Abstract
This study utilized dietary salt loading and ion-poor water (IPW) exposure of rainbow trout (Oncorhynchus mykiss) to further understand the role of fish gill epithelium tight junction (TJ) physiology in salt and water balance. Gill morphology, biochemistry and molecular physiology were examined, with an emphasis on genes encoding TJ proteins. Fish were either fed a control or salt-enriched diet (~10 % NaCl) for 4 weeks prior to IPW exposure for 24 h. Serum [Na+], [Cl−] and muscle moisture content were unaltered by salt feeding, but changed in response to IPW irrespective of diet. Dietary salt loading altered the morphology (reduced Na+–K+-ATPase-immunoreactive cell numbers and surface exposure of mitochondrion-rich cells), biochemistry (decreased vacuolar-type H+-ATPase activity) and molecular physiology (decreased nkaα1a and cftrII mRNA abundance) of the gill in a manner indicative of reduced active ion uptake activity. But in control fish and not salt-fed fish, gill mRNA abundance of nkaα1c increased and nbc decreased after IPW exposure. Genes encoding TJ proteins were typically either responsive to salt feeding or IPW, but select genes responded to combined experimental treatment (e.g. IPW responsive but only if fish were salt-fed). Therefore, using salt feeding and IPW exposure, new insights into what factors influence gill TJ proteins and the role that specific TJ proteins might play in regulating the barrier properties of the gill epithelium have been acquired. In particular, evidence suggests that TJ proteins in the gill epithelium, or the regulatory networks that control them, respond independently to external or internal stimuli.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In aquatic vertebrates such as fishes, mechanisms involved in the regulation of salt and water balance both resemble as well as differ from those found in terrestrial vertebrates. For example, in fishes the intestine and kidney play important roles in the acquisition, retention and/or elimination of ions (Marshall and Grosell 2006; Wood and Bucking 2011). This is driven by systemic needs and largely mirrors the role that these organs fulfill in the regulation of salt and water balance in non-aquatic vertebrates. However, fishes also acquire, retain or eliminate ions directly to or from the surrounding environment across epithelia that interface with water, such as the gills or skin (Evans et al. 2005; Chasiotis et al. 2012a; Glover et al. 2013). Therefore, the interplay of iono/osmoregulatory organs in the maintenance of salt and water balance in fishes is arguably more complex than what is typically found in terrestrial vertebrates.
The diet of terrestrial vertebrates serves as the sole route for the acquisition of salt (NaCl) and in this regard, acute or chronic alterations in dietary salt load can have profound effects on the physiology of these animals (He and MacGregor 2009; Meneton et al. 2005). As a result of this, the deleterious effects of sub- or supra-optimal salt levels in mammalian diets have been the subject of numerous studies (He and MacGregor 2009; Meneton et al. 2005). The acquisition of salt from dietary sources also strongly influences the physiology of fishes, but the ability of these organisms to either eliminate or acquire salts directly to or from surrounding water has, in part, resulted in fewer studies examining the physiological consequences of acute or chronic alterations in dietary NaCl levels in these animals (see Wood and Bucking 2011). Nevertheless, this area has been explored by a number of groups and includes studies aimed at understanding how dietary salt content alters the vertebrate endocrine and cardiovascular systems (Chen et al. 2007; Johnson and Olson 2009a, b; Kelly and Peter 2006; Olson and Hoagland 2008; Perry et al. 2011), as well as studies that document the beneficial effect of manipulated dietary salt levels on parameters such as fish growth and health (Alam et al. 2015; Cnaani et al. 2010; D’Cruz and Wood 1998; Eroldoğan et al. 2005; Garcia et al. 2007; Gatlin et al. 1992; Harpaz et al. 2005; Keshavanath et al. 2012; Salman and Eddy 1988). In addition, the physiological consequences of elevating dietary salt levels prior to the release or transfer of fishes from freshwater (or low salinity) to hyper-osmotic surroundings (e.g. seawater) are reported to positively influence hypo-osmoregulatory ability as well as fish survival (Alam et al. 2015; Al-Amoudi 1987; Basulto 1976; Duston 1993; Pelletier and Besner 1992; Salman and Eddy 1990; Staurnes and Finstad 2000; Zaugg et al. 1983). This is often seen to occur in association with a dietary salt-induced development of phenotypic characteristics which resemble those found in fishes residing in (or acclimated to) SW (Alam et al. 2015; Duston 1993; Fontainhas-Fernandes et al. 2001; Salman and Eddy 1987; Staurnes and Finstad 2000; Trombetti et al. 1996). In particular, adjustments in gill epithelium morphology and transcellular ion transport mechanisms are well documented (Alam et al. 2015; Perry and Rivero-Lopez 2012; Perry et al. 2006). But despite numerous observations that relate manipulated dietary salt levels to altered endpoints of transcellular ion transport, no study has examined how elevated dietary salt content impacts the proteins that influence paracellular ion movement across ionoregulatory epithelia of fishes.
