Abstract
The main goal of this study was to expand our knowledge of what happens to the soil bacterial community in an eroded desert soil when improvement of soil fertility is derived from the application of debris of tertiary wastewater treatment containing immobilized microalgae Chlorella sorokiniana and the plant growth-promoting bacterium (PGPB) Azospirillum brasilense. We hypothesized that an “improved” non-agricultural desert soil will exhibit substantial changes in the structure of the bacterial community in a relatively short time after amendment. To assess the effect of the amendments, microalgae and PGPB alone or combined, on the structure of the rhizosphere bacterial community, changes in species richness and bacterial diversity over time were based on sequence differences in the 16S rRNA gene, performed with PCR–denaturing gradient gel electrophoresis (DGGE) and then analyzed by similarity test and non-metric multidimensional scaling analysis. Root surface colonization and persistence in the rhizosphere of A. brasilense was monitored by fluorescent in situ hybridization and sequencing of DGGE bands. Application of waste debris significantly changed the rhizosphere bacterial population structure, whether comparisons were made over time, between inoculated and non-inoculated soil, and among different inoculated microorganisms. Species richness and diversity increased when the waste debris contained the microalgae–bacteria association and also over time. Even as its secondary role as an inoculant after wastewater treatment, A. brasilense colonized the root surface profusely and persisted within the rhizosphere bacterial community. This study demonstrated that small organic amendment to desert soil significantly changed soil bacterial community compared to the original soil and also 2 months after amendments were added.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Deserts are frequently characterized by low productivity and large patches of bare surfaces. Most nutritional and water resources are concentrated in resource islands of three types: under the canopy of nurse plants, around pioneer plants in rocky environments, and in soil crusts. In these resource islands, proliferation of numerous microorganisms occurs (Bashan and de-Bashan 2010). Nonetheless, compared to the literature on bacteria in agriculture, our understanding of arid zone soil microorganisms and microbial ecology of arid soils is miniscule.
Bacteria in arid lands were studied for decades, mostly related to agricultural crops. Bacteria in native desert habitats are far less studied. Mostly, studies were done in specific environments, such as the diversity of bacterial populations tolerant to desiccation and radiation in the Tataouine Desert in Tunisia (Chanal et al. 2006) or the limited bacterial population and diversity in the hyper-arid Atacama Desert in Chile (Drees et al. 2006; Warren-Rhodes et al. 2006; Gómez-Silva et al. 2008).
Studies of bacterial populations in eroded desert lands and factors controlling them, apart from arid mine tailings that are man-made “soils,” are scarce. Restoration of these soils with plant growth-promoting bacteria (PGPB) that are well known in agriculture (Bashan and de-Bashan 2005; Lugtenberg and Kamilova 2009) was demonstrated only in recent years in vegetation restoration projects in abandoned, dry land habitats (for a review, see de-Bashan et al. 2012 and references therein).
The use of wastewater sludge for improving soil is very common (Smith 1995) and also tested in arid lands (Brendecke et al. 1993). A tertiary wastewater treatment, using immobilized microalgae and PGPB, was proposed, tested on an experimental scale, and is currently in a process of scaling up (de-Bashan et al. 2004; Hernandez et al. 2006). Once employed, this wastewater treatment produces leftover debris. This debris contains large populations of the microalgae Chlorella spp. and the PGPB Azospirillum brasilense. Recently, using this debris, small improvements in the fertility of highly eroded desert soil was detected, and its application significantly improved plant growth (Trejo et al. 2012). Consequently, the central hypothesis of this molecular study was that an “improved” non-agricultural desert soil will exhibit substantial changes in the profile of the bacterial community in a relatively short time after amendment. This will happen because changes in soil fertility, supported by various organic amendments in agricultural soils, are usually associated with effects on the microbial communities of the soil. The main goal of this study was to expand our knowledge of what happens to the soil bacterial community and to the PGPB in an eroded desert soil when improvement of soil fertility is derived from debris of tertiary wastewater treatment containing microalgae and a PGPB.
Denaturing gradient gel electrophoresis (DGGE) fingerprinting is a standard method of characterizing soil bacterial communities, together with T-RFLP and SSCP fingerprints of PCR-amplified 16S rRNA gene fragments. Although each method assesses a different fragment, the general results regarding bacterial communities in the soil are very similar (Smalla et al. 2007). DGGE was employed in several studies of bacteria in deserts (Nagy et al. 2005; Drees et al. 2006; Gothwal et al. 2007; Campbell et al. 2009; Kenzaka et al. 2010).
Four strategies were employed to address our hypothesis: (1) analysis of the structure of the bacterial community under various combinations of amendments. We used PCR-DGGE fingerprinting analyzed by a similarity test and a statistical multivariate analysis (non-metric multidimensional scaling analysis). (2) Partial assessment of bacterial richness and diversity, since the number of bands observed in the PCR-DGGE profile provides an estimate of species richness and that the relative intensity of each band provides an estimate of the relative abundance of each species (Iwamoto et al. 2000). We were also aware of the inherent limitation of this analysis (Bent and Forney 2008), which is incapable of detecting all of the microbial diversity in the soil of the Sonoran Desert. (3) Monitoring PGPB that was applied indirectly with the debris using the molecular detection technique of fluorescent in situ hybridization under confocal laser scanning microscope (FISH; Dazzo et al. 2007). (4) Identification of the applied A. brasilense within the general bacteria community of the rhizosphere by sequencing PCR products from excised bands of PCR-DGGE profiles.
