Abstract
Soil microorganisms with phytase- and 1-aminocyclopropane-1-carboxylate (ACC) deaminase activities are widely studied as plant growth-promoting rhizobacteria (PGPR). Here, we explored the bacterial community structure and occurrence of putative PGPR in plants grown in agro-ecosystems and undisturbed ecosystems from northern, central, and southern Chile. Total rhizobacterial community structure was evaluated by denaturing gradient gel electrophoresis, and dominant bands present in diverse ecosystems were sequenced. Significant differences in total bacterial communities were shown with some bacterial orders (Enterobacteriales, Actinomycetales, and Rhizobiales) being highly similar to both ecosystems. Twenty-nine putative PGPR, showing phytate- and ACC-degrading activities and production of auxin, were selected from across the sites. Based on 16S rRNA gene sequencing, the putative PGPR were characterized as Enterobacteriales (Enterobacter, Serratia, Pantoea, Rahnella, Leclercia), Pseudomonas, and Bacillus, consistent with previously reported PGPR and endophytic bacteria. Beta-propeller phytase genes with similarity to Bacillus were also identified. PGPR from agro-ecosystems appeared to show higher auxin production compared to those from undisturbed ecosystems. This study demonstrates that putative PGPR are widely distributed across Chilean soils. Further understanding of their contribution to the growth and adaptation of plant hosts to local soil conditions may provide opportunity for development of new PGPR in Chilean agriculture.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Global agriculture production is highly dependent on phosphate fertilizers because phosphorus (P) is an essential nutrient for plants and many soils are deficient in P. World consumption of P-based fertilizers has increased from 5 to 30 Mt in recent decades and is estimated to reach between 60 and 70 Mt by 2050 (Steen 1998). However, the production of phosphate fertilizers relies on the availability of phosphate rock, which is a non-renewable resource. According to various estimates, the world reserves of high-quality rock phosphate will deplete within in the 300–400 years (Van Kauwenbergh 2010). In addition to rock phosphate dependence, global agriculture also faces major challenge due to climate change, whereby crop production systems (e.g., wheat) are expected to be subject to increased climate variability, temperature extremes, and incidence of drought (Atkinson and Urwin 2012; Grover et al. 2011). Development of soil microbial and plant-based agronomic strategies to alleviate these threats is therefore a significant priority for more sustainable agriculture (Richardson and Simpson 2011).
Plants live in close association with an enormous diversity and abundance of soil microorganisms that interact with roots and in many cases can promote the growth and fitness of plants (Berendsen et al. 2012). As such, plant growth-promoting rhizobacteria (PGPR) have been proposed as soil inoculants to improve P nutrition and environmental stress tolerance (Bulgarelli et al. 2013; Vacheron et al. 2013). Phosphate-solubilizing and phosphate-mineralizing rhizobacteria have the potential to improve P uptake by plants by increasing the availability of otherwise poorly available forms of P in soil (Bhattacharyya and Jha 2012; Martínez-Viveros et al. 2010). In many soils, including acidic volcanic soils as found widely throughout Chile, phosphates accumulate in organic forms such as inositol phosphates (i.e., phytate or myo-inositol hexakisphosphate) (Mullen 2005; Turner et al. 2002), which must be mineralized by phosphatases (phytases) to release available P for plant nutrition. Phytase-producing rhizobacteria are thus attractive as soil inoculants to increase the availability of P to plants (Richardson 2001). The production of the enzyme 1-aminocyclopropane-1-carboxylate (ACC) deaminase by rhizobacteria is also a desirable PGPR trait (Kang et al. 2010; Nadeem et al. 2010). ACC deaminase produced by PGPR plays an important role in suppressing ethylene production and mediating plant growth and root development in response to environmental stresses, such as drought, salinity, and flooding (Bhattacharyya and Jha 2012; Glick 2004; Martínez-Viveros et al. 2010). However, much of our knowledge on rhizobacterial taxonomic biogeography and functional ecology of PGPR has come from studies on agro-ecosystems (i.e., pasture, crop, and forest tree species), with relatively little known about the occurrence, diversity, and activity of PGPR in soil under natural vegetation.
Microbial communities under natural vegetation may specifically be adapted to their plant host and the local soil conditions. For example, we recently investigated rhizobacterial communities associated with ancient clones of Creosote bush (Larrea tridentata) in the Mohave Desert (California, USA), and showed that the culturable rhizobacterial included PGPR with genes specifically encoding phytase and ACC deaminase activity (Jorquera et al. 2012). Undisturbed ecosystems may therefore harbor wide diversity of bacteria that contribute to the adaptation of plants in nutrient limited and stressful environments and thus be an important source of novel PGPR (Timmusk et al. 2011). Thus, we hypothesized that PGPR are common inhabitants in the rhizosphere of plants grown in Chilean undisturbed ecosystems. In this study, we used both culture-dependent and culture-independent methods to examine bacterial community structure and occurrence of putative PGPR in rhizobacterial communities associated with plants grown in a range of Chilean ecosystems. This included agro-ecosystems from central Chile, which supports crop and livestock production, and a range of undisturbed ecosystem representing diverse environments from northern, central, and southern Chile.
Material and methods
Agro-ecosystems and ecosystems sampling
Rhizosphere samples (including roots and adhering soil) were collected from plants at each site using a cleaned spade to excavate intact roots from soil to a depth of 0–20 cm. Samples were placed into sterile plastic 50 ml Falcon tubes and immediately transported to the laboratory on ice for soil and microbiological analyses. Rhizosphere soil was removed from roots by shaking, homogenously mixed and representative samples from three plants in each ecosystems were processed.