In vertebrate epithelia, the principal barrier of the paracellular pathway is an assemblage of structural proteins located in the apicolateral region of epithelial cells which are known as the tight junction (TJ) complex (Farquhar and Palade 1963; Günzel and Fromm 2012). Extracellular loops of transmembrane TJ proteins form regions of occlusion between adjacent epithelial cells while cytosolic TJ proteins tether the intracellular domain of transmembrane TJ proteins to the cell cytoskeleton (Günzel and Fromm 2012). In terrestrial vertebrates, transmembrane TJ proteins include occludin (Ocln), tricellulin (Tric) as well as a large family of claudin (Cldn) proteins, numbering around 27 (Günzel and Fromm 2012; Günzel and Yu 2013). Tissue specific expression profiles of Cldn proteins are generally accepted to reflect the different barrier properties of vertebrate epithelia as the extracellular domains of Cldns (and the interaction between them) can be either “barrier” or “pore” forming. Therefore, paracellular permeability dynamics of vertebrate epithelia (or any alterations that occur in them) are dependent on the presence and abundance of a distinct assembly of TJ proteins at the TJ complex (Günzel and Fromm 2012; Günzel and Yu 2013). In teleost fishes, occludin and tricellulin have been identified and demonstrated to influence gill epithelium permeability (Chasiotis and Kelly 2008; Chasiotis et al. 2012b; Kolosov and Kelly 2013). However, due to genome duplication and tandem gene duplication events, the teleost Cldn family has approximately doubled in size (Loh et al. 2004; Kolosov et al. 2013). Therefore, making sense of the role that specific Cldns play in modulating epithelial permeability of fishes is challenging. Nevertheless, recent studies have begun to decipher the function of specific Cldn TJ proteins in the regulation of teleost fish TJ permeability, and in particular a number of studies have highlighted the importance of these proteins in the gill epithelium (for review see Chasiotis et al. 2012a; Kolosov et al. 2013). This is because TJ heterogeneity in the gill epithelium of fishes contributes significantly to strategies that permit the maintenance of ionoregulatory homeostasis (Sardet et al. 1979; Evans et al. 2005; Chasiotis et al. 2012a).
With regard to the contribution of specific TJ proteins to gill epithelium permeability and TJ heterogeneity, a primary experimental manipulation that has permitted insight thus far is a comparison of tissues/cells isolated from fish acclimated to environments that vary in ion content, such as FW versus SW or FW versus ion-poor water (IPW) (e.g. Chasiotis et al. 2012a, b; Bui and Kelly 2014). The reason for this is that permeability of the gill epithelium differs greatly in these environments, which is generally accepted to reflect differences in the architecture and physiology of TJs between specific types of gill cell (Evans et al. 2005; Chasiotis et al. 2012a). However, greater insight into the role of TJ proteins in gill epithelium permeability will undoubtedly be provided as the experimental toolbox used to examine this phenomenon continues to diversify. Therefore, given the observations that chronic dietary NaCl elevation can induce alterations in gill epithelium morphology and transcellular ion transport characteristics that are SW-like, and that salt-fed FW fish transferred to hyper-osmotic surroundings perform better than those fed regular diets, it can be hypothesized that alterations in the molecular physiology of the TJ complex will also occur in salt-fed fishes and that salt-feeding could be a useful tool to advance our understanding of TJ protein physiology. In accordance with this, the current study sought first to examine the response of the FW rainbow trout gill to a chronic elevation in dietary salt (NaCl) content with an emphasis on the molecular physiology of the TJ complex. Following chronic dietary NaCl elevation, fish were not abruptly exposed to SW, but instead to IPW (for a 24 h period). It has been reported that too little salt in the diet of FW fishes is deleterious when they are exposed to conditions that severely limit ion acquisition from surrounding water (D’Cruz and Wood 1998). Therefore it can be reasoned that salt feeding FW fish prior to an abrupt exposure to IPW may ameliorate the effects of this challenge on ionoregulatory homeostasis. Furthermore, because exposure to IPW represents a challenge to salt and water balance that is substantially different from that of salt-feeding (e.g. ion loss versus ion loading etc.), the combined use of these experimental variables have the potential to provide additional insight into the molecular physiology of the TJ complex.
