Abstract
Aims
Understanding the factors that influence the diversity of soybean-nodulating rhizobia is important before doing inoculation. Since studies about this topic in tropical regions are limited, this could lay the groundwork for related research particularly on Bradyrhizobium elkanii.
Methods
To determine the genetic diversity of B. elkanii in different regions, we conducted Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) and sequence analysis of 16S rRNA gene, internal transcribed spacer (ITS) region and rpoB gene. Also, sequence analysis of symbiotic nifD and nodD1 genes was conducted.
Results
Analysis of the rpoB gene revealed a higher genetic diversity than the ITS region, and possible endemic B. elkanii strains were observed. Meanwhile, no variation was detected among the strains in both nifD and nodD1 phylogenies. Through rpoB gene analysis, variations in the ITS-rpoB type of B. elkanii strains were distinguished and differentiated with that of the closest reference strains. We identified potential soybean inoculants which possess symbiotic efficiency regardless of the Rj genotypes used, suggesting broad host-range of the strains.
Conclusions
We show how the genetic diversity of soybean-nodulating B. elkanii strains in subtropical and tropical regions might be influenced by temperature and soil pH and, provided some insights between the symbiotic genes and Rj genotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean (Glycine max [L.] Merrill.) is a high protein legume (40%) and can be used as food, animal feed and as an industrial raw material. This legume can establish a symbiotic relationship with the nitrogen-fixing bacteria, known as rhizobia. Recent literatures reported that there are more than 100 species of rhizobia which were isolated from legumes and other sources (Gyaneshwar et al. 2011; Peix et al. 2015) and currently, the major soybean-nodulating rhizobia that were identified are as follows: Bradyrhizobium japonicum, Bradyrhizobium diazoefficiens, Bradyrhizobium elkanii, and Sinorhizobium/Ensifer fredii (Jordan 1982; Scholla and Elkan 1984; Kuykendall et al. 1992; Young 2003; Delamuta et al. 2013). In soybean, some cultivars possess nodulation regulatory genes known as Rj genes, and the genotypes which have been confirmed to exist in nature are non-Rj, rj 1, Rj 2, Rj 3, and Rj 4 (Devine and Kuykendall 1996).
Unlike Japan, wherein soybean plays an important role in daily cuisine, soybean has very minimal role in Filipinos’ diet since 90% of local production and importation are used for animal feed (Manuel et al. 1986). Nevertheless, soybean production in both countries cannot supply its local demand as reflected in the amount of soybean importation (Manuel et al. 1986; Wang 2016). One way to increase soybean yield per unit area is through inoculation. Inoculating useful rhizobia may lead to an increase in the yield of soybean as revealed by several studies (Alves et al. 2003; Njira et al. 2013; Alam et al. 2015; Sanz-Sáez et al. 2015). However, soybean inoculation does not always succeed due to competition between the inoculated and indigenous rhizobia in the soil (Yamakawa et al. 2003). Thus, there is a need to first understand the ecology of indigenous rhizobia in the soil before inoculation should be conducted.
Investigation of diversity and distribution of indigenous soybean rhizobia in Japan identified that B. japonicum strains were dominant in the northern part whereas B. elkanii strains were dominant in the southern part (Suzuki et al. 2008; Saeki et al. 2006, 2008, 2010, 2013) and temperature was the most influential factor in its dominance (Saeki and Shiro 2014). However, no studies about indigenous soybean rhizobia have been done in the Philippines as of this time.
Diversity analyses of soybean rhizobia in Japan were conducted mainly by analyzing the 16S rRNA gene and the ITS region between the 16S and 23S rRNA gene (Saeki et al. 2006, 2008, 2013; Suzuki et al. 2008; Minami et al. 2009; Saeki and Shiro 2014). But, a major disadvantage of 16S rRNA gene in taxonomic studies is that it is often multiple-copy (Vos et al. 2012) and has little resolution below the species level (Germano et al. 2006; Martens et al. 2008). Meanwhile, ITS region and rpoB gene provided better discriminatory power up to species level and below (Martens et al. 2008; Vinuesa et al. 2008; Delamuta et al. 2012; Vos et al. 2012; Degefu et al. 2013; Yan et al. 2014; Guimarães et al. 2015). Therefore, it is better to analyze other genes in addition to ITS region to provide better identification of rhizobial strains. However, taxonomical studies of rhizobia do not necessarily reflect their symbiotic features, particularly their host range, which is an important character of a suitable inoculant. Thus, sequence analysis of symbiotic genes nifD (encoded the α subunit of dinitrogenase) and nodD1 (nodulation regulation protein) was also conducted. Several studies reported that genes located in symbiosis island might not show diversity even among related species in rhizobial genera commonly due to horizontal gene transfer as directed by their location (Minamisawa et al. 2002; Barcellos et al. 2007; Ramirez-Bahena et al. 2009; Ling et al. 2016). But the role of NodD regulator proteins (including nodD1) in activating the transcription of nod genes is known to be a key factor that influences the competitiveness of rhizobia (Maj et al. 2010) due to its assumed specific interaction with flavonoids (Redmond et al. 1986; Zaat et al. 1989). Hence, it is an important genetic marker to be included for evaluation of potential inoculant strains. Meanwhile, the role of nifD in partner quality for Rhizobium was investigated and reported that there might be a causal relationship between the locus and measures of partner quality (Gordon et al. 2016) which in turn, could influence mutualism between macro and microsymbionts for effective N fixation.
Majority of the studies about diversity and distribution of soybean rhizobia were conducted in temperate and subtropical regions of Japan (Ikeda et al. 2008, 2010; Nguyen et al. 2010; Saeki et al. 2006, 2008, 2010; Shiro et al. 2012; Suzuki et al. 2008) but there is limited research about this topic in tropical regions (Loureiro et al. 2007; Sharma et al. 2010; Ansari et al. 2013). Tropical rhizobia represent a key component for the sustainability of tropical soils; and the genus Bradyrhizobium, which is considered to be the ancestral of all nitrogen-fixing rhizobial species, was thought to be originated from the tropics (Delamuta et al. 2012). Even in the subtropical and tropical regions of China, which is said to be the center of diversification of G. max, the diversity of soybean rhizobia has not yet been clearly described (Man et al. 2008). Therefore, this study would be a helpful foundation for future research and studies about the diversity and endemism of soybean rhizobia in subtropical and tropical regions. It aimed to determine the possible endemism and genetic diversity of soybean-nodulating B. elkanii species between three different geographical regions and provide the first report in the Philippines and in Kumamoto, Japan.