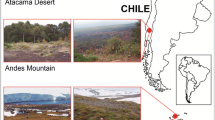
Rhizosphere samples were collected from diverse ecosystems and agro-ecosystems across Chile (Fig. 1), including three samples each from shrubs (Atriplex sp.) grown in the Atacama Desert (AD) (Region of Antofagasta; 22°S, 68°W), perennial weeds (Senecio sp. and Cortaderia sp.) from Conguillio National Park (NP) in the Andes mountains (Region of La Araucanía; 38°S, 71°W) and shrubs (Chuquiraga sp.) from the steppe-like plains in Patagonia (PT) (Region of Magallanes; 53°S; 70°W). Samples from agricultural systems were collected from Region of La Araucanía (38°S; 71°W to72°W) from various sites that included a 10-year-old naturalized pasture (PS1) containing a mixture of perennial ryegrass (Lollium perenne L.), white clover (Trifolium repens L.), and dandelion (Taraxacum officinale); pasture of perennial ryegrass (L. perenne L.) under cow grazing (PS2), from a 2-month oat (Avena sativa L.) crop (CC1) and from a crop soil containing wheat (Triticum aestivum L.) stubble (CC2).
Chemical properties of rhizosphere soils
Soil pH was measured in 1:2.5 soil/deionized water suspensions. Inorganic P was extracted using 0.5 M Na bicarbonate and analyzed using the molybdate-blue method (Murphy and Riley 1962). Exchangeable cations (K, Ca, Mg, and Na) were extracted with 1 M CH3COONH4 at pH 7.0 and analyzed by flame atomic adsorption spectrophotometry (FAAS) (Warncke and Brown 1998). Exchangeable aluminum was extracted with 1 M KCl and analyzed by FAAS (Bertsch and Bloom 1996).
Bacterial community composition in the rhizosphere
DNA was extracted from 0.5 g of soil maintained at −80 °C using a soil DNA Isolation Kit (Ultraclean, Mo-Bio Laboratories) according to manufacturer’s instructions. The whole bacterial community composition in the rhizosphere samples was examined by denaturing gradient gel electrophoresis (DGGE) of partial 16S rDNA genes according to the procedure described by Jorquera et al. (2010). PCR reactions were done with DNA template concentrations of ~20 ng μl−1. Fragments of 16S rRNA genes (regions V6–V8) were amplified by hot start touchdown PCR with primers EUB f933-GC/EUB r1387 (Table 1) using the following conditions: 95 °C for 10 min, annealing at 65 °C followed by 20 cycles for 1 min each with a 0.5 °C decrease each cycle to 55 °C and extension at 72 °C for 3 min. Ten additional cycles were then carried out at 55 °C annealing and 94 °C denaturation for 1 min each and primer extension at 72 °C for 3 min, and a final extension step of 7 min at 72 °C. DGGE analysis was performed using a DCode system (Bio-Rad Laboratories, Inc.). Briefly, 30 μl of PCR products were loaded onto 6 % (w/v) polyacrylamide gel with a 40–70 % denaturing gradient (7 M urea and 40 % formamide) and electrophoresis was run for 12 h at 100 V. The gel was stained with SYBR Gold (Invitrogen, Co.) for 30 min and photographed on a UV transilluminator. Similarities in bacterial community composition were determined following visualization and quantification of dominant bands using Phoretix 1D analysis software (TotalLab Ltd.) and analysis by non-metric multidimensional scaling (nMDS) using PAST freeware (http://folk.uio.no/ohammer/past/) as described by Jorquera et al. (2013).
In addition, dominant bands present in agro-ecosystems and ecosystems, with similar migration in DGGE gels, were chosen for sequencing to determinate common and specific bacterial groups in both ecosystems. Dominant bands in the DGGE gels were excised, purified and reamplified, and run again in DGGE gels to avoid sequencing of multiple amplicons due to close proximity. PCR products were then sequenced by (Macrogen, Inc., Korea). The sequences obtained were compared with those present in the GenBank database from the National Center for Biotechnology Information (NCBI) using megablast tool (http://blast.ncbi.nlm.nih.gov/) for taxonomic assignment.
Isolation of phytate- and ACC-degrading rhizobacteria
One gram of each rhizosphere sample (in triplicate) was suspended in 25 ml of sterile saline solution (8.5 g l−1 of NaCl) and sonicated for 30 s at 130 W (20 kHz). Appropriate serial dilutions were then spread onto both NM-1 agar medium (0.5 g l−1 D-glucose, 0.5 g l−1 polypeptone, 0.5 g l−1 Na-glutamate, 0.5 g l−1 yeast extract, 0.44 g l−1 KH2PO4, 0.1 g l−1 (NH4)2SO4, 0.1 g l−1 MgSO4 · 7H2O, 15 g l−1 agar, and 1 ml vitamin solution containing 1 g l−1 nicotinamide, 1 g l−1 tiamine hydrochloride, 0.05 g l−1 biotin, 0.5 g l−1 4-aminobenzoic acid, 0.01 g l−1 vitamin B12, 0.5 g l−1 D-pantothenic acid hemicalcium salt, 0.5 g l−1 pyridoxamine dihydrochloride, 0.5 g l−1 folic acid; Nakamura et al. 1995), and soil extract (SE) agar medium prepared with 500 ml l−1 of soil extract (1 kg soil from the Region of La Araucanía mixed with 2 l of 50 mM NaOH) according to Hamaki et al. (2005). Both media contained 15 g l−1 agar and were supplemented with 100 μg ml−1 of cycloheximide (Sigma-Aldrich) to prevent fungal growth. The number of total heterotrophic culturable rhizobacteria was estimated at each dilution after incubation for 4–6 days at 30 °C. Approximately 760 representative colony forming units (CFU) were chosen randomly and transferred to screening media for assessment of phytate and ACC-degrading activity.