Materials and methods
Experimental animals
Rainbow trout (Oncorhynchus mykiss, Walbaum 1792; 15–20 g) were obtained from Humber Springs Trout Hatchery (Orangeville, ON, Canada). Fish were maintained in opaque 600 l tanks in aerated, dechlorinated flow-through freshwater (FW, approximate concentration in μmol l−1: Na+ 590, Cl− 920, Ca2+ 760, K+ 43; pH 7.35) under a constant photoperiod (12 h light, 12 h dark). Temperature ranged between 8 and 10 °C and fish were fed daily (~2 % body weight) with commercial trout pellets (Martin Mills Profishent, Elmira, ON, Canada). For experiments, fish were transferred to 200 l opaque tanks (n = 40/tank) and allowed to settle for 1 week before the introduction of experimental diets. Conditions for the acclimation week were identical to those already outlined.
Experimental conditions and tissue sampling
Rainbow trout were fed either a control trout pellet diet (Martin Mills Profishent: no NaCl added, nominal levels of NaCl 0.45 g/100 g) or a salt-enriched diet (10 g of added NaCl per 100 g diet) at a 2 % body weight ration/day for a period of 4 weeks. Both diets were made by grinding the commercial trout pellet diet into powder, and then either reconstituting this powder with or without the addition of NaCl. In the case of a salt-rich diet, this was done by adding 10 g of reagent grade NaCl (BioShop Canada Inc., Burlington, ON, Canada) per 100 g of diet, following which double-distilled water was added and the powder was worked into a ‘dough’. To reform pellets, the ‘dough’ was run through an electronic meat grinder to form long spaghetti-like strands. After these were dried they could be broken up into pellets. Control diets were made in an identical fashion but without the addition of NaCl.
Following a 4 week feeding period, FW-residing control and salt-fed fish were captured, anaesthetized in 0.5 g/l tricaine methasulfonate (MS-222, Syndel Laboratories, Canada) and sampled, while simultaneously a second set of fish were exposed to ion-poor water (IPW, approximate composition of IPW in μmol l−1 was as follows: Na+ 20, Cl− 40, Ca2+ 2, K+ 0.4, pH 6.5) for 24 h and then sampled. IPW was rapidly introduced by partially draining tank water and switching inlet pipe water from FW to IPW. To ensure complete replacement of water, partial draining of the tank was repeated twice more each time the tank filled up. Complete replacement of FW with IPW took ~3 h. For sampling, blood was collected from caudal vessels using a 25G gauge syringe (BD Falcon, BD Biosciences, Mississauga, ON, Canada), after which the spine was transected before collecting gill and muscle (standardized region of epaxial white muscle) tissue. Blood was allowed to clot at room temperature for 30 min and then centrifuged at 10,600×g in a chilled (4 °C) centrifuge for 10 min to collect serum. Serum was stored at −80 °C until further analysis. Gill tissue was collected for molecular (quantitative real-time PCR, qPCR), biochemical (enzyme activity) and morphological [immunohistochemistry (IHC) and scanning electron microscopy (SEM)] analyses. In each case, a standardized gill arch was used and for molecular (first on the left) and biochemical analysis (first on the right). Gill tissue was flash frozen in liquid nitrogen immediately after collection. Flash frozen tissue was then stored at −80 °C until it was used. For molecular analysis, each gill arch collected was placed in a microcentrifuge tube containing 1 ml of ice-cold Trizol reagent prior to freezing. After careful removal, gill samples for SEM were immediately fixed in 2.5 % glutaraldehyde in phosphate-buffered saline (PBS) (4 h at 4 °C) and samples for IHC were immediately fixed in Bouin’s solution for 4 h at room temperature. Fish husbandry, animal experiments and tissue collection methods were conducted in accordance with an approved York University Animal Care Protocol that conformed to the guidelines of the Canadian Council on Animal Care.