Material and methods
Soil source, sampling, and soil fertility
Sorghum (Sorghum bicolor (L.) Moench, cv. Honey Graze, Cal-Oro, Lubbock, TX) plants were grown in highly degraded alluvial desert soil (haplic yermosol; described in Bashan et al. 2000). Artificially prepared domestic wastewater (Gonzalez and Bashan 2000) was used. The wastewater contained no heavy metals, antibiotics, human pathogens, or recalcitrant materials. Debris from a laboratory simulation of tertiary wastewater treatment was performed with the removal of nutrients using alginate beads containing the microalgae Chlorella sorokiniana Shih. et Krauss (UTEX 2805, Austin TX; de-Bashan et al. 2008) and the microalgae growth-promoting bacterium A. brasilense Cd (DSM 1843, Braunschweig, Germany), immobilized singly or together (de-Bashan et al. 2004). After wastewater treatment, alginate beads were recovered and oven-dried at 40–43 °C for 24 h, yielding debris of dry beads containing live A. brasilense. Then, 0.2 g of this debris was added to the soil at a level of 0.3 × 106 CFU bead−1 (A. brasilense), corresponding to 1.5 × 106 CFU pot−1 (120 ml). The debris of dry beads did not contain additional organic material. In this soil, sorghum plants were grown in a shade house at ambient temperature ~29 °C and light intensity of ~1,000 μmol photons m2 s−1 for 20 days (cycle 1). The potted seedlings were irrigated with 38.5 ml distilled water every 3 days to maintain field capacity and then harvested. A second application of new debris was added to the same soil and a new crop of sorghum was grown (cycle 2) for 20 days. The process was repeated one more time (cycle 3). Effects on plant growth (dry weight, shoot and root length) and increases in total C in the soil were measured after each cycle. Microbial C was measured after cycle 3 (Electronic supplementary material (ESM) Fig. S1 and Table S1). An increase in total C and microbial C served as an indicator of improved soil fertility. All of this was conducted in a previous study (Trejo et al. 2012). This experiment was repeated twice, once with tomato plants (unpublished data) and once with sorghum plants (Trejo et al. 2012). Both experiments yielded very similar results. In this study, soil samples from the experiment with sorghum plants were analyzed for changes in the microbial populations.
Extraction of DNA from soil and PCR amplification
Rhizosphere soil samples (soil adhered to root after mild agitation; Hartmann et al. 2008) from the treatment of alginate bead debris containing microalgae and bacteria were collected after each planting cycle. Samples from the remaining treatments were collected only at the third planting cycle. About 3 g of root-free rhizosphere soil was collected in 1.5-ml microcentrifuge tubes (Eppendorf, Hamburg, Germany) and stored at −20 °C until analysis. Cold storage has no effect on the analysis of bacterial community by PCR-DGGE (Campbell et al. 2009). Extraction of DNA from soil was a modification of the method described by de-Bashan et al. (2010a, b), using a kit (Fast DNA SPIN for soils, MP Biomedicals, Santa Ana, CA), according to the manufacturer’s instructions. To remove humic acids, the binding matrix–DNA complex was rinsed with saturated 5.5 M guanidine thiocyanate (Fluka Sigma-Aldrich, Buchs, Switzerland). Each DNA extraction was performed with a 0.6-g soil sample. Three replicates were analyzed for each treatment because of the technical limitation of the DGGE gels (see below).
Polymerase chain reaction
A modification of the PCR procedure described by de-Bashan et al. (2010a, b) was used. The V9 variable region of the 16S rRNA gene was amplified using the bacteria primers 1070F (5′-ATG GCT GTC GTC AGC T-3′) and 1406R (5′-ACG GGC GGT GTG TAC-3′) with a 40-bp GC clamp (Ferris et al. 1996). A modification of the PCR for DGGE by Colores et al. (2000) was used. Details of the modifications are presented in the ESM.
Denaturing gradient gel electrophoresis (PCR-DGGE) analysis
Initially, the community was analyzed at the end of the third cycle of inoculation, comparing inoculated plants with bead debris containing A. brasilense and C. sorokiniana, un-inoculated plants, unplanted soil, and a control of only beads. We also examined the time effect of inoculation with co-immobilized A. brasilense and C. sorokiniana on the rhizosphere bacterial community, comparing the three cycles of growth. A modification of the DGGE of the 16S rRNA gene products by de-Bashan et al. (2010a, b) was performed using a D-Code Universal Mutation Detection System (Bio-Rad Laboratories, Hercules, CA). Details of the modifications are presented in the ESM.
Measuring numerous bands on DGGE gels seldom allows reliable comparison among different gels. The effect of gel-to-gel variation is an inherent difficulty of DGGE analysis and is complicated to address (Tourlomousis et al. 2010). Practically, this limits the number of replicates of different treatments a single gel can handle. Only very recently, a simplified technical improvement to address this difficulty was proposed (Valentín-Vargas et al. 2013). This was not employed in this study which was carried out earlier. Consequently, a combination of several gels, each containing comparable treatments with their respective replicates, were used in this study to measure the effect of a specific treatment on the soil bacterial population (Lynch et al. 2004; Smalla et al. 2007). Each gel contained three or four soil treatments and an external reference ladder (total of 10–13 lanes per gel, as permitted by the equipment we used). Digital images of DGGE were analyzed to generate a densitometric profile of bands using imaging software (Quantity One 4.6.7, Bio-Rad Laboratories).
Identification of A. brasilense in PCR-DGGE profiles
Presumptive bands of A. brasilense Cd were excised from DGGE gels (45–60 % gradient) using sterile razor blades under UV illumination. The excised bands were eluted in 300 μl ultrapure water and incubated at 37 °C for 1 h. Aliquots were diluted 1:10 in ultrapure water; 2 μl of this dilution was used as a template to re-amplify the replicon using the same PCR conditions and DGGE primers described earlier. The size of the PCR product was confirmed on 2 % agarose gel after each round of amplification. Successive PCR-DGGE gels were run to verify the identity and purity of the excised bands by comparing the re-amplified PCR products to the profile of the external reference ladder containing A. brasilense. PCR products that exhibited the highest identity to the Azospirillum band in the DGGE gel were purified using the QIAquick PCR purification kit protocol (Qiagen Sciences) and then submitted for commercial sequencing using primer 1070F (Genewiz, South Plainfield, NJ). The original PGPB inoculum and its corresponding band in the external reference ladder were also sequenced at the same time as the experimental samples. Representative sequences from experimental samples at each cycle of inoculation were deposited in the GenBank database (accession nos. JX133090, JX133091, JX133092, JX133093, JX133094).