Isolates were examined for growth in 5 ml tubes and on agar plates of phytase-screening medium (PSM; Kerovuo et al. 1998) and ACC-degrading medium (DF; Penrose and Glick 2003) supplemented with Na-phytate (C6H18O24 P6 · xNa · yH2O; Sigma-Aldrich, Co.) and ACC (C4H7NO2; Calbiochem®) as source of P and N, respectively. PSM contained the following: 10 g l−1 D-glucose, 4 g l−1 Na-phytate, 2 g l−1 CaCl2, 5 g l−1 NH4NO3, 0.5 g l−1 KCl, 0.5 g l−1 MgSO4 · 7H2O, 0.01 g l−1 FeSO4 · 7H2O, 0.01 g l−1 MnSO4 · H2O. DF medium contained the following: 4 g l−1 KH2PO4, 6 g l−1 Na2HPO4, 0.2 g l−1 MgSO4 · 7H2O, 2 g l−1 gluconic acid, 2 g l−1 citric acid, 3 mM ACC, and 1 ml trace elements containing 0.001 g l−1 FeSO4 · 7H2O, 0.01 g l−1 H3BO3, 0.011 g l−1 MnSO4 · H2O, 0.125 g l−1 ZnSO4 · 7H2O, 0.078 g l−1 CuSO4 · 5H2O, 0.01 g l−1 MoO3.
Detection of phytase and ACC deaminase genes
Approximately 200 isolates showing growth in both PSM and DF media were screened by colony PCR for putative genes encoding β-propeller phytase (BPP) and ACC deaminase (acdS) using primers for diverse bacterial target groups as shown in Table 1. The PCR reactions were first carried out according to protocols described by authors (Table 1) or were modified to obtain single amplicons. PCR products were amplified using GoTaq® Flexi DNA polymerase (Promega, Inc.) or SapphireAmp Fast PCR Master Mix (Takara Bio, Inc.). The presence of amplicons in PCR-positive isolates was confirmed by a second-round PCR using purified templates (Gentra Puregene Kit; Qiagen Inc.). Where available, amplicons were sequenced (Macrogen, Inc., Korea) and translated to protein sequences and compared with those in the GenBank using the blastx tool.
Characterization of putative PGPR
A total of 29 putative PGPR stains that showed growth in both PSM and DF media and were PCR positive for both target genes were selected and identified to the genus level by partial sequencing of their 16S rRNA genes. PCR was conducted with the primers 27f /1492r (Table 1) hot start reaction at 94 °C for 5 min and 35 cycles at 94 °C for 1 min, 52 °C for 1 min, and 72 °C for 2 min. PCR products were purified and sequenced (as above), and compared with those in the GenBank using the megablast tool.
The production of auxin of putative PGPR was determined by colorimetric assay at 530 nm using Salkowski’s reagent as described by Patten and Glick (2002). Bacterial cells were grown with shaking (120 rpm) for 2 days at 30 °C in LB broth supplemented with tryptophan (1 mg ml−1) as auxin precursor. Cells were then centrifuged (1,500 × g for 10 min at 4 °C) and 1 ml of supernatant was combined with 2 ml of Salkowski’s reagent (150 ml of 98 % H2SO4, 7.5 ml of 0.5 M FeCl3 · 6H2O, and 250 ml distilled water) and incubated for 30 min at room temperature. Quantification of auxin was determined relative to pure indole acetic acid (Sigma-Aldrich, Co.).
Nucleotide sequence accession number
The nucleotide sequences (>200 bp) obtained in this study were deposited in the GenBank database under accession numbers from KJ545705 to KJ545753.
Results
Soil chemical properties
Analysis of the ecosystem soils is shown in Table 2. Values of N ranged from 26 to 30 mg kg−1 in agro-ecosystems and from 12 to 18 mg kg−1 in ecosystems. Extractable P and K varied across the sites and were lowest in the Conguillio National Park (NP) soil. As we expected, higher values of organic matter were observed in agro-ecosystems (14–20 %) compared with the undisturbed ecosystems (1–8 %). Soil pH was acidic for rhizosphere soils taken from agro-ecosystems (pH 5.4 to 5.6) and varied from acidic to alkaline (pH 5.8 to 8.9) in the natural ecosystem. Higher levels of Al-base saturation were observed in the NP and CC2 (9.4 and 10.7 %, respectively) compared to negligible levels in the other soils.
Bacterial community composition in the rhizosphere
Community profiles generated from DGGE analysis of 16S rRNA genes of the indigenous rhizobacteria revealed a level of similarity across the ecosystem soils, but with significant differences in presence of dominant bands (Fig. 2a). The nMDS analyses showed mainly three clusters (Fig. 2b), with clear separation of rhizobacterial communities from the Region of La Araucanía (PS2, CC1, CC2, and NP) compared to the Atacama Desert (AD) and Patagonia (PT) regions. An exception to this was the PS1 soil from undisturbed National Park soil from Region of La Araucanía, which grouped more similarly with the Atacama Desert soil.
Rhizobacterial communities associated with plants grown in agro-ecosystems (blue) and undisturbed ecosystems (red). a Community profiles obtained by denaturing gradient gel electrophoresis (DGGE) of bacterial 16 rRNA genes. Arrowheads and numbers indicate major bands that were excised for DNA sequence analysis. b Non-metric multidimensional scaling (nMDS) analysis of DGGE profiles generated by PAST freeware (http://folk.uio.no/ohammer/past/) with the Bray–Curtis method (Somerfield 2008) using stress value of <0.1 and P value of 0.05
DNA sequence analysis of dominant bands discriminated by DGGE indicated a strong presence of Enterobacteriales in all of the rhizosphere soils analyzed (bands no. 5, 11, 13, 14, 15, 20, 21, 22, 25, 31, 33, 34, and 36; Fig. 2a), with exception of the Patagonia soil (Table 3). Citrobacter freundii was identified (100 % sequence similarity) as being present in at least three of the soils from across diverse ecosystems. Sphingobacteriales were similarly observed in samples from the Region of La Araucanía (bands no. 10, 19, and 23) and particularly in the soil from oat crops (CC1; bands no. 1, 2, 3, 6, 7, and 8). Actinomycetales and Rhizobiales were detected in both agro-ecosystems (bands 4, 9, 17, and 18) and undisturbed ecosystems (bands 24, 27, 28, 30, and 32) and Pseudomonales in the PS1 and AD soils (bands no. 16 and 35, respectively).