Serum [Na+] and [Cl−], muscle moisture content and gill enzyme activity
Serum [Na+] was determined using reference and ion-selective microelectrodes (ISMe) that were constructed according to Donini and O’Donnell (2005). Serum [Cl−] was determined using a spectrophotometric protocol described in detail by Zall et al. (1956). To measure muscle moisture content, muscle tissue was dried to a constant weight at 60 °C and moisture content was then calculated as wet weight lost and expressed as a percentage. Gill Na+–K+-ATPase (NKA) activity were measured using methodology described by McCormick (1993). Vacuolar-type H+-ATPase (VA) activity was measured in a similar fashion, but using 25 mM bafilomycin A1 (BioShop Canada Inc, Burlington, ON, Canada) as the enzyme inhibitor.
IHC, SEM and gill surface mitochondrion-rich cell (MRC) morphometric analysis
For IHC analysis of NKA-immunoreactive (NKA-ir) cells in the gill epithelium, samples were processed and prepared as described in detail by Chasiotis and Kelly (2008). NKA α-subunit antibody (α5; Developmental Studies Hybridoma Bank, Iowa City, IA, USA) was used and cell nuclei were stained with 4,6-diamidino-2-phenylindole. IHC images of NKA-ir cells in the gill epithelium were captured using an Olympus DP70 camera (Olympus Canada) coupled to a Reichert Polyvar microscope.
For SEM analysis, fixed gill arches were carefully cut so as to isolate individual filaments. These were then dehydrated through an ascending acetone series (30–100 %) and final drying was achieved using tetramethylsilane (2 × 10 min, Sigma-Aldrich). Individual gill filaments were mounted on SEM stubs and then sputter coated (Hummer VI Au/Pd 40/60). Samples prepared in this way were examined using a Hitachi S-520 scanning electron microscope and images for morphometric analysis were captured using a Hitachi Quartz PCI (Version 6) image capture system. Data for gill MRC morphometric analysis was obtained using images captured with Image J software (National Institutes of Health, Bethesda, MD, USA; http://imagej.nih.gov/ij/, 1997–2011). The number of exposed MRCs, individual MRC surface area, and MRC fractional surface area were determined using methods and calculations as described by Chasiotis et al. (2012b).
RNA extraction, synthesis of cDNA and qPCR analysis
Total RNA was extracted from gill samples in Trizol reagent (Invitrogen Canada) according to manufacturer’s instructions. The concentration of total RNA extracted from each sample was determined spectrophotometrically using Multiskan Spectrum UV/Vis microplate spectrophotometer (Thermo Fisher Scientific, Nepean, ON, Canada). 2 μg of total RNA was treated with DNase I (Amplification Grade, Invitrogen) and used for cDNA synthesis. First-strand cDNA was synthesized using Super-Script III Reverse Transcriptase and Oligo(dT)12–18 primers (Invitrogen Canada). Following this, transcript abundance of genes of interest were determined by qPCR. Genes encoding the following proteins were examined: occludin (Ocln), tricellulin (Tric), Zona Occludens-1 (ZO-1), claudin (Cldn)-5a, -6, -7, -8b, -8c, -8d, -10c, -10d, -10e, -23a, -27b, -28b, -29a, -30, -31, and -32a, as well as isoforms 1a, 1b and 1c of α-subunit of NKA, Na/HCO3 cotransporter (NBC) and cystic fibrosis transmembrane conductance regulator II (CFTRII). GenBank accession numbers, primer sets, annealing temperature and expected amplicon size for each gene of interest examined can be found in Table 1. All primer sets were designed using Primer3 software (v. 0.4.0). qPCR was performed using SYBR Green I Supermix (Bio-Rad Laboratories Canada, Mississauga, ON, Canada), primer sets (see Table 1), and a Chromo4 Detection System (CFB-3240; Bio-Rad Laboratories). The following reaction conditions were used: 1 cycle denaturation (95 °C, 4 min), 40 cycles of: (1) denaturation (95 °C, 30 s), (2) annealing (see Table 1, 30 s), and (3) extension (72 °C, 30 s), with each qPCR run culminating in a melting curve. For qPCR analysis mRNA abundance was normalized using transcript encoding β-actin. The validity of actb as a reference gene for qPCR in this study was established by statistically comparing actb threshold cycle values between experimental groups and confirming that experimental manipulation did not significantly alter actb abundance.
