Abstract
A study on the diversity, phylogeny, and host specificity of soybean (Glycine max L.) and peanut (Arachis hypogaea L.) bradyrhizobia was conducted based on the 16S ribosomal RNA (rRNA) restriction fragment length polymorphisms (RFLPs), 16S rRNA sequencing, and 16S–23S rRNA intergenetic spacer (IGS) RFLP assays. Based on 16S rRNA RFLP assay, tested bradyrhizobia were divided into five genotypes, which could be further clustered into five groups by IGS RFLP assays. According to the 16S rRNA sequencing, strains of IGS-II, IV, and V were phylogenetically related to Bradyrhizobium liaoningense, Bradyrhizobium japonicum, and Bradyrhizobium elkanii, while strains of IGS-Ic and IGS-III related to Bradyrhizobium yuanmingense and Bradyrhizobium canariense, respectively. All isolates could crossly nodulate Phaseolus vulgaris, forming small white nodules. Strains of IGS-II originally isolated from peanut could efficiently nodulate Glycine soja, and two strains isolated from soybean could also nodulate peanut.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean (Glycine max L.) and peanut (Arachis hypogaea L.) are broadly cultivated in many countries and play important roles in the sustainable agriculture and local economy. Associated with legume host, rhizobia is indispensable in biological nitrogen fixation. Although fast-growing rhizobia have been discovered, slow-growing bradyrhizobia are predominant population in soybean and peanut rhizobia. Strains of bradyrhizobia have miscellaneous host specificity and outstanding ecological adaptability because they not only inhabit in soil and rhizosphere but also can inhabit aquatic ecosystems and nodulating Aeschynomene species (So et al. 1994; Willems et al. 2000). They can nodulate legumes, nonlegume Parasponia andersonii as the nitrogen fixation endosymbionts (Barrera et al. 1997; Vinuesa et al. 1998; Pueppke and Broughton 1999; Han et al. 2005), or even in rice as the endophytic bacteria (Chaintreuil et al. 2000).
Until present, six bradyrhizobia species have been identified (Jordan 1982; Kuykendall et al. 1992; Xu et al. 1995; Yao et al. 2002; Rivas et al. 2004; Vinuesa et al. 2005). Among them, Bradyrhizobium japonicum, Bradyrhizobium elkanii, and Bradyrhizobium liaoningense have been originally isolated from soybean. Recent investigations on soybean bradyrhizobia isolated from America (van Berkum and Fuhrmann 2000; de Fátima Loureiro et al. 2007), tropical Africa (Abaidoo et al. 2000), Japan, Southeast Asia (Sameshima et al. 2003), and China (Yang et al. 2006) have demonstrated that they are more diverse than the three species mentioned above. Reported rhizobia nodulating on peanut are all slow-growing bradyrhizobia and described as Bradyrhizobium spp. until now. However, whereas some peanut bradyrhizobia have been identified, others are still uncharacterized (van Rossum et al. 1995; Urtz and Elkan 1996; Zhang et al. 1999; Yang et al. 2005).
China is the origin center of soybean, and peanut is also cultivated since more than 500 years ago. Studies on soybean and bradyrhizobia from cropping zones of China revealed that they hold rich diversity. B. japonicum, B. liaoningense, and B. elkanii were identified but the dominant groups of the isolates from typical geographical regions were phylogenetically divergent from identified species, and their taxonomy statues are not clear (Yang et al. 2005, 2006). Moreover, considering the promiscuous hosts of bradyrhizobia and the similar ecological niches in which soybean and peanut are cultivated, it could be speculated that there is a certain phylogenic connection existing between each other. Systematic analysis can clarify the taxonomy of bradyrhizobia and the groups. In addition, the bradyrhizobia collected could be potentially used as bacterial inoculants for the cultivation of soybean and peanut (Anandham et al. 2007).
For this reason, we have conducted a study on diversity, phylogeny, and host specificity of soybean and peanut bradyrhizobia isolated from different geographical regions of China by using restriction fragment length polymorphisms (RFLP) analysis of 16S and 16S–23S ribosomal RNA (rRNA), internally transcribed spacer polymerase chain reaction (PCR)–RFLP, and 16S rRNA sequencing.