Isolation of putative PGPR
Total numbers of culturable heterotrophic rhizobacteria on NM-1 medium were greater in rhizosphere soils taken from plants grown in the Region of La Araucanía (1 to 10 × 106 CFU g−1 soil) compared with soil from the AD and PT (1 to 5 × 105 CFU g−1 soil) regions (Fig. 3a). Numbers of total bacteria were similarly observed on soil extract agar for the Region of La Araucanía soil (1–4 × 106 CFU g−1 soil), but interestingly bacterial growth was not observed on this medium (which was prepared with soil from La Araucanía region) for either of the Atacama Desert or Patagonia rhizosphere soils. It is also noteworthy that greater diversity in bacterial phenotypes were visualized on NM-1 agar medium in samples from agro-ecosystems compared with soils from undisturbed ecosystems (e.g., compare panels b and c of Fig. 3).
a Counts of total heterotrophic rhizobacteria on NM-1 (light bars) and soil extract agar (solid bars) media for undisturbed ecosystems, Atacama Desert (AD), Patagonia (PT), and Conguillio National Park (NP) and agro-ecosystems cereal crops (CC) and pastures (PS). Bars represent standard error and same letter (lowercase for NM-1 or uppercase for soil extract agar) denotes no statistical difference (P ≤ 0.05, Duncan multiple range test). Example of isolates cultured from rhizosphere soils collected from cereal crops (b) and the Atacama Desert (c)
Isolates showing ability to degrade both phytate and ACC in culture media represented 26 to 30 % of the total number of colony forming units obtained and were observed in rhizosphere soils from across all seven ecosystems with no clear differences according to land use or location. Screening of these isolates by PCR indicated the presence of phytase and ACC deaminase genes by PCR, in many of the isolates with between 2 and 29 % and 9 and 32 % of isolates from PSM media being positive for putative BPP and acdS genes, respectively. Likewise for isolates with ACC-degrading activity, between 0 and 26 % and 6 and 29 % of those screened were positives for BPP and acdS genes. However, sequencing of the PCR amplicons from the majority of these isolates did not show reliable similarity with target genes, with exception of three putative phytase genes generated with primers BPPf/BPPr (Table 2) from cereal crops (CC 3.6 and CC3.18) and Atacama Desert (AD7.21) soils (Fig. 4). These clones showed 94 to 97 % similarity with the 3-phytase gene of Bacillus subtilis subsp. subtilis (accession no. CP004405) and ~74 % with PhyL of Bacillus licheniformis (accession no. AY651979), respectively.
Phylogenetic tree showing affiliation of selected phytase genes found in this study (AD7.21, CC3.6, and CC3.18) in relation to phytase genes previously found in Chilean agro-ecosystems by Jorquera et al. (2011) (MQH-15 and SPT-03) and representative phytase gene sequences taken from GenBank. The neighbor-joining tree was constructed using MEGA6 with the bar indicating 10 % sequence divergence
Characterization of putative PGPR
Phylogenetic characterization of 29 putative PGPR isolates that showed growth in both PSM and DF media identified species that belonged to Enterobacter, Rahnella, Pseudomonas, Bacillus, Serratia, and Pantoea genera (Table 4). Blast analysis of the 16S gene sequences also showed isolates (CC3.12, CC3.13, CC3.16, and CC3.6) from agro-systems with high similarities (99–100 %) with previously indentified phytate-degrading Enterobacter and plant growth-promoting Enterobacter and Rahnella. In undisturbed ecosystems, the occurrence of species with high similarity (93–100 %) to plant growth-promoting Serratia, Bacillus, Pantoea, Enterobacter, and Leclercia genera (isolates AD7.5, AD7.15, AD7.20, AD7.38, PT10.2, PT10.6, and PT2.8) were likewise identified (Table 4).
All of the selected putative PGPR were able to produce auxin in culture media with higher levels generally being observed in isolates from agro-ecosystems (mean of 45.6 μg ml−1) compared with undisturbed natural ecosystems (mean of 17.6 μg ml−1) (Fig. 5).
Discussion
Bacterial community structure in the rhizosphere of plants and presence of putative PGPR based on phytase-degrading and ACC-degrading bacteria were investigated in a range of undisturbed ecosystems and agro-ecosystems across Chile. Sequencing of dominant DGGE bands from 16 rRNA amplicons revealed the occurrence of members belonging to Enterobacteriales, Sphingobacteriales, Actinomycetales, Rhizobiales, and Pseudomonales orders, in both ecosystems, despite contrasting geographical location and diverse climatic environments. These bacterial orders are commonly found in soil (Pastorelli et al. 2013; Ramond et al. 2013) and have been described in the rhizosphere of various shrubs, grasses, and forests (Bardhan et al. 2012; Jorquera et al. 2012; Saul-Tcherkas et al. 2013). Their presence in Chilean soils reported here is also consistent with previous studies (Briceño et al. 2010; Cea et al. 2010; Jorquera et al. 2010). However, they differ from those taxa with high (Gemmatimonadetes, Planctomycetes, Actinobacteria, and Chloroflexi) and low (Acidobacteria and Proteobacteria) abundances reported for bulk soil of Atacama Desert by Drees et al. (2006) and Neilson et al. (2012), evidencing a possible plant-specific selection and stimulation of bacterial groups in the rhizosphere of plants grown in dry and salty ecosystems. In this context, the importance of root exudates (such as flavonoids and vitamins) on selection and activity of soil heterotrophic bacteria in the rhizosphere is widely known (Berendsen et al. 2012; Bulgarelli et al. 2013; Cesco et al. 2012; Palacios et al. 2014). Our results suggest however that some bacterial orders (such as Enterobacteriales, Actinomycetales, and Rhizobiales) may be more dominant across ecosystems whereas other (such as Sphingobacteriales and Pseudomonales) appear to be restricted more to specific regions or rhizosphere types. But, this result should be interpreted with caution because the absence of dominant groups could be due to the incomplete and biased DNA extraction from soils, representing a fraction of the whole community present in rhizosphere samples (Bakken and Frostegård 2006; Lombard et al. 2011; van Elsas and Boersma 2011). Further studies to assess the influence of biogeography and climate on bacteria in Chilean soils are therefore warranted.