Statistical analysis
Statistical analysis was conducted using a two-way ANOVA where two factors were: (1) salt feeding and (2) 24 h IPW exposure. The interaction term of the two-way ANOVA allowed for the analysis of the effect of inclusion of extra salt in the fish diet on the effects of subsequent IPW exposure. Every statistical test was coupled with an equal variance test and a normality test. None of the data were transformed. The need for non-parametric tests was not detected and they were therefore not used in this study. The protocol was coupled with a Holm-Sidak multiple comparison test in SigmaPlot (Version 11) statistical software. Statistical significance was based on the observation of a fiduciary limit of p < 0.05.
Results
Muscle moisture content and serum [Na+] and [Cl−]
Systemic endpoints of salt and water balance (i.e. serum [Na+] and [Cl−], and muscle moisture content) did not significantly alter in response to salt feeding (Fig. 1). In contrast, and irrespective of dietary salt content, serum [Na+] and [Cl−] significantly decreased 24 h following IPW exposure and muscle moisture content significantly increased (Fig. 1).
Systemic endpoints of salt and water balance in rainbow trout fed a salt-rich diet (salt-fed, ~10 % NaCl) and exposed to ion-poor water (IPW) for 24 h. Serum a [Na+], b [Cl−] and c muscle moisture content do not significantly alter in response to salt feeding, but all exhibit significant alterations in response to IPW. Data are presented as mean values ± SEM (n = 8–9). Dagger symbol indicates significant difference between fish held in freshwater (FW) and those exposed to IPW for 24 h
Gill epithelium MRC surface morphometrics and NKA-ir cells
Dietary salt loading significantly decreased NKA-ir cell numbers in the gill epithelium of rainbow trout (Fig. 2a, b, e). This was particularly noticeable on and at the base of the secondary lamellae where NKA-ir cells in salt-fed fish more or less disappeared (Fig. 2b). Upon exposure to IPW for 24 h, no significant change in the number of NKA-ir cells in the gill epithelium of trout was seen in either control or salt-fed fish (Fig. 2).
Effect of elevated dietary salt content (salt-fed, ~10 % NaCl) and 24 h ion-poor water (IPW) exposure on Na+–K+-ATPase (NKA) immunoreactive (NKA-ir) cells in the gill epithelium of rainbow trout. Representative images of NKA-ir cells are shown for the gills of a control freshwater (FW), b salt-fed FW, c control IPW and d salt-fed IPW fish. The total number of NKA-ir cells, (per unit length), in all treatments were determined and are shown in e. In a–d, each panel shows the phase contrast view of a gill section (upper inset) with its corresponding immunofluorescence view (lower inset). NKA-ir cells are visualised (in green) with FITC-conjugated anti-NKA antibody. Cell nuclei are stained with DAPI (blue). Scale bar 50 μm. A salt feeding-induced reduction in the presence of NKA-ir cells in the secondary gill lamellae can be seen when comparing a and b. But in d it can be seen that in salt-fed fish exposed to IPW, NKA-ir cells in the secondary lamellae begin to reappear. Data in e are expressed as mean values ± SEM (n = 3). Asterisk indicates significant difference between control and salt-fed fish (color figure online)
Salt-loading also resulted in a decrease in the number of MRCs exposed at the surface of the gill epithelium (Fig. 3a, b, e). Although individual MRC surface area did not significantly alter in salt-fed fish (Fig. 3f), subtle changes as well as the decrease observed in the number of MRCs exposed at the surface of the gill epithelium was enough to see a decrease in MRC fractional surface area of salt-fed fish (Fig. 3g). Following 24 h in IPW, a modest but significant decrease in the number of MRCs exposed at the surface of the gill epithelium was observed in control fish but not in salt-fed fish (Fig. 3e). In FW control and salt-fed fish, the apical surface of MRCs was typically seen to be covered in short microvilli, but following IPW exposure these were no longer visible (Fig. 3a–d). Nevertheless, control fish exhibited a large increase in MRC surface area that, despite a reduced number of exposed MRCs, resulted in an increase in MRC fractional surface area (Fig. 3f, g). In salt-fed fish, there was also a significant increase in individual MRC surface area (Fig. 3f), but this was less pronounced than what was seen in control fish and consequently, no significant increase in MRC fractional surface area was observed in salt-fed fish exposed to IPW for 24 h (Fig. 3g).