Materials and methods
Soil collection
Soil samples were collected from three field sites (Kumamoto: Kumamoto Prefectural Agricultural Research Center, Goshi, Kumamoto, Japan and Okinawa: University of the Ryukyu, Nishihara, Okinawa, Japan and Nueva Ecija: Central Luzon State University, Nueva Ecija, Philipppines) previously planted with soybean and/or other legumes. The surface was cleared with litters before obtaining a bar of soil with dimension of 20 cm depth and 2 to 3 cm thickness and weighed approximately 1 kg. Half of the 1 kg soil sample was air dried and pulverized for soil pH and electrical conductivity (EC) analyses by water extraction method (1:2.5 soil: water for pH and 1:5 soil: water for EC) whereas the remaining 0.5 kg was freshly used for soybean cultivation. Data of annual average temperature from Nueva Ecija, Philippines was obtained from Philippine Atmospheric Geophysical and Astronomical Services Administration (PAGASA) Central Luzon State University (CLSU) station while temperature data from Kumamoto and Okinawa, Japan were obtained from Japan Meteorological Agency website at http://www.data.jma.go.jp/. All data were averages from this last decade.
Isolation of indigenous soybean rhizobia
Three soybean cultivars of three Rj genotypes, Bragg (BM) or Akishirome (AK) as non-Rj, CNS (CM) or Bonminori (BO) as Rj 2 Rj 3, and Hill (HM) or Fukuyutaka (FK) as Rj 4 were used to isolate the indigenous soybean rhizobia. Each soybean cultivar was planted in 1-l culture pots (n = 3). Culture pots were filled with vermiculite containing N-free nutrient solution (Saeki et al. 2000) at 40% (vol/vol) water content then, were autoclaved at 121 °C for 20 min. Soybean seeds were surface-sterilized by soaking in 70% ethanol for 30 s then, in a diluted sodium hypochlorite solution (0.25% available chlorine) for 3 min. Afterwards, the seeds were washed with sterile distilled water. Soil sample (2 to 3 g) was placed on the vermiculite at a depth of 2 to 3 cm, the seeds were then sown on the soil, and the pot was weighed. Plants were grown for 4 weeks in a growth chamber (day, 28 °C for 16 h; night, 23 °C for 8 h), and were supplied weekly with sterile distilled water until the initial weight of the pot was reached.
After 4 weeks, 24 nodules were randomly collected from the soybean roots per Rj genotype and sterilized by soaking them in 70% ethanol for 3 min and in a diluted sodium hypochlorite solution (0.25% available chlorine) for 30 min; then washed with sterile distilled water. Each nodule was homogenized in sterile distilled water, streaked onto a yeast extract mannitol agar (YMA; Vincent 1970) plate medium, and incubated for about 1 week in the dark at 28 °C. A single colony was streaked onto YMA plate containing 0.002% (wt/wt) bromothymol blue (Keyser et al. 1982) to determine the genus then, incubated as described above.
Inoculation test
From the primary 16S rRNA gene and ITS region RFLP analysis of all collected isolates, representative isolates were selected and tested for their capability to form nodules on host soybean by inoculation test with the three Rj genotypes of soybean cultivars used in this study. Each isolate was cultured in YM broth (Vincent 1970) inside a dark shaker with continuous agitation at 28 °C for 1 week. Afterwards, the cultures were diluted with sterile distilled water to approximately 106 cells ml−1. Then soybean seeds were sown as described above but without soil and inoculated with 1 ml aliquot of each isolate per seed, replicated thrice. Nodule formation was assessed after 4 weeks in growth chamber under similar conditions mentioned above. Control pots (un-inoculated) for both Japanese (AK, BO, FK) and US (BM, CM, HM) cultivars were also prepared under similar conditions.
The nodule number and its dry weight for each Rj genotype as well as the dry weight of shoot were obtained for symbiotic analysis. Oven drying was done at 70 °C for 48 h. Shoot was finely ground into 2 mm size prior to Nitrogen analysis. Total N was analyzed by automatic high sensitive NC Analyzer Sumigraph NC-220F (Sumika Chemical Analysis Service. Ltd., Tokyo, Japan). Amount of N fixed was computed from the difference between the shoot N content of the isolates with that of the control plants. The symbiotic efficiency of the isolates was obtained by the following formula: (mg N fixed/mg dry nodule) × 100 (Risal et al. 2010). Statistical analysis was conducted employing R software (v. 3.3.2) and means of three replicates were compared by Tukey’s HSD test at P < 0.05. The comparison among means were conducted only between each isolate within the same Rj genotype and not between each Rj genotype.
DNA extraction
Each isolate was cultured in HEPES-MES (HM) broth culture (Cole and Elkan 1973; Sameshima et al. 2003) for 6 days at 28 °C with continuous agitation at 120 rpm, and the bacteria cells cultured in the HM medium were collected by centrifugation and washed with sterile distilled water. Extraction of DNA was done by using BL buffer as described (Minami et al. 2009) from the method reported by Hiraishi et al. (1995). Similar method was done for the DNA extraction of reference strains.
PCR amplification of 16S rRNA gene, ITS region, rpoB gene and symbiotic genes nifD and nodD1
Amplification of target genes were conducted using Ex Taq DNA polymerase (TaKaRa Bio, Otsu, Shiga, Japan) and previously designed primers (Table S1). The PCR cycle consisted of a pre run at 94 °C for 5 min, 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min, with a final post run extension at 72 °C for 10 min. This cycle was used for all the five (5) target genes except that: for 16S rRNA gene, denaturation was done for 1 min while for rpoB gene, annealing was done at 60 °C for 1 min; then annealing was conducted at 57 °C for 1 min for both nifD and nodD1 genes.
RFLP analysis
The RFLP analysis of the ITS region was performed using the restriction enzymes HaeIII, HhaI, MspI and XspI (TaKaRa Bio) whereas for rpoB gene, HaeIII, MspI and AluI (TaKaRa Bio) were used. Bradyrhizobium USDA strains B. japonicum 4, 6T, 38, 122, 123, 124, 129, 135, B. diazoefficiens 110 T, B. elkanii 31, 46, 76T, 94, and 130 and B. liaoningense 3622T (Saeki et al. 2004) were used as reference strains for the RFLP analysis of 16S rRNA gene, ITS region and rpoB gene. A 2.5 μl aliquot of the PCR product was digested with the restriction enzymes at 37 °C for 16 h in a 10 μl reaction mixture. The restriction fragments were separated on 3 or 4% agarose gels in TBE buffer by means of electrophoresis and visualized with ethidium bromide.
Selection of representative isolates for sequence analysis
After collecting all the isolates that formed nodules with soybean, Bradyrhizobium species were differentiated from each other. This was done first by observing the differences in the colony morphology then confirmed by primary RFLP analysis of the 16S rRNA gene and ITS region (data not shown). Since almost all isolates collected from Nueva Ecija, Philippines belonged to Be 76 cluster, then only those isolates from Kumamoto and Okinawa, Japan, which also belonged to Be76 cluster were considered. Then, RFLP analysis of ITS region and rpoB gene were conducted for the selected isolates and based from the band pattern, random samples were further selected for sequence analysis of 16S rRNA, ITS region, rpoB, nifD and nodD1 genes.