Fluorescence in situ hybridization
These procedures followed Trejo et al. (2012). Briefly, the three root zones (root tip, elongation zone, and lateral and root hair zone), all areas colonized by Azospirillum spp. (Bashan et al. 2004), were prepared as described in de-Bashan et al. (2010a) and Trejo et al. (2012). Specific detection of A. brasilense on roots was done by FISH with the following oligonucleotide probes: an equimolar mix of probes EUB-338-I (5′-GCTGCCTCCCGTAGGAGT-3′; Amann et al. 1990), EUB-338-II (5′-GCAGCCACC CGTAGG TGT-3′), and EUB-338-III (5′-GCTGCCACCCGTAGGTGT-3′; Daims et al. 1999) that are specific for the domain bacteria and the probe Abras-1420 (5′-CCACCTTCGGGTAAAGCCA-3′; Stoffels et al. 2001) specific for A. brasilense. Probes were labeled with either fluorochrome Cy3 or Cy5 (Interactive Division, Thermo Electron, Ulm, Germany). Hybridization was performed according to Stoffels et al. (2001) and de-Bashan et al. (2010b). Visualization was performed as described by Rothballer et al. (2003), with confocal laser scanning microscopy (LSM 510, Axiovert 100M, Zeiss, Jena, Germany). A three-channel observation technique was used at excitations of 543 and 633 nm, corresponding to the dyes Cy3 (red) and Cy5 (blue); a third color channel (488 nm, green) was used to visualize the autofluorescence of the root. This technique can use overlapping of images, resulting in differentiation among the observed objects: root surfaces were green, bacteria in general were red, and Azospirillum cells were magenta. The images were analyzed with specialized software (LSM 510 4.2, Carl Zeiss, Oberkochen, Germany).
Statistical analysis
Each gel, with its respective treatments, was run several times; one representative gel was considered for each comparison of treatments. The band profiles obtained from DGGE gels were analyzed for similarity using the Dice coefficient. A dendrogram was built from the weighted pair group matching average. Similarity varies from 0 to 1, where 1 indicates 100 % similarity.
Additionally, the observed similarities between the profiles of DGGE were analyzed using Kruskal’s non-metric multidimensional scaling (NMDS; Venables and Ripley 2002) using a computing software (Statistica 8.0, StatSoft, Tulsa, OK). Kruskal’s stress coefficient was used to reflect the goodness of fit of the model. Values of Kruskal’s stress <0.1 are considered a good fit.
Bacterial richness considered each band as an individual operative taxonomic unit (Kisand and Wikner 2003). This was obtained from the Band Type Report of the Quantity One 4.6.7 imaging software (Bio-Rad Laboratories) that provides the number of bands detected in the DGGE profiles. Bacterial diversity was calculated by analyzing the relative intensity of each peak (corresponding to a defined band) in the densitometric profile with Shannon’s diversity index (Iwamoto et al. 2000), calculated using the formula: H = −ΣP i log10 P i, where P i is the importance, probability of the bands in a gel lane and is calculated as P i = n i/N, where n i is the intensity of a peak and N is the sum of all peak intensities of bands (Iwamoto et al. 2000). Data were analyzed using one-way ANOVA and then by Tukey’s post hoc analysis at P < 0.05 using statistical software (Statistica 6.0, StatSoft).
Results
Effect of the application of jointly immobilized A. brasilense and C. sorokiniana on the structure of the rhizosphere bacterial community
The bacterial rhizosphere community was monitored from plant samples harvested randomly from three pots in each treatment of the experiment with plants (Trejo et al. 2012; ESM Fig. S1). Figure 1 shows the results of the PCR-DGGE based on 16S rDNA fragments. The dendrogram built with the Dice coefficient measures the similarity among the banding patterns of replicate samples from each treatment. Four distinctive clusters (Fig. 1a) separate the community of inoculated plant from the controls that were not inoculated. Three-dimensional NMDS analysis of the PCR-DGGE profiles of the treatments confirmed that there was a significant effect of inoculation on the structure of the bacterial community. Similar to the results obtained from the dendrogram, each of the treatment replicates in the NMDS analysis were clustered, forming four distinctive groups that were significantly different from each other (stress factor S = 0.0027; Fig. 1b). One group contained the untreated (control) soil samples. The second group contained the second control samples that were not inoculated. The third group contained the inoculated samples. The fourth group contained only empty beads.
Comparison of the rhizosphere bacterial community structure of six treatments: soil supplemented with immobilized C. sorokiniana and planted; soil supplemented with immobilized A. brasilense and planted; soil supplemented with jointly immobilized C. sorokiniana–A. brasilense and planted; no inoculated plant (plant only); no planted soil (initial soil); and soil supplemented with empty alginate beads (alginate only). a–d Comparison at the third cycle of inoculation. e, f Comparison of the three cycles of growth for soils supplemented with co-immobilized C. sorokiniana–A. brasilense and planted. a, b Comparison among soils treated with jointly immobilized microorganisms and soils without supplementation of microorganisms. c, d Comparison among soils treated with microorganisms, alone or jointly immobilized. a, c, e Dendrograms from cluster analyses by Dice coefficient. b, d, f 3D configuration derived from Kruskal’s NMDS of bands. Data with the same symbols represent the replicates within each treatment
There were also differences in community structure when comparing the effect of inoculation with debris containing: (1) jointly immobilized microorganisms; (2) only the bacteria, and (3) only the microalgae. The overall results of the similarity analysis show three distinctive clusters of PCR-DGGE samples (Fig. 1c). These clusters were similar to the results obtained from the NMDS analysis (S = 0.000036; Fig. 1d): one group corresponds only to A. brasilense immobilized alone, the second group corresponds only to C. sorokiniana immobilized alone, and the third group corresponds to the two microorganisms jointly immobilized.