Effect of elevated dietary salt content (salt-fed, ~10 % NaCl) and 24 h ion-poor water (IPW) exposure on the ultrastructure and morphometrics of mitochondrion-rich cells (MRCs) at the surface of the rainbow trout gill epithelium. Representative scanning electron micrograph images of the gill epithelium surface of a control freshwater (FW), b salt-fed FW, c control IPW and d salt-fed IPW fish are shown. Electron micrographs were used to determine e the number of exposed MRCs at the surface of the gill epithelium, f individual MRC surface area and g MRC fractional surface area. Morphometric data presented in e–g are expressed as mean values ± SEM (n = 5). For morphometric data, asterisk indicates significant difference between control and salt-fed fish and dagger symbol indicates significant difference between fish held in FW and those exposed to IPW for 24 h. In a–d, black arrowheads point to MRCs and scale bars 10 µm. Both salt feeding and IPW exposure alter MRC morphometrics (e–g), but only IPW exposure appears to alter MRC surface ultrastructure (a–d)
Gill enzyme activity and mRNA abundance of transcellular ion transport proteins
Gill NKA activity was not significantly altered by dietary salt-loading or 24 h following IPW exposure (Fig. 4a). In contrast, VA enzyme activity exhibited a marked significant reduction in activity in the gills salt-fed fish (Fig. 4b). This trend was also apparent in fish exposed to IPW, where it was significant but less marked (Fig. 4b). 24 h following IPW exposure, VA activity was not significantly different from values seen in FW control or salt-fed animals (Fig. 4b).
Biochemical and molecular endpoints of transcellular ion transport in the gill of rainbow trout fed a salt-rich diet (salt-fed, ~10 % NaCl) and exposed to ion-poor water (IPW) for 24 h. Measurements of gill a Na+–K+-ATPase (NKA) and b V-type H+-ATPase (VA) enzyme activity are shown for control and salt-fed fish held in freshwater (FW) and after 24 h IPW exposure. For the same groups, gill transcript abundance of c NKAα1a subunit (nka α1a), d NKAα1b subunit (nka α1b), e NKAα1c subunit (nka α1c), f sodium-bicarbonate co-transporter (nbc) and g cystic fibrosis transmembrane conductance regulator (cftrII) are shown. Data are presented as mean values ± SEM (n = 8–10). Asterisk indicates significant difference between control and salt-fed fish and dagger symbol indicates significant difference between fish held in FW and those exposed to IPW for 24 h. Gene of interest transcript abundance was normalized using actb as a validated non-responsive reference gene and expressed as a percentage relative to values determined for FW control fish which were assigned a value of 100
The mRNA abundance of nka α1a was significantly lower in salt-fed fish in both FW and in IPW, but unaltered 24 h following IPW exposure (Fig. 4c). In contrast, neither salt feeding nor IPW exposure altered the transcript abundance of gill nka α1b (Fig. 4d). When fish were held in FW, salt feeding had no significant effect on nka α1c mRNA abundance (Fig. 4e). However, 24 h following IPW exposure, nka α1c mRNA abundance was significantly elevated in control fish and unaltered in salt-fed fish, meaning that in IPW control fish gill nka α1c mRNA abundance was significantly greater than nka α1c transcript abundance in salt-fed fish gills (Fig. 4e). Transcript abundance of gill Na/HCO3 co-transporter (nbc) was greatly reduced by salt feeding (Fig. 4f). Following IPW exposure, nbc mRNA abundance was reduced in control fish, but was unaltered and remained low in salt-fed fish (Fig. 4f). Finally, gill cystic fibrosis transmembrane conductance regulator II (cftrII) mRNA abundance was significantly reduced by salt feeding, but was unaffected by IPW exposure (Fig. 4g).