Sequence analysis
The PCR amplified products were purified according to the protocol of NucleoSpin® Gel and PCR Clean-up (Macherey-Nagel, Germany). The DNA concentration of the purified product was determined by using NanoDrop 2000 Spectrophotometer (Thermo Scientific, U.S.A.).
Preparation of samples for sequence analysis from purified DNA followed the protocol for the premixed template and primer of the company (EUROFINS GENOMICS). After preparation, samples were sent to the company for sequence analysis. The sequence primers (Table S1) used were all designed for this study and calculated using OligoEvaluator™ by Sigma-Aldrich® Co. LLC.
Sequence alignment and construction of phylogenetic trees
To search the homology of sequences, Basic Local Alignment Search Tool (BLAST) program in DNA Databank of Japan (DDBJ) was used. Those sequences of type strains having 100% similarity with our isolates were retrieved from BLAST database. The alignment also included sequences of Bradyrhizobium genospecies for 16S rRNA gene and ITS region which were previously determined (Saeki et al. 2004; van Berkum and Fuhrmann 2000). Alignment of sequences obtained were performed using ClustalW. Phylogeny was determined by the Neighbor-Joining (Saitou and Nei 1987) method for the 16S rRNA, ITS region, rpoB, nifD and nodD1 genes. Genetic distances were calculated using Kimura 2-parameter model (Kimura 1980) in the Molecular Evolutionary Genetic Analysis (MEGA v7) software (Kumar et al. 2016). Phylogenetic trees were bootstrapped with 1000 replications of each sequence to evaluate the reliability of the tree topology. All the nucleotide sequences determined in this study were deposited in DDBJ under accession numbers LC167347 to LC167402; LC167474 to LC167485; LC168752 to LC168753; LC217878 to LC217896 and LC218023 to LC218041 at http://www.ddbj.nig.ac.jp/ and listed in the supplementary information (Table S3).
Results
Soil pH and EC
The soils collected from Kumamoto, Japan and Nueva Ecija, Philippines were both slightly acidic (6.23 and 6.21, respectively) while Okinawa soil was very strongly acidic (4.79). The EC (dS/m) for Kumamoto, Okinawa, and Nueva Ecija were as follows: 0.088, 0.072 and 0.046, respectively, which were all within the acceptable range of EC for soybean (Bernstein et al. 1955). The annual average temperature from Nueva Ecija, Philippines and Kumamoto and Okinawa, Japan were 26.8 °C, 15.8 °C and 23.3 °C, respectively.
Isolation of soybean rhizobia and selection of representative isolates
A total of 216 isolates were obtained from the three locations (72 isolates per location) and their nodulation capability were confirmed through inoculation test. The number of isolates which belonged to Be76 cluster collected from 216 samples were 21, 42, and 71 from Kumamoto, Okinawa, and Nueva Ecija, respectively. The rest of the isolates belonged to B. japonicum USDA6T and B. diazoefficiens USDA110T and other minor B. elkanii strains. Therefore, 20 isolates which belonged to Be76 cluster only from each location were selected for final RFLP analysis of ITS region and rpoB gene which totaled to 60 isolates. Afterwards, 6 isolates from each location where randomly selected based from the different rpoB gene type that was observed from RFLP analysis. However, since some Okinawa samples showed similar ITS-rpoB type from RFLP analysis with both Kumamoto and Nueva Ecija, we selected 7 isolates from this location to represent the differences. Thus, 19 isolates were used for sequence analysis of ITS, rpoB, nifD and nodD1 whereas 12 isolates were used for 16S rRNA gene.
Nodulation and symbiotic analysis
The oven dry weight of shoot and nodules were obtained as well as nodule number for the three Rj genotypes of soybean cultivars that we used (Table S2). All the isolates were able to form nodules on both non-Rj and Rj 4 genotypes. Four (4) Kumamoto isolates (HFK2, HFK10, HFK12, HBO21) and 2 out 7 Okinawa isolates (OAK10, OFK6) did not form nodules with Rj 2 Rj 3. In contrast, all isolates from Nueva Ecija formed nodules with all the Rj genotypes used in this study. Accordingly, control plants did not produce any nodule. The highest number of nodules produced were obtained from OBO4, OFK8, PBM1 and PHM4 isolates regardless of the Rj genotpyes which were significantly different than the other isolates. HFK2, HFK10 and HBO21 showed comparably high number of nodules for both non-Rj and Rj 4 genotypes.
The amount of N fixed showed significant differences among the isolates (Fig. 1a). For non-Rj genotype, HFK2, OBO4 and PHM1 fixed the highest N; HBO14 and PHM have the highest fixed N for Rj 2 Rj 3; and for Rj 4, HFK2 fixed the highest amount of N. Some isolates did not form nodules with soybean cultivars harboring Rj 2 Rj 3 (HFK2, HFK10, HFK12, HBO21, OAK10, OFK6) which indicated the absence of symbiosis (Fig. 1b). In general, higher amount of N fixed and symbiotic efficiency were observed with plants that produced higher number of nodules for particular Rj genotype.
RFLP analysis of ITS region and rpoB gene
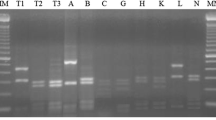
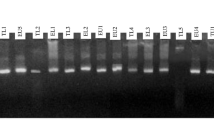
Figure 2 showed the dendrogram of the preliminary RFLP analysis of 60 B. elkanii isolates from the three locations that elucidated the clusters formed from ITS region (Fig. 2a) and rpoB gene (Fig. 2b). All the 60 B. elkanii isolates belonged to Be76 cluster for the ITS region-RFLP analysis but were divided into two clusters (Be76 and Be46) for the rpoB gene-RFLP analysis. The RFLP band patterns of all the 60 isolates for ITS region were similar to each other on all four restriction enzymes (HaeIII, HhaI, MspI and XspI) indicating that they all belonged to only one ITS type. Therefore, only the band patterns of 1 isolate per location (HBO14 – Kumamoto, OAK10 – Okinawa and PBM1 – Nueva Ecija) and B. elkanii reference strains were shown and it was clear that all isolates belonged to Be76 cluster (Fig. 3a). Meanwhile, the RFLP band patterns of the 60 isolates for rpoB gene digested with 3 restriction enzymes (HaeIII, MspI and AluI) showed two distinct band patterns similar to clusters Be76 and Be46 (data not shown). All Kumamoto isolates and three Okinawa isolates (represented by OAK11) have identical band patterns with Be76 cluster whereas, all Nueva Ecija isolates and the remaining Okinawa isolates have identical band patterns with Be46 cluster (data not shown). Therefore, one isolate per location which represented the band patterns clearly were chosen and plotted against B. elkanii reference strains (Fig. 3b) indicating the two distinct band patterns for rpoB gene.