We also examined the time effect of inoculation with leftover debris containing A. brasilense and C. sorokiniana on the rhizosphere bacterial community, comparing the three cycles of growth. Once again, the similarity analysis (Fig. 1e) and NMDS (S = 0.0027; Fig. 1f) indicated three distinctive groups in each cycle that revealed a significant effect of treatment during the three cycles. These results suggest a shift in the community structure over time.
Effect of adding debris containing jointly immobilized A. brasilense and C. sorokiniana on rhizosphere bacterial richness and diversity
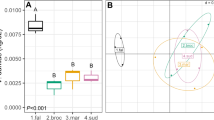
Richness of bacterial species in the rhizosphere community of plants inoculated with bead debris containing A. brasilense and C. sorokiniana at the end of the third cycle (60 days after the first inoculation) was significantly higher than the controls which were not inoculated (P = 0.0006; Fig. 2a). Species richness after the application of bead debris with jointly immobilized microorganisms or bead debris containing only C. sorokiniana was also higher than on plants inoculated only with bead debris containing A. brasilense (P = 0.002; Fig. 2c). The effect of time of inoculation with bead debris containing A. brasilense and C. sorokiniana was indicated by the increased richness of species. A significant (P = 0.03) increase in the number of species occurred in the second and third cycles compared to the first cycle (Fig. 2e).
Comparison of the species richness and diversity of the bacterial community of the six treatments: soil supplemented with immobilized C. sorokiniana and planted; soil supplemented with immobilized A. brasilense and planted; soil supplemented with jointly immobilized C. sorokiniana–A. brasilense and planted; non-inoculated plant; no planted soil; and soil supplemented with empty alginate beads. Species richness (a) and diversity (b) comparison among soils treated with jointly immobilized microorganisms and soils without supplementation of microorganisms. Species richness (c) and diversity (d) comparison among soils treated with microorganisms, alone or jointly immobilized. Species richness (e) and diversity (f) comparison among soil supplemented with jointly immobilized microorganisms during a time course of three cycles of growth. In each subfigure, columns denoted with different lowercase letter differ significantly at P < 0.05 by one-way ANOVA and Tukey’s post hoc analyses. Bars represent SE. Absence of a bar, negligible SE
The Shannon diversity index (H′) was calculated to assess changes in bacterial diversity on the rhizosphere of the plants based on the intensity of the DGGE banding patterns. The diversity index of the bacteria in plants inoculated with bead debris containing A. brasilense and C. sorokiniana is significantly higher (P = 0.0007) than the values obtained for the three controls: (1) plants that were not inoculated, (2) soil that was not treated, and (3) soil containing beads without immobilized microorganisms (Fig. 2b). The diversity index is significantly higher (P = 0.002) in rhizosphere bacteria of plants inoculated with jointly immobilized microorganisms or only with C. sorokiniana than the diversity index of the bacterial community on plant roots inoculated only with A. brasilense (Fig. 2d). Similar to the richness of species, the Shannon diversity index indicates that the diversity of bacteria increased with time when plants were inoculated; diversity in cycle 3 was higher than in cycles 1 and 2 (P = 0.02; Fig. 2f).
Identification of A. brasilense within the rhizosphere bacterial community
Purified bands of putative A. brasilense were obtained after the second purification step of the bands of PCR-DGGE in treatments involving the application of debris with this PGPB. When comparing the sequences of these bands with those of the positive control in the external reference ladder or the sequence of pure culture of A. brasilense Cd, we found 100 % homology among all tested sequences (ESM Table S2). This confirmed the identity for the recovered bands (representing the PGPB) with the original inoculum. Further analysis compared each of the excised band sequences with the GenBank database and showed 99 % identity with A. brasilense Cd.
Persistence of the original inoculum within the bacterial community of the rhizosphere of sorghum plants was detected at the end of each cycle after the application of debris containing A. brasilense and C. sorokiniana. Similarly, the PGPB was identified after cycle 3 application of debris only containing A. brasilense.
Colonization of sorghum root by A. brasilense derived from bead debris
FISH analysis assayed the colonization of A. brasilense on the roots of sorghum growing in soil amended with bead debris. Colonized roots were detected during the three growth cycles in all three zones of most root segments (Fig. 3). Colonization was primarily found in the apical root zone (Fig. 3b, c, f, arrows) and the root elongation zone (Fig. 3d, e); if plants had not been inoculated, only unidentified bacteria were detected (Fig. 3a).
Colonization of roots of sorghum by A. brasilense Cd detected by FISH probes and observed under confocal laser scanning microscopy. a Non-inoculated control. b–c, f Root tip. d, e Elongation zone. FISH experiments were performed with Abras-1420-Cy5 (blue) specific for A. brasilense and with the probe made of a mix of EUB-338-I, II, III-Cy3 (red) specific for the domain bacteria. The composed RGB images result in a magenta color for A. brasilense Cd cells (marked with yellow arrows), which indicates co-labeling by both probes. The third color channel (green) was used to visualize autofluorescence and the structure of roots. Red labeling corresponds to cells of unidentified bacteria (indicated with white arrows)
Discussion
Earlier plant experiments demonstrated that there was a small improvement in soil fertility from the application of small amounts of debris containing microorganisms immobilized in alginate beads derived from simulation of tertiary wastewater treatment. Small increases in total C and microbial C and significant improvement in plant growth were observed. These effects were significantly greater compared to effects from untreated, eroded soil (Trejo et al. 2012). This molecular study focuses on the structure of the bacterial community in this amended desert soil.