Materials and methods
Bacterial strains and DNA extraction
Plant sample collection and rhizobia isolation were conducted mainly according to the procedures described by Vincent (1970). Plants were collected from 12 long-time soybean and peanut cropping regions, Dedu (E 126.17, N 48.50), Baoqin (E 132.17, N 46.33), Zhengzhou (E 113.42, N 34.44), Shijiazhuang (E 114.28, N 38.02), Guangxi (E 108. 20, N 22.48), Wuming (E 108.27, N 23.17), Hongan (E 114.61, N 31.29), Jingzou (E 112.17, N 30.16), Henan (E 113.42, N 34.44), Anhiu (E 117.27, N 31.86), Wuchang (E 114.33, N 30.35), and Shandong (E 117.00, N 36.38). Root nodules randomly picked up from plants were surface sterilized in 2% sodium hypochlorite, crushed, and streaked on yeast extract mannitol agar medium plates. Individual isolates were established from a single colony. The nodulation capacity of isolated strains was confirmed by plant nodulation test on their original cultivar. Twenty-four slow-growing strains from soybean and 23 from peanut were randomly selected for the analysis described below. The strains were tested and their geographical origins are listed in Table 1.
Strains cultured in yeast extract mannitol liquid medium at 28°C for 48h were harvested. The pellets were washed twice with Tris–ethylenediaminetetraacetic acid (EDTA) buffer (Tris 10mM, EDTA 1mM, NaCl 50mM, pH 8.0) and then total DNA was extracted by the hexadecyltrimethyl ammonium bromide method (Wilson et al. 1989).
PCR–RFLP of 16S rRNA gene
Primers fD1 (5′-CCCGGGATCCAAGCTTAAGGAGGTGATCCAGCC-3′) and rD1 (5′-CCGAATTCGTCGACAACAGAGTTTGATCCTGGCTCAG-3′) corresponding to 16S rRNA gene positions 8–27 and 1524–1540 of Escherichia coli, respectively, were used for PCR amplification using Thermocycler controller (M J Research Inc.) with the following procedure: 94°C 4min for initial denaturation, 32 cycles of 94°C 1min, 56°C 1min, and 72°C 2min, 72°C 6min for the last prolongation. One-hundred-microliter volume contained 1 × buffer, 2.5mM MgCl2, 0.2mM deoxyribonucleotide triphosphate, primer fD1 0.5μM, primer rD1 0.5μM, 2U Taq DNA polymerase, and 20ng total DNA as template. Five units of restriction endonucleases, DdeI, HaeIII, HhaI, HinfI, MspI, and RsaI (Promega) were used separately to digest 100-ng PCR product separately. The restriction fingerprints were recorded by Gel Image System (Kodak, Inc.) after separating the DNA fragments on 3% (w/v) agarose gels at 80–85mV for approximately 2.5h.
16S rRNA gene sequencing and evolutionary analysis
PCR products of 16S rRNA gene of representative strains were purified and cloned into the pGEM-T vector (Promega). T7 promoter primer (5′-TAA TAC GAC TCA CTA TAG GG-3′), SP6 promoter primer (5′-CAT ACG ATT TAG GTG ACA CTA TAG-3′), and a series of medium primers were used to sequence the double strands of 16S rRNA gene by ABI3730 DNA sequence analyzer.
The sequences were aligned using the Clustal W Multiple Alignment program of BioEdit Sequence Alignment Editor Software package (North Carolina State University). Aligned sequences were analyzed using the Molecular Evolutionary Genetics Analysis 2 software, version 2.1, which was further used to produce bootstrap dendrogram reflecting the distance between test representatives and reference strains by neighbor-joining method according to the model of Kimura (1980) 2-parameter.
16S–23S rRNA IGS PCR–RFLP
Primer of pHr (5′-TGCGGCTGGATCACCTCCTT-3′) corresponding to positions 1518–1541 of 16S rRNA gene and p23SRO1 (5′-GGCTGCTTCTAAGCCAAC-3′) corresponding to positions 1069–1052 of 23S rRNA gene of E. coli were used for PCR amplification. PCR was carried out in a 100-μl reaction volume and the amplification procedure was as described previously (Deya et al. 1995). An aliquot of 100-ng PCR products were digested with 5U restriction endonucleases, HaeIII, HhaI, HinfI, and MspI (Promega) in each reaction. The RFLP patterns were recorded as described above.
Clustering analysis
The band patterns of RFLP fingerprints were converted into a two-dimensional binary matrix through a binary scoring system (1 for the presence of a band and 0 for the absence). Then, the similarity of strains tested was evaluated by simple matching coefficient. With the assistance of the NTSYS software package (Applied Biostatistic Inc.), a dendrogram was constructed from the distance matrix by the means of Unweighted Pair Group Method with Arithmetic mean (UPGMA) algorithm, and the Mantel test (Mantel 1967) was also conducted for linear correlation between geographic origin and genetic distance.