Schematic representation of gel electrophoresis patterns based on a ITS region and b rpoB gene PCR-RFLP analysis of representative isolate per location and 6 B. elkanii reference strains. Sizes (bp) are indicated in the column. Smaller fragment sizes were not shown due to difficulty in recognition on the gel
These results showed that although the RFLP analysis of ITS region indicated that all the isolates have Be76 ITS type, the analysis of rpoB gene indicated that Okinawa (except for three isolates represented by OAK11) and Nueva Ecija samples have different rpoB type than their ITS type.
Sequence analysis of 16S rRNA, ITS region and rpoB gene
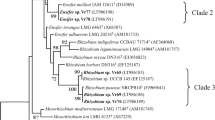
The phylogenetic tree for ITS region showed that all the isolates were grouped into Be76 cluster which included B. elkanii USDA76T, 31 and 130 with bootstrap support of 43 to 99% (Fig. 4a). Other Bradyrhizobium strains obtained from BLAST database which included Bradyrhizobium sp. CB1809, Bradyrhizobium sp. Glm-3, Bradyrhizobium sp. WB1, B. elkanii LMG 6134, B. elkanii NBRC 14791 and B. elkanii UM19 showed 100% sequence homology with B. elkanii USDA76T whereas B. elkanii LMG 6135 and B. elkanii MAS8 showed 99–100% sequence homology with B. elkanii USDA31 and 130. For simplicity, we refer to this cluster as Be76 cluster. This result was similar with RFLP analysis of ITS region and indicated that all the isolates belonged to cluster Be76.
Phylogenetic tree based on sequence analysis of a 16S–23S rRNA gene ITS region and b rpoB housekeeping gene. The tree was constructed using the Neighbor-Joining method with the Kimura 2-parameter (K2P) distance correlation model and 1000 bootstrap replications in MEGA v.7 software. The first letter of isolates’ name indicates the location as follows: H - Kumamoto; O - Okinawa; P - C. Luzon, Philippines
Meanwhile, sequence analysis of the rpoB gene revealed three (3) distinct groups under Be46 and Be76 clusters (Fig. 4b). Group I is composed of five (5) isolates from Okinawa and Group II is composed of all the six (6) isolates from Nueva Ecija along with one (1) isolate from Okinawa (OAK7). These two groups belong to Be46 cluster. Group III is composed of all the six (6) isolates from Kumamoto along with one (1) isolate from Okinawa (OAK11) and it belongs to Be76 cluster. Additionally, Fig. 4 showed the first phylogeny of Bradyrhizobium USDA strains with specific serogroups from rpoB gene sequence analysis. This phylogeny is similar with the band patterns of the rpoB gene obtained from RFLP treatment which indicated its usefulness (Fig. S1). On the other hand, the phylogenetic tree of the 16S rRNA gene of the 12 representative isolates clearly separated the groups of B. elkanii strains from B. japonicum and S. fredii (Fig. S2).
Sequence analysis of symbiotic genes nifD and nodD1
The 19 representative isolates which were used for the ITS region and rpoB gene were classified phylogenetically based from the DNA fragments of nifD and nodD1 genes. The results are shown as supplementary material (Fig. S3) as we did not detect diversity among the isolates. All the isolates from the three locations have homogenous nucleotide (nt) sequences for both nifD (785 nt) and nodD1 (717 nt). It is evident that all the isolates were grouped under B. elkanii in nifD wherein separation of B. elkanii and B. japonicum strains was distinguished. For nodD1, all the isolates showed 100% similarity with B. elkanii USDA94 and B. elkanii M13 and was differentiated from B. japonicum and B. diazoefficiens strains. We were not able to find other B. elkanii strains in DDBJ database for the nodD1 which had at least 97% similarity with any of our isolates. In relation, we used B. elkanii USDA94 nodD2 nucleotide sequence as the outgroup.
Summary of RFLP and sequence analysis
The results from RFLP and sequence analysis were summarized (Table 1) and showed that from the 19 representative isolates used in this study, ITS-rpoB types 31′-46′ and 31–46 were found to be possibly endemic in Okinawa, Japan and Nueva Ecija, Philippines, respectively. We also observed in this study that some isolates have ITS type which was different from its rpoB type and these isolates were found in Okinawa, Japan and Nueva Ecija, Philippines.
We were able to observe the genetic diversity of the isolates through sequence analysis better than by just RFLP analysis. Two ITS-rpoB types were observed from Nueva Ecija which were Be31-Be46 and Be76-Be46 whereas, three ITS-rpoB types were observed from Okinawa as follows: Be76-Be76’, Be76-Be46, and Be31′-Be46’. All Kumamoto isolates have the same ITS-rpoB types which was Be76-Be76’. Highest diversity among the isolates was observed in Okinawa, followed by Nueva Ecija then, Kumamoto. Meanwhile, no diversity was observed for both nifD and nodD1 genes but both have similar results with 16S rRNA gene showing high homogeneity with B. elkanii USDA strains and B. elkanii M13 strain, which was isolated from Vigna radiata plant in Nepal.
Discussion
Genetic diversity of indigenous soybean rhizobia by RFLP analysis
Previous studies conducted in this laboratory already revealed that Okinawa, Japan was dominated by B. elkanii particularly Be76 cluster (Saeki et al. 2006, 2008) and in our present study, we obtained similar result. Meanwhile, this is the first report that identified the indigenous soybean-nodulating B. elkanii strains from Nueva Ecija, Philippines and Kumamoto, Japan.
As previously reported, the diversity of soybean rhizobia is influenced by several factors such as soil acidity, salinity, geographic location and environmental gradients (Suzuki et al. 2008; Zhang et al. 2011; Adhikari et al. 2012; Shiro et al. 2013; Yan et al. 2014; Zhao et al. 2014; Htwe et al. 2015). Similarly, the most influential factors in our study might as well be temperature and soil pH. Probably, this is not the case for salinity level since it was almost similar for all the study sites. It is observed that in the RFLP analysis of Okinawa isolates, where the soil was strongly acidic, all isolates with Be76 ITS type were divided into Be76 and Be46 type in rpoB gene. In case of Nueva Ecija isolates, all Be76 ITS type became Be46 type in rpoB gene. Additionally, Okinawa, Japan is considered as a subtropical region whereas Nueva Ecija, Philippines is a tropical region. On the other hand, Kumamoto isolates obtained from a temperate region, remained to have the same Be76 ITS type with rpoB gene type. Since Kumamoto, Japan and Nueva Ecija, Philippines have both the same degree of soil acidity, the main difference between the two location is temperature, which might have caused the change in rpoB type. The effect of different temperature regimes on the changes of population occupancy of bradyrhizobia in Japan was evaluated and revealed that cluster Be76 was dominant in the middle (25 °C) and high (35 °C) temperatures (Saeki et al. 2010). Therefore, our results seem to support the idea that Be76 is a dominant cluster at higher temperatures which can be found in subtropical and tropical climates of the three locations in this study by RFLP analysis of ITS region.