The main goal of this study was to expand our knowledge of what happens to the soil bacterial community and to the PGPB in an eroded desert soil when improvement of soil fertility is derived from bead debris of tertiary wastewater treatment containing microalgae and a PGPB. Application (=an indirect inoculation) of leftover dry bead debris to the desert soil planted with sorghum significantly shifted the rhizobacterial community after three successive applications compared to untreated soil, soil with plants that were not inoculated, or soil having plants receiving only one microorganism. Similar detectable change of bacterial community structure based on sequence differences in the 16S rRNA gene and analyzed by PCR-DGGE was observed when seedlings of the desert quail bush (Atriplex lentiformis) were inoculated with A. brasilense Sp6 or Bacillus pumilus ES4 during experiments in the phytostabilization of two toxic mine tailings (de-Bashan et al. 2010a, b). Baudoin et al. (2009) document a transient, but statistically significant, change in the structure of the rhizobacteria communities when Azospirillum lipoferum CRT1 was inoculated on maize using automated ribosomal intergenic spacer analysis (ARISA). Yet, two other studies using A. brasilense examining maize grown in pots in a controlled greenhouse showed the contrary. Analysis of the bacterial community structure in these experiments, using the fingerprinting methods DGGE and ARISA, showed that inoculation with A. brasilense strains Cd and Sp 245 had no effect or very marginal effect on the size or structure of the bacterial communities. Furthermore, the age of the plants was far more significant in affecting bacterial communities (Herschkovitz et al. 2005; Lerner et al. 2006). Other parameters of importance affecting the structure of the bacterial community are the microbial species and their genomic traits (Barret et al. 2011). Using DGGE, another study showed similar trends (no effect) with tomatoes inoculated with A. brasilense combined with B. subtilis in a peat-based substrate (Felici et al. 2008).
Our study further measured DGGE patterns to partially assess bacterial richness and diversity as intrinsic descriptors of the rhizosphere community. Richness assessment in complex microbial communities, such as soil, required extensive sampling using diversity indices through the analysis of clone libraries (Bent and Forney 2008). However, because our main aim was to measure whether there is any effect of the application of PGPB on the soil bacterial community and not measure the total diversity and richness of a soil of the Sonoran Desert per se, partial analyses that detect dominant populations in the soil, as performed in this study, are meaningful. In the soil, the structural and functional diversity of bacterial populations is affected by root exudates, flavonids, rhizodeposition, soil amendments, and the characteristics of the soils (Gomes et al. 2001; Nannipieri et al. 2008a, b; Cesco et al. 2012). Our results demonstrated that three successive applications of small amounts of bead debris containing the mix of A. brasilense and C. sorokiniana or only C. sorokiniana favored bacterial richness, which may have occurred by growth of the bacterial population after the first application of debris. This first cycle represents the highest bacterial richness, while no significant changes occurred in the second or the third cycle of inoculation. The increase in bacterial richness could be explained by the increase of organic matter, as indicated by higher organic C and higher bacterial C in this poor soil (Trejo et al. 2012).
Even though richness of the bacterial community increased after the application of amendments, our study raised further questions regarding changes in the composition of the bacterial community. These will require further refinements of microbial ecology analysis, such as determination of the 16S rRNA gene copy numbers that might elucidate increased numbers for the amendment treatments and replacement of the commonly used Sybr Green staining that may not be sensitive enough for a soil environment with possibly AgNO3 or ethidium bromide as stains.
Small increases in bacterial diversity after the three applications of A. brasilense and C. sorokiniana indicate that the abundance of bacterial species is becoming more evenly distributed, adapting to improved organic matter in the soil. The bead debris may have led to higher bacterial diversity in the rhizobacterial community of inoculated plants, considering that heterogeneity of microenvironments in the rhizosphere presumably results in the formation of many different in situ niches supporting an abundant and diverse bacterial assemblage in these root microenvironments (Wang et al. 2007). Additionally, increased diversity and richness after the third application suggests that the soil environment becomes less restrictive than the initial eroded soil. Debris with microalgae and/or PGPB has a stronger effect on the community than expected because it promotes microbial succession more efficiently than applications of debris without microorganisms or when the plants were grown without amendments.
Apart from compost (not part of this study), soils receiving large amounts of organic waste of various sources or pioneer plants that add large amounts of C support greater biomass of the microbial populations (Van Veen and Kuikman 1990; Luna-Suárez et al. 2000; Bastida et al. 2008, 2009). Microalgae are excellent for transporting nutrients because they contain important quantities of essential nutrients, especially N and P (about 10 and 1 %, respectively, on a dry weight basis), quantities that are several-fold higher than most terrestrial plants (Benemann et al. 2003). Nutrients recovered from wastewater treatment in the residues of the algal biomass could have premium value as organic fertilizers (Benemann et al. 2003). Chlorella spp. are not known to promote plant growth via active production of substances affecting plant metabolisms. Even though Chlorella sp. can survive, in small numbers, in the soil for prolonged periods (Trainor and Gladych 1995), the population of this aquatic microalga diminished rapidly when applied to soil. Thus, we theoretically assumed that the effect recorded on the microbial community is through the addition of organic matter by the decomposing cells. Azospirillum spp., on the other hand, is known to survive for limited time in the bulk soil (Bashan 1999) and far longer in the rhizosphere of many plants and in different soils (Bashan et al. 1995). Consequently, we had followed its incorporation into the rhizosphere community of the plant in this study. Although the use of microalgae as a soil amendment is not widespread, microalgal biomass as a soil conditioner is a promising biotechnological venue (Pulz and Gross 2004).
In general, to verify that the effects on plant growth are derived from inoculation with PGPB, it is necessary to demonstrate that plants inoculated with a PGPB strain show root colonization by the specific strain (Lugtenberg and Kamilova 2009). Different methods for identifying root colonization, specifically by Azospirillum, have been developed, including traditional microbiology, immunology, molecular insertion of specific marker genes (Jacoud et al. 1998), insertion of the gfp green fluorescent protein (Ramos et al. 2002; Bacilio et al. 2004), lacZ markers (Arsene et al. 1994), and FISH (Rothballer et al. 2003). FISH monitored with confocal laser microscopy is a powerful tool to detect PGPB in soils and rhizosphere samples (Assmus et al. 1995). In our study, A. brasilense cells were specifically detected by the FISH technique on colonized root tips and elongation zone areas, which are known locations for colonization by Azospirillum (Bashan and Levanony 1989a, b; Bashan et al. 2004).