The nMDS analysis indicated a general difference in rhizobacterial community composition from plants grown in the Region of La Araucanía (NP, PS2, CC1, and CC2) compared to those from more extreme environments in the Atacama Desert or Patagonia. This result is also supported by differences in total counts of heterotrophic bacteria in these environments and it is intriguing as to why communities from the Atacama Desert and Patagonia were not culturable on soil extract agar prepared from soil obtained from the Region of Araucanía in central Chile. The abundance, composition, and activity of rhizobacterial populations may be affected by a wide variety of factors including climate, plant-specific selection, and/or soil properties, such as nutrient available (N, P, and K), organic matter content, pH, and Al (Marschner et al. 2001; Mendes et al. 2014). In particular, soil carbon availability and soil pH have been shown to be major factors in influencing the composition and activity of bacterial communities (Bausenwein et al. 2008; Fierer and Jackson 2006) and are thus considered to be fundamental drivers of bacterial biogeography (Bissett et al. 2010). In the present study, the closeness in rhizobacterial community composition between 10-year naturalized pasture (PS1) and the Atacama Desert is noteworthy, despite the extreme difference in climate and geographical location (~1,800 km distance) of these two environments. Based on data obtained, this might be explained by (i) the occurrence of rhizobacterial groups, which were not highly dominant in other samples as revealed by DGGE (such as the Pseudomonales), and (ii) resilience capacity in PS1, but further analysis to verify this is required.
It is widely assumed that management of PGPR in agro-ecosystems provides an attractive proposition for improving the P nutrition and stress tolerance in agricultural plants. Numerous studies have described the beneficial effect of PGPR through traits such as phosphate-solubilizing and phytate-mineralizing activities, ACC deaminase and auxin production (Bhattacharyya and Jha 2012; Glick 2004; Kang et al. 2010; Martínez-Viveros et al. 2010; Nadeem et al. 2010; Richardson and Simpson 2011). However, their abilities are commonly tested in vitro and does not guarantee their beneficial effect in plants under field conditions. For example, media supplemented with insoluble tricalcium phosphates are commonly used to select efficient phosphate-solubilizing rhizobacteria, but when they are applied for improving of P nutrition in plants, only few isolates appear to be true P-solubilizers in soils (Bashan et al. 2013a, b). In Chilean agro-ecosystems, the occurrence of rhizobacteria having such PGPR traits has similarly been reported (Acuña et al. 2013; Jorquera et al. 2008; Martínez et al. 2011; Shoebitz et al. 2009). However, few studies have specifically examined natural and undisturbed ecosystems as a potential source of PGPR. Timmusk et al. (2011) reported that rhizosphere of wild barley (Hordeum spontaneum) grown in a “stressful” south-facing slope contains significantly higher populations of ACC deaminase-producing, P-solubilizing, and osmotic stress-tolerant bacteria compared with north-facing slope in northern Israel. Similarly, Mayak et al. (2004a, b) isolated ACC deaminase-producing rhizobacteria from dry salty environments and their inoculation increased the resistance of tomatoes and pepper plants seedlings to salt and water stress, hence alleviating the suppression of growth. The long-term selection and co-evolution of rhizobacterial communities with the roots of plants in co-adapted environments may provide a unique opportunity to obtain novel isolates of PGPR and to investigate how certain bacteria may assist plant growth and provide increased tolerance to abiotic stress. In this study, we isolated diverse culturable bacteria with identifiable PGPR traits from native plants grown in a range of undisturbed Chilean ecosystems, which included the Atacama Desert, the Andes mountains, and steppe-like plains of Patagonia. PGPR adapted to plant hosts in these local soil conditions may play a significant role in adaptation and growth of their respective plant hosts as previously demonstrated by Jorquera et al. (2012) with rhizobacteria isolated from shrubs grown in Mohave Desert (California), which were shown subsequently to also increase the growth of cucumber plants.
The identity of putative PGPR from the different ecosystems was investigated by 16S rRNA gene sequencing, which indicated the predominance of the Enterobacteriaceae family (Enterobacter, Rahnella, Serratia, Pantoea, and Leclercia) with high similarity to reported plant growth-promoting and antagonistic bacteria, including some endophytic bacteria; thus, we cannot discard the possibility that some of the isolated putative PGPR can act as endophytes in plants. While this occurrence of enterobacteria is a likely result of the selection protocol used, we do not disregard the higher abundance of other putative PGPR belonging to bacterial groups, such as Pseudomonas and Bacillus. Enterobacteriales, Pseudomonales, and Bacilli with PGPR traits are commonly reported for rhizosphere soils (Bulgarelli et al. 2013; Vacheron et al. 2013), including those from Chilean pastures and crops (Acuña et al. 2013; Jorquera et al. 2010; Martínez et al. 2011). Interestingly, higher auxin production was detected in putative PGPR from agro-ecosystems compared with undisturbed ecosystems. This result could be attributed to higher N available in soil of agro-ecosystems as shown in Table 1. In context, long-term N fertilization has previously been shown to affect both total rhizobacterial community composition and the population densities of auxin-producing rhizobacteria in pasture from the Region of La Araucanía (Jorquera et al. 2013; Martínez et al. 2011). These studies also demonstrated that organic acids, naturally found in the plant rhizosphere, can stimulate the auxin production by rhizosphere Bacilli strains isolated from Chilean pastures (Acuña et al. 2011). It can be postulated that rhizobacteria present in extreme environments may be better adapted to low nutrient availability resulting in differing auxin production. However, reasons why putative PGPR from undisturbed ecosystems produce lower amounts of auxin remain unclear.