Host specificity test
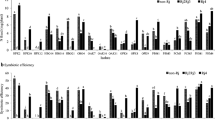
Seeds of Glycine soja, A. hypogaea, Phaseolus vulgaris, and Vicia sativa were surface sterilized in a 2% sodium hypochlorite solution for 3min and washed with sterile water six times. After germination, three seedlings were transplanted into Leonard jars with vermiculite–perlite substrate and nitrogen-free low-nutrient solution for nodulation assays. Suspensions of strains tested were inoculated into the plant roots, respectively. B. japonicum USDA6, B. elkanii USDA76, Rhizobium leguminosarum USDA2370, and Rhizobium etli CFN42 were used as the control. Plants were grown in greenhouse for 28 days. The symbiotic efficiency of the nodules was estimated from the pink color of nodules and significant differences in plant growth compared with the control plants.
Results
16S rRNA PCR–RFLP patterns
A dendrogram was generated based on the combined 16S rRNA restriction patterns by UPGMA algorithm (Fig. 1). Strains tested and reference strains were clustered into seven genotypes (Table 1). Genotype I consisted of GXD1 isolated from soybean, JZ1 isolated from peanut, B. japonicum, and B. liaoningense. Strains of genotype II was solely isolated from peanut (HA1) in Hongan, China. B. japonicum USDA110 was solely clustered into genotype III. Soybean strain (BQ3 and DD3) isolated from North and Northeast China clustered into genotype IV. The RFLP patterns showed that this group was genetically different from B. japonicum, B. liaoningense, and B. elkanii. Genotype V comprised strains isolated from peanut (SD5 and WC4) and clustered with Bradyrhizobium yuanmingense CCBAU10071, which belongs to genotype VI. Three strains isolate from soybean (HAS5) and B. elkanii belongs to genotype VII.
Distance analysis
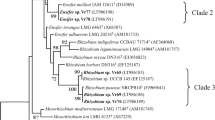
A bootstrap phylogenetic dendrogram was generated by neighbor-joining method (Fig. 2). With high confidence, the representative strains and the reference strains were divided into five groups. Strains JZ1 and GXD1 clustered with B. liaoningense 2281. Similar with 16S rRNA RFLP, strain WC4 and SD5 isolated from peanut clustered with B. yuanmingense. Strain BQ3 and DD3 were clustered with Bradyrhizobium canariense. Strain HA1 clustered with B. japonicum USDA110 and USDA122. It was noticed that B. japonicum and B. liaoningense sharing the same genotype in 16S rRNA RFLP were divided into two groups in this assay, thus reflecting the difference of the two procedures on the resolution capacity. HAS5 from soybean was clustered with the reference strains of B. elkanii.
Phylogenetic relationship between the representatives of isolated bradyrhizobia and reference strains of Bradyrhizobium based on aligned sequences of 16S rRNA. Kimura-2 distances were derived from a distance matrix to construct an optimal unrooted tree using the neighbor-joining method. Bootstrap values, expressed as a percentage of 1,000 replications, are given at the branching. Numbers in parentheses are the accession numbers of the sequences used. Open line box indicate the clusters with the similarity higher than 99%
16S–23S rRNA IGS PCR–RFLP
The 16S–23S rRNA intergenetic spacer (IGS) region contained the intergenetic space between 16S rRNA and 23S rRNA and 1-kb size of the 5′-end sequence of 23S rRNA. All strains tested produced a single band ranging from 1,900 to 1,950bps, which could be partially explained by the varying of the conserved block region in the IGS region.
Tested strains were clustered into five groups by considering 80% of similarity. Strains represented by JZ1 and GXD1 was separately clustered into IGS-Ia and IGS-Ib, and strains represented by WC4 and SD5 were clustered into IGS-Ic. Peanut strains from Hongan were solely clustered into IGS-II, while soybean strains from same region were clustered into IGS-IV and IGS-V. BQ3 and DD3 were clustered into IGS-III (Fig. 3).