We observed that by analyzing the polymorphisms of rpoB gene, possible endemic strains of soybean rhizobia in Okinawa and Nueva Ecija isolates were detected which were not distinguished in the ITS region. Also, we observed the existence of strains with similar ITS type but completely different rpoB type as detected from Okinawa and Nueva Ecija isolates. This observation could be due to the influence of temperature and soil acidity, although more detailed analyses should be conducted to verify this. The observed inconsistency between the ITS and rpoB type within the same strains could be likely due to recombination events which is widespread within bacteria, more often to members of the same microbiological species (Didelot and Maiden 2010). As recently investigated for Rhizobium species associated with Phaseolus vulgaris, higher nucleotide diversity is introduced from recombination events rather than mutation (Carrascal et al. 2016).
Genetic diversity of indigenous soybean rhizobia as revealed by sequence analysis of ITS and rpoB
We showed the usefulness of sequence analysis for ITS region to distinguish Bradyrhizobium species even at the strain level (Fig. 4a). However, the ITS region failed to detect a clear endemism and genetic diversity of soybean rhizobia in this study. Considering the previous studies on the diversity of soybean rhizobia in temperate regions as earlier cited, it is possible that the evolution rate of ITS region of soybean rhizobia in subtropical and tropical regions was not similarly affected by environmental conditions, particularly temperature. Thus, we have elucidated that for subtropical and tropical regions, the use of ITS region solely was not enough to detect diversity among B. elkanii strains.
On the other hand, sequence analysis of the rpoB gene (Fig. 4b) provided a better discrimination among the strains and revealed that the genetic diversity of indigenous soybean rhizobial isolates varied geographically and we believe that this variation could be due to temperature as well as soil pH. High discriminatory power of rpoB gene was already proven (Vinuesa et al. 2008; Rivas et al. 2009; Degefu et al. 2013; Yan et al. 2014; Guimarães et al. 2015). Also, the existence of Okinawa isolates (OAK7 and OAK11) in Nueva Ecija and Kumamoto groups indicated that distribution of soybean rhizobia could be affected by the change in temperature as well as soil pH. It was previously stated that physical proximity, genetic distance and environmental changes are some of the factors for the occurrence of recombination events (Didelot and Maiden 2010) and in our case, it might be the differences in temperature and soil pH. Thus, we demonstrated that rpoB gene is useful for the analysis of diversity and detection of potential endemic strains of soybean rhizobia in these three locations, particularly for B. elkanii.
Genetic diversity of indigenous soybean rhizobia based from symbiotic genes
In contrast with the ITS region and rpoB gene, sequence analysis of nifD and nodD1 genes did not provide diversity among the isolates. The very high similarity (99–100%) observed between the isolates and B. elkanii strains for both nifD and nodD1 and its congruence with 16S rRNA gene phylogeny might be an indication that the evolution of symbiotic genes from the isolates of Kumamoto and Okinawa, Japan and Nueva Ecija, Philippines have progressed similarly with their conserved genes. Previous studies (Minamisawa et al. 2002; Barcellos et al. 2007; Ling et al. 2016) stated that horizontal gene transfer seldom occur for symbiotic genes in rhizobial genera that commonly causes the conformity in phylogenetic analyses. Although we cannot say that this is the same case with our study because we did not perform an analysis that will support this. However, our result is also similar with earlier report (Risal et al. 2010) stating the similarity of phylogenies obtained from conserved 16S rRNA gene region and symbiotic genes nifD and nodD1 for Nepalese isolates. Thus, we suggest that symbiotic genes may not be enough as indicators for genetic diversity observations, particularly for B. elkanii. This idea is supported by a previous study stating that even distinct rhizobial species can share similar symbiotic genes and because they are located in easily interchangeable elements like the symbiosis island (Ramirez-Bahena et al. 2009). Another possible reason for the similar phylogenies we obtained from nifD and nodD1 genes might be due to gene exchange and internal genetic rearrangements that could have occurred after the co-transfer of nod and nif genes as previously reported (Laguerre et al. 2001).
On the contrary, the incongruent phylogenies of nifD and nodD1 with that of rpoB gene in our study is possibly due to lateral gene transfer as previously observed (Martinez-Romero and Caballero-Mellado 1996; Laguerre et al. 2001; Tian et al. 2010). Hence, there are cases wherein symbiotic genes, particularly nodulation genes, have independent phylogenies from other taxonomic markers such as chromosomal genes (Tian et al. 2010). In this study, we analyzed nodD sequence as a representative gene of common nod gene. The common nod genes including nodD, A, B, C are concerned with the construction of based structure of Nod factor. The Nod factor is related with host specificity between rhizobia and leguminous species, not with the compatibility between rhizobia and Rj-genotype varieties. The responsible gene in bradyrhizobia for incompatibility with Rj-genotype soybean is not clarified yet with some candidate genes (Tsurumaru et al. 2008; Yasuda et al. 2016). For incompatibility with Rj 2-genotype soybean, Tsurumaru et al. (2008) reported that some bradyrhizobial mutants could break the incompatibility with Rj 2-genotype, and the breaking genes were not common nod genes. Though the responsible gene for the incompatibility is not elucidated yet, the gene may be important not only for compatibility but also for genomic diversity.
Symbiotic efficiency of the isolates
One very important feature of an inoculant is its efficiency in symbiosis with the host. Here, we observed that most of the isolates have broad host-range, which is a positive characteristic for a potential inoculant, particularly for Nueva Ecija isolates where 100% possessed this quality. The fact that the phylogeny of nodD1 did not detect any differences among the isolates, it might be possible that this nodulation regulator protein had no or little correlation with the Rj genotypes although it might have influenced the broad host-range in some isolates. Not a single isolate in this study was host-specific which could be generally due to the role of nodD1 as a nodulation regulator. These isolates also showed varied symbiotic efficiency, which were significantly different between isolates within the same Rj genotype. Although there was no indication of this result in the phylogeny of nifD gene, it might be a possibility that nifD gene had no direct or little relationship with the Rj genotypes. Nevertheless, it is worthy to note that some isolates from Philippines (PBM1, PCM5, PHM1 and PHM4) and Southern Japan (HBO14, HBO16, OBO4, OAK11) maybe further studied for their potential as inoculant in relation to host-range and symbiotic efficiency.