The occurrence of rhizobacteria with phytase- and ACC-degrading activity in all rhizosphere soils analyzed is consistent with the widespread presence of PGPR across diverse environments. With respect to the detection of putative BPP genes by PCR, the amplicons sequencing only confirmed the presence of rhizobacteria having phytase genes with similarity to known Bacillus. Irrespective though, the phytases identified were distinct from those previously reported (Bacillus MQH-15, Bacillus MQH-19, and Paenibacillus SPT-03) in Chilean soils (Jorquera et al. 2011). Rhizosphere harboring phytase genes with similarity to Bacillus have also been reported in plants grown in undisturbed ecosystems (Jorquera et al. 2012). However, and despite extensive optimization of PCR reactions for the primer sets used here, sequence results showed numerous unspecific amplification with poor or no identity to known BPP genes. It is suggested that the current database of bacterial phytases is insufficient to provide adequate coverage of phytase genes across bacterial phyla (Lim et al. 2007). Based on our inability in the present study to also reliable amplify ACC deaminase genes using well-described primers, we suggest that a similar situation occurs for this gene. In previous studies, we have successfully detected the occurrence of genes encoding BPP and acdS in environmental isolates using the same primers and optimization of PCR reactions (Jorquera et al. 2011, 2012). The low detection of BPP genes and the inability to detect genes encoding ACC deaminase by PCR in the present study may thus be due to either the use of different bacterial selection protocols or that the sequence of BPP and ACC deaminase genes in rhizobacteria from the Chilean ecosystems we sampled were sufficiently different to those previously reported and available in public databases. Clearly, further studies are required to investigate the diversity and abundance of BPP and ACC deaminase genes in various ecosystems and their contribution to plant growth promotion.
Conclusions
Despite significant differences in rhizobial community composition across ecosystems and land-use systems of northern, central, and southern regions of Chile, we showed that the rhizosphere of plants collected from these diverse environments harbored common orders of rhizobacteria, such as Enterobacteriales. Moreover, the presence of bacterial isolates with phytate- and ACC-degrading activities was demonstrated in both agro-ecosystems and undisturbed ecosystems. Selected putative PGPR showing both phytate- and ACC-degrading activities and auxin production (higher in agro-ecosystems) were similarly characterized as belonging to the Enterobacteriales, with high similarity to previously described PGPR and other plant growth-promoting endophytic bacteria. This study demonstrates that bacteria having PGPR traits are widely distributed in the rhizosphere of plants from vastly different agro-ecosystem and climatic regions. We suggest that undisturbed ecosystems may therefore provide novel sources of PGPR for development as soil inoculants for overcoming P deficiency and other abiotic plant stresses.
References
Acuña J, Jorquera M, Martínez OA, Menezes-Blackburn D, Fernández MT, Marschner P, Greiner R, Mora ML (2011) Indole acetic acid and phytase activity produced by rhizosphere bacilli as affected by pH and metals. J Soil Sci Plant Nutr 11:1–12
Acuña JJ, Jorquera MA, Barra PJ, Crowley DE, Mora ML (2013) Selenobacteria selected from the rhizosphere as a potential tool for Se biofortification of wheat crops. Biol Fertil Soils 49:175–185
Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63:3523–3543
Bakken LR, Frostegård Å (2006) Nucleic acid extraction from soil. In: Smalla K, Nannipieri P (eds) Series on soil biology: nucleic acids and proteins in soil. Springer, Berlin, 50–73 pp
Bardhan S, Jose S, Jenkins MA, Webster CR, Udawatta RP, Stehn SE (2012) Microbial community diversity and composition across a gradient of soil acidity in spruce–fir forests of the southern Appalachian Mountains. Appl Soil Ecol 61:60–68
Bashan Y, Kamnev AA, de-Bashan LE (2013a) A proposal for isolating and testing phosphate-solubilizing bacteria that enhance plant growth. Biol Fertil Soils 49:1–2
Bashan Y, Kamnev AA, de-Bashan LE (2013b) Tricalcium phosphate is inappropriate as a universal selection factor for isolating and testing phosphate-solubilizing bacteria that enhance plant growth: a proposal for an alternative procedure. Biol Fertil Soils 49:465–479
Bausenwein U, Gattinger A, Langer U, Embacher A, Hartmann HP, Sommer M, Munch JC, Schloter M (2008) Exploring soil microbial communities and soil organic matter: variability and interactions in arable soils under minimum tillage practice. Appl Soil Ecol 40:67–77
Berendsen RL, Pieterse CMJ, Bakker PA (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486
Bertsch PM, Bloom PR (1996) Aluminum. In: Bigham JM (ed) Methods of soil analysis, part 3—chemical methods. Soil Science Society of America, Madison, 526-527 pp
Bhattacharyya PN, Jha DK (2012) Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol 28:1327–1350
Bissett A, Richardson AE, Baker G, Wakelin S, Thrall PH (2010) Life history determines biogeographical patterns of soil microbial communities over multiple spatial scales. Mol Ecol 19:4315–4327
