Tukey HSD test (P ≤ 0.05) considering the two rootstocks independently. C, control plants; I, inducer-treated plants; M, AMF mixed inoculum-treated plants; M+I AMF mixed inoculum + inducer-treated plants for 1103P and SO4 selected rootstocks (PNG 13337 kb)

Fig. S2
NMDS of root-associated fungal communities. NMDS algorithm based on Bray-Curtis distances matrixes was used to reduce into a bi-dimensional scaling data obtained for and fungi community (n=3) (PNG 7484 kb)

Fig. S3
Distinct root-associated fungal community structure among treatments. Relative abundances of bacterial class (a) and genera (b) among treatments. Only genera representing at least the 1% over the total number of classified amplicons were retained (n = 3). C, control plants; I, inducer-treated plants; M, AMF mixed inoculum-treated plants; M+I AMF mixed inoculum + inducer-treated plants for 1103P and SO4 selected rootstocks (PNG 14150 kb)

Fig. S4
Relative abundances of fungal pathogens genera. C, control plants; I, inducer-treated plants; M, AMF mixed inoculum-treated plants; M+I AMF mixed inoculum + inducer-treated plants for 1103P and SO4 selected rootstocks (n = 3) (PNG 5432 kb)
Table S1
Oligonucleotides used in this study (DOC 73 kb)
Table S2
Analysis of variance (ANOVA) outcomes of target genes, metabolites and nitrogen content in leaf and root tissues. Different letters within each column indicate significant differences according to Tukey HSD test (P ≤ 0.05). Rootstock (R), inducer (I) and Myc (M) main effects were compared using the Student’s t test (P ≤ 0.05) (XLSX 55 kb)
Table S3
General feature from sequencing results of MiSeq Illumina using specific 16S or ITS primers together with PNA (DOCX 31 kb)
Table S4
Shannon index for bacterial (16S) communities sampled among the different treatments (DOCX 22 kb)
Table S5
Shannon index for fungal (ITS) communities sampled among the different treatments (DOCX 22 kb)
Table S6
Summary of bacterial and fungal community composition among treatments (XLSX 375 kb)
Table S7
Statistical analysis of the bacterial community among the different treatments (XLSX 99 kb)
Table S8
Statistical analysis of the fungal community among the different treatments (XLSX 82 kb)
Rights and permissions
About this article
Cite this article
Nerva, L., Giudice, G., Quiroga, G. et al. Mycorrhizal symbiosis balances rootstock-mediated growth-defence tradeoffs. Biol Fertil Soils 58, 17–34 (2022). https://doi.org/10.1007/s00374-021-01607-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-021-01607-8