Plant specificity test
Beside nodulating their original host, all isolates could nodulate P. vulgaris, forming small white nodules, which did not show nitrogen fixation. The IGS-II strains originally isolated from peanut could efficiently nodulate G. soja, which further demonstrated that this group was related to B. japonicum and B. liaoningense originally isolated from soybean (Table 2). Two IGS-Ia strains isolated from peanut could also nodulate soybean, and two strains isolated from soybean could also nodulate peanut. The cross nodulation capacity between these groups reflected their tightly phylogenic relationship. However, IGS-III, IV, and V strains did not show the cross nodulation capacity between their original hosts.
Discussion
Diversity and phylogeny of soybean and peanut bradyrhizobia
As a heterogeneous group, bradyrhizobia is always undergoing differentiation. In this study, tested rhizobia isolated from 13 geographical regions could be divided into five groups. Some strains isolated from peanut and soybean and reference strains of B. japonicum and B. liaoningense form a complex group. Several of them have the cross nodulation capacity. Further assays by 16S rRNA gene sequencing and IGS RFLP showed that this group could be further divided into three subgroups. In addition, these data suggest that the main bradyrhizobia population in China is divergent from the population in Japan and North America; a similar phenomenon was also found in strains isolated from other regions (Abaidoo et al. 2000; Sameshima et al. 2003).
Group III consisted of strains isolated from soybean in North and Northeast China. Four assays coincidently revealed that this group was phylogenetically divergent from other soybean rhizobia groups. Phylogeny analysis based on the 16S rRNA gene sequencing revealed that this group was phylogenetically related to B. canariense, which originally isolated from endemic genistoid legumes and was regarded as sister species of B. japonicum. Strains in Group IV were isolated from peanut. Phylogenetical analysis revealed that this group was related to B. yuanmingense isolated from Lespedeza.
Relationship between host plant and rhizobia
The establishment of the nodulation and nitrogen fixation is a complicated interaction or “dialogue” between rhizobia and legume plants. Among these procedures, the molecular recognition between rhizobia and host plants is a critical step in determining the host range of rhizobia (Freiberg et al. 1997; Perry et al. 2007). Some strains showed strict bacterial host specificity such as Mesorhizobium huakuii, which can only nodulate the legume species Astragalus sinicus (Cheng et al. 2006), while some rhizobia represented by strain NGR234 can nodulate a very broad host range including nonlegume Parasponia (Pueppke and Broughton 1999).
In this study, the cross nodulation of strains from soybean and peanut was observed. Strains from Hongan, China can crossly nodulate peanut and G. soja clustered into a solo group. 16S rRNA sequencing assay revealed that this group is still related to B. japonicum.
In addition, the predominant peanut bradyrhizobia are related to B. yuanmingense, which is isolated from Lespedeza. Recent study found that strains isolated from lima bean (Phaseolus lunatus) in Peru and strains isolated from the genus Pachyrhizus are related to B. yuanmingense (Rodríguez-Navarro et al. 2004; Ormeño-Orrillo et al. 2006).
Relationship between the biodiversity and geographical origin
Geographical origin is another important factor in affecting the composition and biodiversity of indigenous rhizobia. Strains restricted to an ecological niche generally hold special phenotypic and genetic characteristics and delineated according to their geographical origins (Xu et al. 1995; Vinuesa et al. 2005). The results of Mantel test between genetic distance generated from IGS RFLP patterns and geographical distance (r = 0.612, P = 0.297) indicated they are directly related. The geographical delimitation of rhizobia was distinctly displayed by strains isolated from Hongan in Central China, a watershed between the humid subtropical climate and inner-land climate. All peanut strains isolated from this region clustered into a solo group.
Soybean bradyrhizobia mainly isolated from North and Northeast China are related to B. canariense, which was initially isolated from Canary Island, Morocco (Vinuesa et al. 2005). Bradyrhizobia related to this species were isolated from a variety of legumes such as yellow serradella (Ornithopus compressus), lupins, and serrada in Italy, Western Australia, and South Africa (Stepkowski et al. 2005; Safronova et al. 2007). This study first reported that the main group of soybean bradyrhizobia in the cropping zone of North and Northeast China was related to B. canariense.
Soybean and peanut bradyrhizobia are not only important rhizosphere bacteria for the plant growth and sustainable agriculture but also an indispensable link in rhizobia phylogeny. The comparative study has resolved the diversity and phylogeny status of bradyrhizobia in main geographical regions of China. The results of this study may be useful and be compared with those involving bradyrhizobia isolated from other geographical regions of the world. The strains collected may be the potential resource for the production of inoculants to be used for increasing yields of peanut and soybean.