Blaha D, Prigent-Combaret C, Mirza MS, Moënne-Loccoz Y (2006) Phylogeny of the 1-aminocyclopropane-1-carboxylic acid deaminase-encoding gene acdS in phytobeneficial and pathogenic Proteobacteria and relation with strain biogeography. FEMS Microbiol Ecol 56:455–470
Briceño G, Jorquera MA, Demanet R, Mora ML, Durán N, Palma G (2010) Effect of cow slurry amendment on atrazine dissipation and bacterial community structure in an agricultural Andisol. Sci Total Environ 408:2833–2839
Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838
Cea M, Jorquera M, Rubilar O, Langer H, Tortella G, Diez MC (2010) Bioremediation of soil contaminated with pentachlorophenol by Anthracophyllum discolor and its effect on soil microbial community. J Hazard Mater 181:315–323
Cesco S, Mimmo T, Tonon G, Tomasi R, Pinton R, Terzano R, Neumann G, Weisskopf L, Renella G, Landi L, Nannipieri P (2012) Plant-borne flavonoids released into the rhizosphere: impact on soil bio-activities related to plant nutrition. A review. Biol Fertil Soils 48:123–150
Drees KP, Neilson JW, Betancourt JL, Quade J, Henderson DA, Pryor BM, Maier RM (2006) Bacterial community structure in the hyperarid core of the Atacama Desert, Chile. Appl Environ Microbiol 72:7902–7908
Duan J, Müller KM, Charles TC, Vesely S, Glick BR (2009) 1-Aminocyclopropane-1-carboxylate (ACC) deaminase genes in rhizobia from southern Saskatchewan. Microb Ecol 57:423–436
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631
Glick BR (2004) Bacterial ACC deaminase and the alleviation of plant stress. Adv Appl Microbiol 56:291–312
Grover M, Ali SZ, Sandhya V, Rasul A, Venkateswarlu B (2011) Role of microorganisms in adaptation of agriculture crops to abiotic stresses. World J Microbiol Biotechnol 27:1231–1240
Hamaki T, Suzuki M, Fudou R, Jojima Y, Kajiura T, Tabuchi A, Sen K, Shibai H (2005) Isolation of novel bacteria and actinomycetes using soil-extract agar medium. J Biosci Bioeng 99:485–492
Huang H, Shi P, Wang Y, Luo H, Shao N, Wang G, Yang P, Yao B (2009) Diversity of beta-propeller phytase genes in the intestinal contents of grass carp provides insight into the release of major phosphorus from phytate in nature. Appl Environ Microbiol 75:1508–1516
Iwamoto T, Tani K, Nakamura K, Suzuki Y, Kitagawa M, Eguchi M, Nasu M (2000) Monitoring impact of in situ biostimulation treatment on groundwater bacterial community by DGGE. FEMS Microbiol Ecol 32:129–141
Jha CK, Annapurna K, Saraf M (2012) Isolation of rhizobacteria from Jatropha curcas and characterization of produced ACC deaminase. J Basic Microbiol 52:285–295
Jorquera MA, Hernández MT, Rengel Z, Marschner P, Mora ML (2008) Isolation of culturable phosphobacteria with both phytate-mineralization and phosphate-solubilization activity from the rhizosphere of plants grown in a volcanic soil. Biol Fertil Soils 44:1025–1034
Jorquera MA, Hernández M, Martínez O, Marschner P, Mora ML (2010) Detection of aluminium tolerance plasmids and microbial diversity in the rhizosphere of plants grown in acidic volcanic soil. Eur J Soil Biol 46:255–263
Jorquera MA, Crowley DE, Marschner P, Greiner R, Fernández MT, Romero D, Menezes-Blackburn D, Mora ML (2011) Identification of β-propeller phytase-encoding genes in culturable Paenibacillus and Bacillus spp. from the rhizosphere of pasture plants on volcanic soils. FEMS Microbiol Ecol 75:163–172
Jorquera MA, Shaharoona B, Nadeem SM, Mora ML, Crowley DE (2012) Plant growth-promoting rhizobacteria associated with ancient clones of creosote bush (Larrea tridentata). Microb Ecol 64:1008–1017
Jorquera MA, Martínez OA, Marileo LG, Acuña JJ, Saggar S, Mora ML (2013) Effect of nitrogen and phosphorus fertilization on the composition of rhizobacterial communities of two Chilean Andisol pastures. World J Microbiol Biotechnol 30:99–107
Kamala-Kannan S, Lee KJ, Park SM, Chae JC, Yun BS, Lee YH, Park YJ, Oh BT (2010) Characterization of ACC deaminase gene in Pseudomonas entomophila strain PS-PJH isolated from the rhizosphere soil. J Basic Microbiol 50:200–205
Kang BG, Kim WT, Yun HS, Chang SC (2010) Use of plant growth-promoting rhizobacteria to control stress responses of plant roots. Plant Biotechnol Rep 4:179–183
Kerovuo J, Lauraeus M, Nurminen P, Kalkkinen N, Apajalahti J (1998) Isolation, characterization, molecular gene cloning, and sequencing of a novel phytase from Bacillus subtilis. Appl Environ Microbiol 64:2079–2085
Lim BL, Yeung P, Cheng C, Hill JE (2007) Distribution and diversity of phytate-mineralizing bacteria. ISME J 1:321–330
Lombard N, Prestat E, van Elsas JD, Simonet P (2011) Soil-specific limitations for access and analysis of soil microbial communities by metagenomics. FEMS Microbiol Ecol 78:31–49
Marschner P, Yang CH, Lieberei R, Crowley DE (2001) Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem 33:1437–1445
Martínez OA, Jorquera MA, Crowley DE, Mora ML (2011) Influence of nitrogen fertilisation on pasture culturable rhizobacteria occurrence and the role of environmental factors on their potential PGPR activities. Biol Fertil Soils 47:875–885
Martínez-Viveros O, Jorquera MA, Crowley DE, Gajardo G, Mora ML (2010) Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J Soil Sci Plant Nutr 10:293–319