Additional evidence of the persistence of A. brasilense within the rhizosphere bacterial community was provided by the sequencing and identification of putative bands of Azospirillum in DGGE gels. The identity of presumptive bands was a 100 % match to the original inoculum, which was present after the third application of inoculation. As was reported by de-Bashan et al. (2010a, b), sequences of the V9 variable region of the 16S rDNA is useful in identifying A. brasilense in the rhizosphere bacterial community.
In summary, this study demonstrated that small improvements in soil fertility from the application of small amounts of waste bead debris from experimental, simulated tertiary wastewater treatment significantly changed the bacterial soil community compared to the original soil and continued for more than 2 months. This amendment improved biological quality by promoting a richer soil bacterial community, which supports the hypothesis of this study. A limitation of this study is the relative short-term growth period of plants (60 days) that was employed. We cannot rule out that the measured effects on the bacterial community are transient and will not have long-term effects on the fertility of the soil.
Taking this molecular study and its predecessor plant study (Trejo et al. 2012) together, several issues are pending. Since our specific objective was to demonstrate the proof of concept of a new idea, scaling up, use of different soils, potential secondary soil contamination when the waste beads came from heavily contaminated industrial wastewater, a strain-specific approach for analyzing the different rhizosphere communities, comparison to application of compost or fertilizers, issues of costs and obstacles to implementation, potential savings, and potential suitability in non-degraded soils have not yet been investigated.
References
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925
Arsene F, Katupitiya S, Kennedy IR, Elmerich C (1994) Use of LacZ fusions to study the expression of nif genes of Azospirillum brasilense in association with plants. Mol Plant–Microbe Int 7:748–757
Assmus B, Hutzler P, Kirchhof G, Amann RI, Lawrence JR, Hartmann A (1995) In situ localization of Azospirillum brasilense in the rhizosphere of wheat with fluorescence labeled, rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl Environ Microbiol 61:1013–1019
Bacilio M, Rodriguez H, Moreno M, Hernandez J-P, Bashan Y (2004) Mitigation of salt stress in wheat seedlings by a gfp-tagged Azospirillum lipoferum. Biol Fertil Soils 40:188–193
Barret M, Morrissey JP, O’Gara F (2011) Functional genomics analysis of plant growth-promoting rhizobacterial traits involved in rhizosphere competence. Biol Fertil Soils 47:729–743
Bashan Y (1999) Interactions of Azospirillum spp. in soils: a review. Biol Fertil Soils 29:246–256
Bashan Y, de-Bashan LE (2005) Bacteria/plant growth-promotion. In: Hillel D (ed) Encyclopedia of soils in the environment, vol. 1. Elsevier, Oxford, UK, pp 103–115
Bashan Y, de-Bashan LE (2010) Microbial populations of arid lands and their potential for restoration of deserts. In: Dion P (ed) Soil biology and agriculture in the tropics, Soil Biology Series 21, chapter 6. Springer, Berlin, pp 109–137
Bashan Y, Levanony H (1989a) Factors affecting adsorption of Azospirillum brasilense Cd to root hairs as compared with root surface of wheat. Can J Microbiol 35:936–944
Bashan Y, Levanony H (1989b) Wheat root tips as a vector for passive vertical transfer of Azospirillum brasilense Cd. J Gen Microbiol 135:2899–2908
Bashan Y, Puente ME, Rodriguez-Mendoza MN, Toledo G, Holguin G, Ferrera-Cerrato R, Pedrin S (1995) Survival of Azospirillum brasilense in the bulk soil and rhizosphere of 23 soil types. Appl Environ Microbiol 61:1938–1945
Bashan Y, Davis EA, Carrillo-Garcia A, Linderman RG (2000) Assessment of VA mycorrhizal inoculum potential in relation to the establishment of cactus seedlings under mesquite nurse-trees in the Sonoran Desert. Appl Soil Ecol 14:165–176
Bashan Y, Holguin G, de-Bashan LE (2004) Azospirillum–plant relationships: physiological, molecular, agricultural, and environmental advances (1997–2003). Can J Microbiol 50:521–577
Bastida F, Kandeler E, Moreno JL, Ros M, Garcıa C, Hernandez T (2008) Application of fresh and composted organic wastes modifies structure, size and activity of soil microbial community under semiarid climate. Appl Soil Ecol 40:318–329
Bastida F, Perez-de-Mora A, Babic K, Hai B, Hernandez T, Garcia C, Schloter M (2009) Role of amendments on N cycling in Mediterranean abandoned semiarid soils. Appl Soil Ecol 41:195–205
Baudoin E, Nazaret S, Mougel C, Ranjard L, Moënne-Loccoz Y (2009) Impact of inoculation with the phytostimulatory PGPR Azospirillum lipoferum CRT1 on the genetic structure of the rhizobacterial community of field-grown maize. Soil Biol Biochem 41:409–413
Benemann JR, van Olst JC, Massingil MJ, Carlberg JA, Weissman JC, Brune DE (2003) The controlled eutrophication process: using microalgae for CO2 utilization and agricultural fertilizer recycling. Proceedings of Greenhouse Gas Control Technologies, 6th International Conference, Kyoto, Japan, pp 1433–1438
Bent SJ, Forney LJ (2008) The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. The ISME J 2:689–695
Brendecke JW, Axelson RD, Pepper IL (1993) Soil microbial activity as an indicator of soil fertility: long-term effects of municipal sewage sludge on an arid soil. Soil Biol Biochem 25:751–758