References
Abaidoo RC, Keyser HH, Singleton PW, Borthakur D (2000) Bradyrhizobium spp. (TGx) isolates nodulating the new soybean cultivars in Africa are diverse and distinct from bradyrhizobia that nodulate North American soybeans. Int J Syst Evol Microbiol 50:225–234
Anandham R, Sridar R, Nalayini P, Poonguzhali S, Madhaiyan M, Sa T (2007) Potential for plant growth promotion in groundnut (Arachis hypogaea L.) cv. ALR-2 by co-inoculation of sulfur-oxidizing bacteria and Rhizobium. Microbiol Res 162:139–153
Barrera LL, Trujillo ME, Goodfellow M, Garcia FJ, Hernamdez-Lucas L, Davila G, van Berkum P, Martinez-Romero E (1997) Biodiversity of bradyrhizobia nodulating Lupinus spp. Int J Syst Bacteriol 47:1086–1091
Chaintreuil C, Giraud E, Prin Y, Lorquin J, Ba A, Gillis M, de Lajudie P, Dreyfus B (2000) Photosynthetic bradyrhizobia are natural endophytes of the African wild rice Oryza breviligulata. Appl Environ Microbiol 66:5437–5447
Cheng GJ, Li YG, Zhou JC (2006) Cloning and identification of opa22, a new gene involved in nodule formation by Mesorhizobium huakuii. FEMS Microbiol Lett 257:152–157
de Fátima Loureiro M, Kaschuk G, Alberton O, hungria M (2007) Soybean (Glycine max (L.) Merrill) rhizobial diversity in Brazilian oxisols under various soil, cropping, and inoculation managements. Biol Fertil Soils 43:665–674
Deya AAM, Odelson DA, Hickey RF, Tidje JM (1995) Bacterial community fingerprinting of amplified 16S and 16S–23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA). In: Akkermans DL, van Elsas JD, de Bruijn FJ (eds) Molecular microbial ecology manual. vol. 3.3.2. Kluwer Academic, Dordrecht, pp 1–8
Freiberg C, Fellay R, Bairoch A, Broughton WJ, Rosenthal A, Perret X (1997) Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394–401
Han SZ, Wang ET, Chen WX (2005) Diverse bacteria isolated from root nodules of Phaseolus vulgaris and species within the genera Campylotropis and Cassia grown in China. Syst Appl Microbiol 28:265–276
Jordan DC (1982) Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing root nodule bacteria from leguminous plants. Int J Syst Bacteriol 32:136–139
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kuykendall LD, Saxena B, Devine EE, Udell SE (1992) Genetic diversity in Bradyrhizobium japonicum Jordan 1982, and a proposal for Bradyrhizobium elkanii sp. nov. Can J Microbiol 38:501–505
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Ormeño-Orrillo E, Vinuesa P, Zúñiga-Dávila D, Martínez-Romero E (2006) Molecular diversity of native bradyrhizobia isolated from lima bean (Phaseolus lunatus L.) in Peru. Syst Appl Microbiol 29:253–262
Perry LG, Alford ER, Horiuchi J, Paschke MW, Vivanco JM (2007) Chemical signals in the rhizosphere root–root and root–microbe communication. In: Pinton R, Varanini Z, Nannipieri P (eds) The rhizosphere biochemistry and organic substances at the soil–plant interface. 2nd edn. CRC, Boca Raton, pp 297–330
Pueppke SG, Broughton WJ (1999) Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol Plant Microbe Interact 12:293–318
Rivas R, Willems A, Palomo JL, Garcia-Benavides P, Mateos PF, Martinez-Molina E, Gillis M, Velazquez E (2004) Bradyrhizobium betae sp. nov. isolated from roots of Beta vulgaris affected by tumour-like deformations. Int J Syst Evol Microbiol 54:1271–1275
Rodríguez-Navarro DN, Camacho M, Leidi EO, Rivas R, Velázquez E (2004) Phenotypic and genotypic characterization of rhizobia from diverse geographical origin that nodulate Pachyrhizus species. Syst Appl Microbiol 27:737–745
Safronova V, Chizhevskaya E, Bullitta S, Andronov E, Belimov A, Charles TC, Lindström K (2007) Presence of a novel 16S-23S rRNA gene intergenic spacer insert in Bradyrhizobium canariense strains. FEMS Microbiol Lett 269:207–212
Sameshima R, Isawa T, Sadowsky MJ, Hamada T, Kasai H, Shutsrirung A, Mitsui H, Minamisawa K (2003) Phylogeny and distribution of extra-slow-growing Bradyrhizobium japonicum harboring high copy numbers of RSa, RSb and IS1631. FEMS Microbiol Ecol 44:191–202