Mayak S, Tirosh T, Glick BR (2004a) Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol Biochem 42:565–572
Mayak S, Tirosh T, Glick BR (2004b) Plant growth-promoting bacteria that confer resistance to water stress in tomatoes and peppers. Plant Sci 166:525–530
Mendes LW, Kuramae EE, Navarrete AA, van Veen JA, Tsai SM (2014) Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. doi:10.1038/ismej.2014.17
Mullen MD (2005) Phosphorus in soils: biological interactions. In: Hillel D, Rosenzweig C, Powlson D, Scow K, Singer M, Sparks D (eds) Encyclopedia of soils in the environment, vol 3, Academic. Elsevier Ltd., Oxford, pp 210–215
Murphy J, Riley JP (1962) A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27:31–36
Nadeem SM, Zahir ZA, Naveed M, Ashraf M (2010) Microbial ACC-deaminase: prospects and applications for inducing salt tolerance in plants. CRC Crit Rev Plant Sci 29:360–393
Nakamura K, Hiraishi A, Yoshimi Y et al (1995) Microlunatus phosphovorus gen. nov., sp. nov., a new gram-positive polyphosphate-accumulating bacterium isolated from activated sludge. Int J Syst Bacteriol 45:17–22
Neilson JW, Quade J, Ortiz M, Nelson WM, Legatzki A, Tian F, LaComb M, Betancourt JL, Wing RA, Soderlund CA, Maier RM (2012) Life at the hyperarid margin: novel bacterial diversity in arid soils of the Atacama Desert, Chile. Extremophiles 16:553–566
Palacios O, Bashan Y, de Bashan LE (2014) Proven and potential involvement of vitamins in interactions of plants with plant-growth-promoting bacteria—an overview. Biol Fertil Soils 50:415–432
Pastorelli R, Piccolo R, Simoncini S, Landi S (2013) New primers for denaturing gradient gel electrophoresis analysis of nitrate-reducing bacterial community in soil. Pedosphere 23:340–349
Patten CL, Glick BR (2002) Role of Pseudomonas putida indole acetic acid in development of the host plant root system. Appl Environ Microbiol 68:3795–3801
Peace TA, Brock KV, Stills HF Jr (1994) Comparative analysis of the 16S rRNA gene sequence of the putative agent of proliferative ileitis of hamsters. Int J Syst Bacteriol 44:832–835
Penrose DM, Glick BR (2003) Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant 118:10–15
Ramond JB, Tshabuse F, Bopda CW, Cowan DA, Tuffin MI (2013) Evidence of variability in the structure and recruitment of rhizospheric and endophytic bacterial communities associated with arable sweet sorghum (Sorghum bicolor (L) Moench). Plant Soil 372:265–278
Richardson AE (2001) Prospects for using soil microorganisms to improve the acquisition of phosphorus by plants. Aust J Plant Physiol 28:897–906
Richardson AE, Simpson RJ (2011) Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol 156:989–996
Saul-Tcherkas V, Unc A, Steinberger Y (2013) Soil microbial diversity in the vicinity of desert shrubs. Microb Ecol 65:689–699
Shoebitz M, Ribaudo CM, Pardo MA, Cantore ML, Ciampi L, Curá JA (2009) Plant growth promoting properties of a strain of Enterobacter ludwigii isolated from Lolium perenne rhizosphere. Soil Biol Biochem 41:1768–1774
Somerfield PJ (2008) Identification of the Bray–Curtis similarity index: comment on Yoshioka (2008). Mar Ecol Prog Ser 372:303–306
Steen I (1998) Phosphorus availability in the 21st century: management of a non-renewable resource. Phosphorus Potassium 217:25–31
Timmusk S, Paalme V, Pavlicek T, Bergquist J, Vangala A, Danilas T, Nevo E (2011) Bacterial distribution in the rhizosphere of wild barley under contrasting microclimates. PLoS ONE 6:e17968. doi:10.1371/journal.pone.0017968
Turner BL, Papházy MJ, Haygarth PM, McKelvie ID (2002) Inositol phosphates in the environment. Philos Trans R Soc Lond B Biol Sci 357:449–469
Tye AJ, Siu FKY, Leung TYC, Lim BL (2002) Molecular cloning and the biochemical characterization of two novel phytases from B. subtilis 168 and B. licheniformis. Appl Microbiol Biotechnol 59:190–207
Vacheron J, Desbrosses G, Bouffaud ML, Touraine B, Moënne-Loccoz Y, Muller D, Legendre L, Wisniewski-Dyé F, Prigent-Combaret C (2013) Plant growth-promoting rhizobacteria and root system functioning. Front Plant Sci 4:1–19
van Elsas JD, Boersma FGH (2011) A review of molecular methods to study the microbiota of soil and the mycosphere. Eur J Soil Biol 47:77–87
Van Kauwenbergh SJ (2010) World phosphate rock reserves and resources. International Fertilizer Development Centre, 48 pp. Muscle Shoals, Alabama, USA
Warncke D, Brown JR (1998) Potassium and other basic cations. In: Brown JR (ed) Recommended chemical soil test procedures for the North Central Region. NCR Publication No. 221. Missouri Agricultural Experiment Station, Columbia, 31–33 pp
Acknowledgments
Chilean Government financed this study through project Fondecyt No. 1120505 and 1141247. The authors would like thank to Dr. Howard Langer and M.Sc. Loreto Manosalva for valuable logistical support during sampling of Conguillio National Park and Patagonia, respectively.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jorquera, M.A., Inostroza, N.G., Lagos, L.M. et al. Bacterial community structure and detection of putative plant growth-promoting rhizobacteria associated with plants grown in Chilean agro-ecosystems and undisturbed ecosystems. Biol Fertil Soils 50, 1141–1153 (2014). https://doi.org/10.1007/s00374-014-0935-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-014-0935-6