Campbell JH, Clark JS, Zak JC (2009) PCR-DGGE comparison of bacterial community structure in fresh and archived soils sampled along a Chihuahuan Desert elevational gradient. Microb Ecol 57:261–266
Cesco S, Mimmo T, Tonon G, Tomasi N, Pinton R, Terzano R, Neumann G, Weisskopf L, Renella G, Landi L, Nannipieri P (2012) Plant-borne flavonoids released into the rhizosphere: impact on soil bio-activities related to plant nutrition. A review. Biol Fertil Soils 48:123–149
Chanal A, Chapon V, Benzerara K, Barakat M, Christen R, Achouak W, Barras F, Heulin T (2006) The desert of Tataouine: an extreme environment that hosts a wide diversity of microorganisms and radiotolerant bacteria. Environ Microbiol 8:514–525
Colores GM, Macur RE, Ward DM, Inskeep WP (2000) Molecular analysis of surfactant-driven microbial population shifts in hydrocarbon-contaminated soil. Appl Environ Microbiol 66:2959–2964
Daims H, Brühl A, Amann R, Schleifer K-H, Wagner M (1999) The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444
Dazzo FB, Schmid M, Hartmann A (2007) Immunofluorescence microscopy and fluorescence in situ hybridization combined with CMEIAS and other image analysis tools for soil and plant-associated microbial autecology. In: Hurst CJ, Crawford RL, Garland JL, Lipson DA, Mills AL, Stetzenbach LD (eds) Manual of environmental microbiology, 3rd edn. American Society for Microbiology, Washington, DC, pp 712–733
de-Bashan LE, Hernandez JP, Morey T, Bashan Y (2004) Microalgae growth-promoting bacteria as “helpers” for microalgae: a novel approach for removing ammonium and phosphorus from municipal wastewater. Water Res 38:466–474
de-Bashan LE, Trejo A, Huss VAR, Hernandez J-P, Bashan Y (2008) Chlorella sorokiniana UTEX 2805, a heat and intense, sunlight-tolerant microalga with potential for removing ammonium from wastewater. Bioresour Technol 99:4980–4989
de-Bashan LE, Hernandez J-P, Bashan Y, Maier RM (2010a) Bacillus pumilus ES4: candidate plant growth-promoting bacterium to enhance establishment of plants in mine tailings. Environ Exp Bot 69:343–352
de-Bashan LE, Hernandez JP, Nelson KN, Bashan Y, Maier RM (2010b) Growth of quailbush in acidic, metalliferous desert mine tailings: effect of Azospirillum brasilense Sp6 on biomass production and rhizosphere community structure. Microbial Ecol 60:915–927
de-Bashan LE, Hernandez J-P, Bashan Y (2012) The potential contribution of plant growth-promoting bacteria to reduce environmental degradation—a comprehensive evaluation. Appl Soil Ecol 61:171–189
Drees KP, Neilson JW, Betancourt JL, Quade J, Henderson DA, Pryor BM, Maier RM (2006) Bacterial community structure in the hyperarid core of the Atacama Desert, Chile. Appl Environ Microbiol 72:7902–7908
Felici C, Vetton L, Giraldi E, Forino LMC, Toffanin A, Tagliasacchi AM, Nuti M (2008) Single and co-inoculation of Bacillus subtilis and Azospirillum brasilense on Lycopersicon esculentum: effects on plant growth and rhizosphere microbial community. Appl Soil Ecol 40:260–270
Ferris MJ, Muyzer G, Ward DM (1996) Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol 62:340–346
Gomes NCM, Heuer H, Schönfeld J, Costa R, Mendonça-Hagler L, Smalla K (2001) Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil 232:167–180
Gómez-Silva B, Rainey FA, Warren-Rhodes KA, McKay CP, Navarro-González R (2008) Atacama Desert soil microbiology. In: Dion P, Nautiyal CS (eds) Microbiology of extreme soils, vol. 13. Springer, Berlin, pp 117–132
Gonzalez LE, Bashan Y (2000) Increased growth of the microalga Chlorella vulgaris when coimmobilized and cocultured in alginate beads with the plant growth-promoting bacterium Azospirillum brasilense. Appl Environ Microbiol 66:1527–1531
Gothwal RK, NigamVK MMK, Sasmal D, Ghosh P (2007) Extraction of bulk DNA from Thar Desert soils for optimization of PCR-DGGE based microbial community analysis. Electron J Biotechnol (Chile) 10:400–408
Hartmann A, Rothballer M, Schmid M (2008) Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312:7–14
Hernandez JP, de-Bashan LE, Bashan Y (2006) Starvation enhances phosphorus removal from wastewater by the microalga Chlorella spp. co-immobilized with Azospirillum brasilense. Enzyme Microb Tech 38:190–198
Herschkovitz Y, Lerner A, Davidov Y, Okon Y, Jurkevitch E (2005) Azospirillum brasilense does not affect population structure of specific rhizobacterial communities of inoculated maize (Zea mays). Environ Microbiol 7:1847–1852
Iwamoto T, Tani K, Nakamura K, Suzuki Y, Kitagawa M, Eguchi M, Nasu M (2000) Monitoring impact of in situ biostimulation treatment on groundwater bacterial community by DGGE. FEMS Microbiol Ecol 32:129–141
Jacoud C, Faure D, Wadoux P, Bally R (1998) Development of a strain-specific probe to follow inoculated Azospirillum lipoferum CRT1 under field conditions and enhancement of maize root development by inoculation. FEMS Microbiol Ecol 27:43–51
Kenzaka T, Sueyoshi A, Baba T, Li P, Tani K, Yamaguchi N, Nasu M (2010) Soil microbial community structure in an Asian dust source region (loess plateau). Microbes Environ 25:53–57
Kisand V, Wikner J (2003) Combining culture-dependent and -independent methodologies for estimation of richness of estuarine bacterioplankton consuming riverine dissolved organic matter. Appl Environ Microbiol 69:3607–3616