So RB, Ladha JK, Young JP (1994) Photosynthetic symbionts of Aeschynomene spp. Form a cluster with bradyrhizobia on the basis of fatty acid and rRNA analyses. Int J Syst Bacteriol 44:392–403
Stepkowski T, Moulin L, Krzyzańska A, McInnes A, Law IJ, Howieson J (2005) European origin of Bradyrhizobium populations infecting lupins and serradella in soils of Western Australia and South Africa. Appl Environ Microbiol 71:7041–752
Urtz BE, Elkan GH (1996) Genetic diversity among Bradyrhizobium isolates that effectively nodulate peanut (Arachis hypogaea). Can J Microbiol 42:1121–1130
van Berkum P, Fuhrmann JJ (2000) Evolutionary relationships among the soybean bradyrhizobia reconstructed from 16S rRNA gene and internally transcribed spacer region sequence divergence. Int J Syst Evol Microbiol 50:2165–2172
van Rossum D, Schuurmans FP, Gillis M, Tcha AM, van Verseveld HW, Stouthamer AH, Boogerd FC (1995) Genetic and phenetic analyses of Bradyrhizobium strains nodulating peanut (Arachis hypogaea L.) roots. Appl Environ Microbiol 61:1599–1609
Vincent JM (1970) A manual for the practical study of root-nodule bacteria. International biological programme handbook. Blackwell Scientific, Oxford, pp 73–97
Vinuesa P, Rademaker JLW, de Bruijn FJ, Werner D (1998) Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S–23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl Environ Microbiol 64:2096–2104
Vinuesa P, Leon-Barrios M, Silva C, Willems A, Jarabo-Lorenzo A, Perez-Galdona R, Werner D, Martinez-Romero E (2005) Bradyrhizobium canariense sp. nov., an acid-tolerant endosymbiont that nodulates endemic genistoid legumes (Papilionoideae: Genisteae) from the Canary Islands, along with Bradyrhizobium japonicum bv. genistearum, Bradyrhizobium genospecies alpha and Bradyrhizobium genospecies beta. Int J Syst Evol Microbiol 55:569–575
Willems A, Doignon-Bourcier F, Coopman R, Hoste B, de Lajudie P, Gillis M (2000) AFLP fingerprint analysis of Bradyrhizobium strains isolated from Faidherbia albida and Aeschynomene species. Syst Appl Microbiol 23:137–147
Wilson K (1989) Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Srtruhl K (eds) Current protocols in molecular Biology. Greene Publishing Associates/Wiley-Interscience, New York, pp 2.4.1–2.4.5
Xu LM, Ge C, Cui Z, Li J, Fan H (1995) Bradyrhizobium liaoningense sp. nov., isolated from the root nodules of soybeans. Int J Syst Bacteriol 45:706–711
Yang JK, Xie FL, Zou J, Zhou Q, Zhou JC (2005) Polyphasic characteristics of bradyrhizobia isolated from nodules of peanut (Arachis hypogaea) in China. Soil Biol Biochem 37:141–153
Yang JK, Zhang WT, Yuan TY, Zhou JC (2006) Genotypic characteristics of the rrn operon and genome of indigenous soybean bradyrhizobia in cropping zones of China. Can J Microbiol 9:968–976
Yao ZY, Kan FL, Wang ET, Wei GH, Chen WX (2002) Characterization of rhizobia that nodulate legume species of the genus Lespedeza and description of Bradyrhizobium yuanmingense sp. nov. Int J Syst Evol Microbiol 52:2219–2230
Zhang X, Nick G, Kaijalainen S, Terefewirjm Z, Paulin L, Tighe SW, Graham PH, Lindstrom K (1999) Phylogeny an diversity of Bradyrhizobium strains isolated from the root nodules of peanut (Arachis hypogaea) in Sichuan, China. Syst Appl Microbiol 22:378–386
Acknowledgements
This work was granted by Chinese High-tech Developing Program 2007AA05Z417, Chinese Microbe Resource Project 2005DKA21208-6 and Grant of State Key Laboratory of Agricultural Microbiology, HAU, China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, J.K., Zhou, J.C. Diversity, phylogeny and host specificity of soybean and peanut bradyrhizobia. Biol Fertil Soils 44, 843–851 (2008). https://doi.org/10.1007/s00374-008-0269-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-008-0269-3