Genetic diversity and detection of some endemic soybean rhizobia
Our research group already established that for diversity investigation of soybean rhizobia in temperate regions, ITS region provided high diversity (Saeki et al. 2006, 2008, 2010; Shiro et al. 2013). However, for subtropical and tropical regions, we suggest that rpoB gene should at least be included in addition to ITS region as one of the target genes. We proposed the use of the following Bradyrhizobium USDA reference strains (B. japonicum 4, 6T, 38, 122, 123, 124, 129, 135, B. diazoefficiens 110 T, B. elkanii 31, 46, 76T, 94, and 130 and B. liaoningense 3622T) in the analysis of rpoB gene for soybean rhizobia. This study was able to distinguish that the ITS-rpoB type of B. elkanii isolates from Kumamoto and Okinawa, Japan and Nueva Ecija, Philippines were not 100% identical to the closest reference strains, which were USDA 76T and USDA46. Thus, we reported the presence of possible endemic strains of B. elkanii that nodulate soybean in Okinawa, Japan and Nueva Ecija, Philippines and that genetic diversity of the isolates studied might have varied with temperature and soil pH as revealed by sequence analysis of rpoB gene. We also proposed that the symbiotic genes nifD and nodD1 were possibly not correlated with the compatibility of the Rj genotypes used in this study, although more detailed analyses are recommended to confirm this statement.
The significant results of this study were: first, the production of the first phylogenetic tree of Bradyrhizobium USDA reference strains for rpoB gene with specific serogroups; second, first study that reported the existence of different rpoB gene type from the ITS type within the same strain for B. elkanii; and last, the first study that detected and reported the presence of possible endemic soybean rhizobia in Nueva Ecija, Philippines and Okinawa, Japan. Additionally, the strains that have possibly broad host-range compatibility and could be efficient microsymbionts of soybean were identified in the three locations that could be further studied for their efficiency and effectiveness as suitable inoculants. The information obtained in this research might help inoculation strategy to be more successful specifically in Nueva Ecija, Philippines and Kumamoto, Japan since the indigenous soybean rhizobia have been identified. But of course, more locations should be considered particularly in the Philippines that could represent the whole country.
References
Adhikari D, Kaneto M, Itoh K, Suyama K, Pokharel BB, Gaihre YK (2012) Genetic diversity of soybean-nodulating rhizobia in Nepal in relation to climate and soil properties. Plant Soil 357:131–145
Alam F, Bhuiyan MA, Alam SS, Waghmode TR, Kim PJ, Lee YB (2015) Effect of Rhizobium sp. BARIRGm901 inoculation on nodulation, nitrogen fixation and yield of soybean (Glycine max) genotypes in gray terrace soil. Biosci Biotechnol Biochem 79:1660–1668
Alves BJR, Boddey RM, Urquiaga S (2003) The success of BNF in soybean in Brazil. Plant Soil 252:1–9
Ansari PG, Rao DLN, Pal KK (2013) Diversity and phylogeny of soybean rhizobia in central India. Ann Microbiol 64:1553–1565
Barcellos FG, Menna P, Batista JS, Hungria M (2007) Evidence of horizontal transfer of symbiotic genes from a Bradyrhizobium japonicum inoculant strain to indigenous Diazotrophs Sinorhizobium (Ensifer) fredii and Bradyrhizobium elkanii in a Brazilian savannah soil. Appl Environ Micobiol 73:2635–2643
Bernstein L, MacKenzie AJ, Krantz BA (1955) Salt tolerance of field crops - soybeans. In: United States salinity laboratory report to collaborators. Riverside, CA, pp 35–36
Carrascal OMP, Vanlnsberghe D, Juarez S, Polz MF, Vinuesa P, Gonzalez V (2016) Population genomics of the symbiotic plasmids of sympatric nitrogen-fixing Rhizobium species associated with Phaseolus vulgaris. Environ Microbiol 18:2660–2676
Cole MA, Elkan GH (1973) Transmissible resistance to penicillin G, neomycin, and chloramphenicol in Rhizobium japonicum. Antimicrob Ag Chemother 4:248–253
Degefu T, Wolde-Meskel E, Frostegård Å (2013) Phylogenetic diversity of Rhizobium strains nodulating diverse legume species growing in Ethiopia. Syst Appl Microbiol 36:272–280
Delamuta JRM, Ribeiro RA, Menna P, Bangel EV, Hungria M (2012) Multilocus sequence analysis (MLSA) of Bradyrhizobium strains: revealing high diversity of tropical Diazotrophic symbiotic bacteria. Braz J Microbiol 43:698–710
Delamuta JR, Ribeiro RA, Ormeño-Orrillo E, Melo IS, Martinez-Romero E, Hungria M (2013) Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int J Syst Evol Microbiol 63:3342–3351
Devine TE, Kuykendall LD (1996) Host genetic control of symbiosis in soybean (Glycine max L.) Plant Soil 186:173–187
Didelot X, Maiden MCJ (2010) Impact of recombination on bacterial evolution. Trends Microbiol 18:315–322
Germano MG, Menna P, Mostasso FL, Hungria M (2006) RFLP analysis of the rRNA operon of a Brazilian collection of bradyrhizobial strains from 33 legume species. Int J Syst Evol Microbiol 56:217–229
Gordon BR, Klinger CR, Weese DJ, Lau JA, Burke PV, Dentinger BTM, Heath KD (2016) Decoupled genomic elements and the evolution of partner quality in nitrogen-fixing rhizobia. Ecol Evol 6:1317–1327
Guimarães AA, Florentino LA, Almeida KA, Lebbe L, Silva KB, Willems A, Moreira FM (2015) High diversity of Bradyrhizobium strains isolated from several legume species and land uses in Brazilian tropical ecosystems. Syst Appl Microbiol 38:433–441
Gyaneshwar P, Hirsch AM, Moulin L, Chen W, Elliott GN, Bontemps C, de los Santos PE, Gross E, dos Resi FB Jr, Sprent JI, Young JPW, James EK (2011) Legume-nodulating Betaproteobacteria: diversity, host range, and future prospects. MPMI 24:1276–1288
Hiraishi A, Kamagata Y, Nakamura K (1995) Polymerase chain reaction amplification and restriction fragment length polymorphism analysis of 16S rRNA genes from methanogens. J Ferment Bioeng 79:523–529
Htwe AZ, Yamakawa T, Sarr PS, Sakata T (2015) Diversity and distribution of soybean-nodulating bradyrhizobia isolated from major soybean-growing regions in Myanmar. Afr J Microbiol Res 9:2183–2196