Lerner A, Herschkovitz Y, Baudoin E, Nazaret S, Moënne-Loccoz Y, Okon Y, Jurkevitch E (2006) Effect of Azospirillum brasilense inoculation on rhizobacterial communities analyzed by denaturing gradient gel electrophoresis and automated ribosomal intergenic spacer analysis. Soil Biol Biochem 38:1212–1218
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Ann Rev Microbiol 63:541–556
Luna-Suárez S, Frias-Hernández JT, Olalde-Portugal V, Dendooven L (2000) Catclaw (Mimosa buincifiera): a pest or a means to restore soil fertility in heavily eroded soil from the central highlands of Mexico? Biol Fertil Soils 32:109–113
Lynch JM, Benedetti A, Insam H, Nuti MP, Smalla K, Torsvik V, Nannipieri P (2004) Microbial diversity in soil: ecological theories, the contribution of molecular techniques and the impact of transgenic plants and transgenic microorganisms. Biol Fertil Soils 40:363–385
Nagy ML, Perez A, Garcia-Pichel F (2005) The prokaryotic diversity of biological soil crusts in the Sonoran Desert (Organ Pipe Cactus National Monument, AZ). FEMS Microbiol Ecol 54:233–245
Nannipieri P, Ascher J, Ceccherini MT, Guerri G, Renella G, Pietramellara G (2008a) Recent advances in functional genomics and proteomics of plant associated microbes. In: Nautiyal CS, Dion P (eds) Molecular mechanisms of plant and microbe coexistence. Springer, Heideleberg, pp 215–241
Nannipieri P, Ascher J, Ceccherini MT, Landi L, Pietramellara G, Renella G, Valori F (2008b) Effects of root exudates on microbial diversity and activity in rhizosphere soils. In: Nautiyal CS, Dion P (eds) Molecular mechanisms of plant and microbe coexistence. Springer, Heideleberg, pp 339–365
Pulz O, Gross W (2004) Valuable products from biotechnology of microalgae. Appl Microbiol Biotechnol 65:635–648
Ramos HJO, Roncato-Maccari LDB, Souza EM, Soares-Ramos JRL, Hungria M, Pedrosa FO (2002) Monitoring Azospirillum–wheat interactions using the gfp and gusA genes constitutively expressed from a new broad-host range vector. J Biotechnol 97:243–252
Rothballer M, Schmid M, Hartmann A (2003) In situ localization and PGPR-effect of Azospirillum brasilense strains colonizing roots of different wheat varieties. Symbiosis 34:261–279
Smalla K, Oros-Sichler M, Milling A, Heuer H, Baumgarte S, Becker R, Neuber G, Kropf S, Ulrich A, Tebbe CC (2007) Bacterial diversity of soils assessed by DGGE, T-RFLP and SSCP fingerprints of PCR-amplified 16S rRNA gene fragments: do the different methods provide similar results? J Microbiol Meth 69:470–479
Smith SR (1995) Agricultural recycling of sewage sludge and the environment. CAB International, Wallingford, UK
Stoffels M, Castellanos T, Hartmann A (2001) Design and application of new 16SrRNA-targeted oligonucleotide probes for the Azospirillum–Skermanella–Rhodocista-cluster. Syst Appl Microbiol 24:83–97
Tourlomousis P, Kemsley EK, Ridgway KP, Toscano MJ, Humphrey TJ, Narbad A (2010) PCR-denaturing gradient gel electrophoresis of complex microbial communities: a two-step approach to address the effect of gel-to-gel variation and allow valid comparisons across a large dataset. Microb Ecol 59:776–786
Trainor FR, Gladych R (1995) Survival of algae in a desiccated soil: a 35-year study. Phycologia 34:191–192
Trejo A, de-Bashan LE, Hartmann A, Hernandez J-P, Rothballer M, Schmid M, Bashan Y (2012) Recycling waste debris of immobilized microalgae and plant growth-promoting bacteria from wastewater treatment as a resource to improve fertility of eroded desert soil. Environ Exp Bot 75:65–73
Valentín-Vargas A, Chorover J, Maier RM (2013) A new standard-based polynomial interpolation (SBPIn) method to address gel-to-gel variability for the comparison of multiple denaturing gradient gel electrophoresis profile matrices. J Microbiol Meth 92:73–177
Van Veen JA, Kuikman PJ (1990) Soil structural aspects of decomposition of organic matter by micro-organisms. Biogeochemistry 11:213–233
Venables WN, Ripley BD (2002) Modern applied statistics with S, 4th edn. Springer, New York
Wang M, Chen J-K, Li B (2007) Characterization of bacterial community structure and diversity in rhizosphere soils of three plants in rapidly changing salt marshes using 16S rDNA. Pedosphere 17:545–556
Warren-Rhodes KA, Rhodes KL, Pointing SB, Ewing SA, Lacap DC, Gómez-Silva B, Amundson R, Friedmann EI, McKay CP (2006) Hypolithic cyanobacteria, dry limit of photosynthesis, and microbial ecology in the hyperarid Atacama Desert. Microb Ecol 52:389–398
Acknowledgments
At CIBNOR, we thank Cristina Galaviz, Manuel Moreno, and Juan-Pablo Hernandez for technical assistance and Ira Fogel for editorial improvements. We thank Anton Hartmann and Michael Schmid of Helmholtz Zentrum, Munich, Germany, for the use of their confocal laser microscope. This study was supported by Consejo Nacional de Ciencia y Tecnologia of Mexico (CONACYT Basic Science-2009, 2011, grants 130656 and 164548) and time for writing by The Bashan Foundation, USA.
Author information
Authors and Affiliations
Corresponding author
Additional information
This study is dedicated to the memory of the German/Spanish mycorrhizae researcher Dr. Horst Vierheilig (1964–2011) of CSIC, Spain.
Rights and permissions
About this article
Cite this article
Lopez, B.R., Bashan, Y., Trejo, A. et al. Amendment of degraded desert soil with wastewater debris containing immobilized Chlorella sorokiniana and Azospirillum brasilense significantly modifies soil bacterial community structure, diversity, and richness. Biol Fertil Soils 49, 1053–1063 (2013). https://doi.org/10.1007/s00374-013-0799-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-013-0799-1