Ikeda S, Rallos LEE, Okubo T, Eda S, Inaba S, Mitsui H, Minamisawa K (2008) Microbial community analysis of field-grown soybeans with different nodulation phenotypes. Appl Environ Microbiol 74:5704–5709
Ikeda S, Okubo T, Kaneko T, Inaba S, Maekawa T, Eda S et al (2010) Community shifts of soybean stem-associated bacteria responding to different nodulation phenotypes and N levels. ISME J 4:315–326
Jordan DC (1982) Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. Nov., a genus of slow-growing, root nodule bacteria from leguminous plants. Int J Syst Bacteriol 32:136–139
Keyser HH, Bohlool BB, Hu TS, Weber DF (1982) Fast-growing rhizobia isolated from root nodules of soybean. Science 215:1631–1632
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kuykendall LD, Saxena B, Devine TE, Udell SE (1992) Genetic diversity in Bradyrhizobium Jordan 1982 and a proposal for Bradyrhizobium elkanii sp. nov. Can J Microbiol 38:501–505
Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N (2001) Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiol 147:981–993
Ling J, Wang H, Wu P, Li T, Tang Y, Naseer N, Zheng H, Masson-Boivin C, Zong Z, Zhu J (2016) Plant nodulation inducers enhance horizontal gene transfer of Azorhizobium caulinodans symbiosis island. Proc Natl Acad Sci U S A 113:13875–13880
Loureiro M, Kaschuk G, Alberton O, Hungria M (2007) Soybean [Glycine max (L.) Merrill] rhizobial diversity in Brazilian oxisols under various soil, cropping, and inoculation managements. Biol Fertil Soils 43:665–674
Maj D, Wielbo J, Marek-Kozaczuk M, Skorupska A (2010) Response to flavonoids as a factor influencing competitiveness and symbiotic activity of Rhizobium leguminosarum. Microbiol Res 165:50–60
Man CX, Wang H, Chen WF, Sui XH, Wang ET, Chen WX (2008) Diverse rhizobia associated with soybean grown in the subtropical and tropical regions of China. Plant Soil 310:77–87
Manuel PC, Huelgas R, Espanto LH (1986) Adoption of soybean in Lupao, Nueva Ecija, Philippines. UN/ESCAP CGPRT Centre, regional coordination Centre for Research and Development of coarse grains, roots and tuber crops in the humid tropics of Asia and the Pacific. CGPRT no. 7: 62 pp
Martens M, Dawyndt P, Coopman R, Gillis M, De Vos P, Willems A (2008) Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int J Syst Evol Microbiol 58:200–214
Martinez-Romero E, Caballero-Mellado J (1996) Rhizobium phylogenies and bacterial genetic diversity. Crit Rev Plant Sci 15:113–140
Minami M, Yamakawa T, Yamamoto A, Akao S, Saeki Y (2009) Estimation of nodulation tendency among Rj-genotype soybeans using the bradyrhizobial community isolated from an andosol. Soil Sci Plant Nutr 55:65–72
Minamisawa K, Itakura M, Suzuki M, Ichige K, Isawa T, Yuhashi K, Mitsui H (2002) Horizontal transfer of nodulation genes in soils and microcosms form Bradyrhizobium japonicum to B. elkanii. Microbes Environ 2:82–90
Njira KOW, Nalivata PC, Kanyama-Phiri GY, Lowole MW (2013) An assessment for the need of soybean inoculation with Bradyrhizobium japonicum in some sites of Kasungu district, Central Malawi. Int J Curr Microbiol App Sci 2:60–72
Nguyen MT, Akiyoshi K, Nakatsukasa M, Saeki Y, Yokoyama K (2010) Multiple occupancy of nodules by nodulating rhizobia on field-grown soybeans with attendance of Sinorhizobium spp. Soil Sci Plant Nutr 56:382–389
Peix A, Ramirez-Bahena MH, Velazquez E, Bedmar EJ (2015) Bacterial associations with legumes. Crit Rev Plant Sci 34:17–42
Ramirez-Bahena MH, Peix A, Rivas R, Camacho M, Rodriguez-Navarro DN, Mateos PF, Martinez-Molina E, Willems A, Velazquez E (2009) Bradyrhizobium pachyrhizi sp. nov. and Bradyrhizobium jicamae sp. nov., isolated from effective nodules of Pachyrhizus erosus. Int J Syst Evol Microbiol 59:1929–1934
Redmond M, Batley M, Djordjevic MA, Innes RW, Kuempel PL, Rolfe BG (1986) Flavones induce expression of nodulation genes in Rhizobium. Nature 323:632–634
Risal CP, Yokoyama T, Ohkama-Ohtsu N, Djedidi S, Sekimoto H (2010) Genetic diversity of native soybean bradyrhizobia from different topographical regions along the southern slopes of the Himalayan Mountains in Nepal. Syst Appl Microbiol 33:416–425
Rivas R, Martens M, de Lajudie P, Willems A (2009) Multilocus sequence analysis of the genus Bradyrhizobium. Syst Appl Microbiol 32:101–110
Sameshima R, Isawa T, Sadowsky MJ, Hamada T, Kasai H, Shutsrirung A et al (2003) Phylogeny and distribution of extra-slow-growing Bradyrhizobium japonicum harboring high copy numbers of RSα, RSβ and IS1631. FEMS Microbiol Ecol 44:191–202
Sanz-Sáez A, Heath K, Burke P, Ainsworth E (2015) Inoculation with an enhanced N2O-fixing Bradyrhizobium japonicum strain (USDA110) does not alter soybean (Glycine max Merr.) response to elevated [CO2]. Plant Cell Environ 38:2589–2602
Saeki Y, Akagi I, Takaki H, Nagatomo Y (2000) Diversity of indigenous Bradyrhizobium strains isolated from three different Rj-soybean cultivars in terms of randomly amplified polymorphic DNA and intrinsic antibiotic resistance. Soil Sci Plant Nutr 46:917–926
Saeki Y, Aimi N, Hashimoto M, Tsukamoto S, Kaneko A, Yoshida N et al (2004) Grouping of Bradyrhizobium USDA strains by sequence analysis of 16S rDNA and 16S-23S rDNA internal transcribed spacer region. Soil Sci Plant Nutr 50:517–525
Saeki Y, Aimi N, Tsukamoto S, Yamakawa T, Nagatomo Y, Akao S (2006) Diversity and geographical distribution of indigenous soybean-nodulating bradyrhizobia in Japan. Soil Sci Plant Nutr 52:418–426
Saeki Y, Minami M, Yamamoto A, Akao S (2008) Estimation of the bacterial community diversity of soybean-nodulating bradyrhizobia isolated from Rj-genotype soybeans. Soil Sci Plant Nutr 54:718–724
Saeki Y, Ozumi S, Yamamoto A, Umehara Y, Hayashi M, Sigua GC (2010) Changes in population occupancy of bradyrhizobia under different temperature regimes. Microbes Environ 25:309–312
Saeki Y, Shiro S, Tajima T, Yamamoto A, Sameshima-Saito R, Sato T, Yamakawa T (2013) Mathematical ecology analysis of geographical distribution of soybean-Nodulating Bradyrhizobia in Japan. Microbes Environ 28:470–478
Saeki Y, Shiro S (2014) Comparison of soybean-Nodulating Bradyrhizobia community structures along north latitude between Japan and USA, advances in biology and ecology of nitrogen fixation, prof. Takuji Ohyama (Ed.) InTech, pp. 195–224
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Scholla MH, Elkan HG (1984) Rhizobium fredii sp. nov. a fast-growing species that effectively nodulates soybeans. Int J Syst Bacteriol 34:484–486
Sharma M, Srivastava K, Sharma S (2010) Biochemical characterization and metabolic diversity of soybean rhizobia isolated from Malwa region of Central India. Plant Soil Environ 56:375–383
Shiro S, Yamamoto A, Umehara Y, Hayashi M, Yoshida N, Nishiwaki A et al (2012) Effect of Rj genotype and cultivation temperature on the community structure of soybean-nodulating bradyrhizobia. Appl Environ Microbiol 78:1243–1250
Shiro S, Matsuura S, Saiki R, Sigua GC, Yamamoto A, Umehara Y et al (2013) Genetic diversity and geographical distribution of indigenous soybean-nodulating bradyrhizobia in the United States. Appl Environ Microbiol 79:3610–3618
Suzuki K, Oguro H, Yamakawa T, Yamamoto A, Akao S, Saeki Y (2008) Diversity and distribution of indigenous soybean - nodulating rhizobia in the Okinawa islands, Japan. Soil Sci Plant Nutr 54:237–246
Tian CF, Young JPW, Wang ET, Tamimi SM, Chen WX (2010) Population mixing of Rhizobium leguminosarum bv. viciae nodulating Vicia faba: the role of recombination and lateral gene transfer. FEMS Microbiol Ecol 73:563–576
Tsurumaru H, Yamakawa T, Tanaka M, Sakai M (2008) Tn5 mutants of Bradyrhizobium japonicum is-1 with altered compatibility with Rj2-soybean cultivars. Soil Sci. Plant Nutr 54:197–203
van Berkum P, Fuhrmann JJ (2000) Evolutionary relationships among the soybean bradyrhizobia reconstructed from 16S rRNA gene and internally transcribed spacer region sequence divergence. Int J Syst Evol Microbiol 50:2165–2172
Vincent JM (1970) A manual for the practical study of the root-nodule bacteria. Blackwell Scientific, Oxford
Vinuesa P, Rojas-Jiménez K, Contreras-Moreira B, Mahna SK, Prasad BN, Moe H et al (2008) Multilocus sequence analysis for assessment of the biogeography and evolutionary genetics of four Bradyrhizobium species that Nodulate soybeans on the Asiatic continent. Appl Environ Microbiol 74:6987–6996
Vos M, Quince C, Pijl AS, de Hollander M, Kowalchuk GA (2012) A comparison of rpoB and 16S rRNA as markers in pyrosequencing studies of bacterial diversity. PLoS One 7:1–8
Wang J (2016) Analysis of the factors influencing Japan’s soybean import trade: based on gravity model. Am J Ind Bus Manag 6:109–116
Yamakawa T, Hussain AKMA, Ishizuka J (2003) Soybean preference for Bradyrhizobium japonicum for nodulation. Soil Sci Plant Nutr 49:835–841
Yan J, Han XZ, Ji ZJ, Li Y, Wang ET, Xie ZH, Chen WF (2014) Abundance and diversity of soybean-Nodulating rhizobia in black soil are impacted by land use and crop management. Appl Environ Microbiol 80:5394–5402
Yasuda M, Miwa H, Masuda S, Takebayashi Y, Sakakibara H, Okazaki S (2016) Effecter-triggered immunity determines host genotype-specific incompatibility in legume-rhizobium symbiosis. Plant Cell Physiol 57:1791–1800
Young JM (2003) The genus name Ensifer Casida 1982 takes priority over Sinorhizobium Chen et al. 1988, and Sinorhizobium morelense Wang et al. 2002 is a later synonym of Ensifer adhaerens Casida 1982. Is the combination ‘Sinorhizobium adhaerens’ (Casida 1982) Willems et al. 2003 legitimate? Request for an opinion. Int J Syst Evol Microbiol 53:2107–2110
Zaat SA, Schripsema J, Wijffelman CA, van Brussel AAN, Lugtenberg BJJ (1989) Analysis of the major inducers of the Rhizobium nodA promoter from Vicia sativa root exudate and their activity with different nodD genes. Plant Mol Biol 13:175–188
Zhang YM, Li Y Jr, Chen WF, Wang ET, Tian CF, Li QQ, Zhang YZ, Sui XH, Chen WX (2011) Biodiversity and biogeography of rhizobia associated with soybean plants grown in the North China plain. Appl Environ Microbiol 77:6331–6342
Zhao L, Fan M, Zhang D, Yang R, Zhang F, Xu L, Wei X, Shen Y, Wei G (2014) Distribution and diversity of rhizobia associated with wild soybean (Glycine soja Sieb. & Zucc.) in Northwest China. Syst Appl Microbiol 37:449–456
Acknowledgements
This study was supported by JSPS KAKENHI (Grant-in-Aid for Scientific Research (B) no. 26310313) and the Japanese Government (MEXT) Scholarship program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Katharina Pawlowski.
Electronic supplementary material
Table S1
List of primers used in this study for the PCR amplification and sequence analysis of 16S rRNA, ITS, rpoB, nifD and nodD1. (PDF 73 kb)
Table S2
Shoot and nodule parameters of 19 representative B. elkanii isolates employing three Rj genotypes. Mean comparison was conducted in triplicates only between isolates within the same Rj genotype. (PDF 89 kb)
Table S3
List of accession numbers for selected Bradyrhizobium USDA reference strains and isolates from the sequence analysis of 16S rRNA gene, 16S–23S rRNA gene ITS region, rpoB housekeeping gene and symbiotic genes nifD and nodD1. (PDF 79 kb)
Figure S1
Indication of the usefulness of rpoB-RFLP analysis as shown by the similarity in the phylogeny and RFLP band patterns. (PDF 58 kb)
Figure S2
Phylogenetic tree based on sequence analysis of 16S rRNA gene. The tree was constructed using the Neighbor- Joining method with the Kimura 2-parameter (K2P) distance correlation model and 1000 bootstrap replications in MEGA v.7 software. (PDF 138 kb)
Figure S3
Phylogenetic tree based on sequence analysis of (A) nifD gene and (B) nodD1 gene. The tree was constructed using the Neighbor-Joining method with the Kimura 2-parameter (K2P) distance correlation model and 1000 bootstrap replications in MEGA v.7 software. The first letter of isolates’ name indicates the location as follows: H - Kumamoto; O - Okinawa; P - C. Luzon, Philippines. (PDF 235 kb)
Rights and permissions
About this article
Cite this article
Mason, M.L.T., Matsuura, S., Domingo, A.L. et al. Genetic diversity of indigenous soybean-nodulating Bradyrhizobium elkanii from southern Japan and Nueva Ecija, Philippines. Plant Soil 417, 349–362 (2017). https://doi.org/10.1007/s11104-017-3263-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-017-3263-4