Transcript abundance of TJ proteins in gill tissue
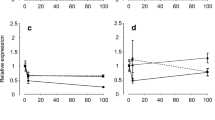
Of the 19 genes encoding TJ proteins examined in the current study, only two (ocln and cldn-5a) were not significantly altered by dietary salt feeding or following 24 h IPW exposure (Figs. 5a, 6). Transcript abundance of tric decreased as a result of salt loading, but no significant alterations were seen in tric mRNA abundance 24 h following IPW exposure (Fig. 5b). In contrast, ZO-1 mRNA abundance was unaltered by salt feeding and in control fish, was elevated 24 h following IPW exposure (Fig. 5c). Salt-fed fish did not exhibit an increase in ZO-1 mRNA abundance following 24 h IPW exposure (Fig. 5c).
Transcript abundance of a occludin (ocln), b tricellulin (tric), and c zonula occludens 1 (ZO-1) tight junction proteins in the gill of rainbow trout fed a salt-rich diet (salt-fed, ~10 % NaCl) and exposed to ion-poor water (IPW) for 24 h. Data are presented as mean values ± SEM (n = 8–10). Asterisk indicates significant difference between control and salt-fed fish and dagger symbol indicates significant difference between fish held in FW and those exposed to IPW for 24 h. Gene of interest transcript abundance was normalized using actb as a validated non-responsive reference gene and expressed as a percentage relative to values determined for FW control fish which were assigned a value of 100
Transcript abundance of claudin (cldn) TJ protein isoforms in the gill of rainbow trout fed a salt-rich diet (salt-fed, ~10 % NaCl) and exposed to ion-poor water (IPW) for 24 h. Data are presented as mean values ± SEM (n = 8–10). Asterisk indicates significant difference between control and salt-fed fish and dagger symbol indicates significant difference between fish held in FW and those exposed to IPW for 24 h. Gene of interest transcript abundance was normalized using actb as a validated non-responsive reference gene and expressed as a percentage relative to values determined for FW control fish which were assigned a value of 100
In the gills of fish held in FW, salt-feeding resulted in an increase in the mRNA abundance of cldn-6, -8d, -10d and -10e (Fig. 6). After 24 h in IPW, transcript abundance of cldn-6, -7, -8b, -8d, -23a, -27b, -28b, -29a, -30, -31, and -32a was elevated in the gill tissue of control fish. Of these IPW-responsive cldns, it was found that cldn-7, -23a, -27b, -28b, -30, and -32a were also elevated in the gills of salt-fed fish 24 h following IPW exposure, but not cldn-6, -8b, -8d, -29a, and -31. In the case of cldn-6, and -8d, salt feeding had already significantly elevated mRNA levels and no further increase was seen in response to IPW (Fig. 6). In control fish gills, no transcript encoding a TJ protein was found to be reduced 24 h following IPW exposure (Fig. 6). But when fish had been salt-fed, cldn-10c and -10e mRNA abundance were found to be reduced 24 h following IPW exposure (Fig. 6). In the case of cldn-10e, an IPW-induced reduction stood out against salt-induced elevation in FW. Therefore, 24 h following IPW exposure, cldn-10e mRNA abundance in salt-fed fish was not significantly different from levels found in control fish. In contrast, 24 h following IPW exposure, cldn-8b, -10c, -10d, -29a, -30, and -31 all differed significantly between control fish and salt-fed fish (Fig. 6). In this regard, only cldn-30 mRNA abundance was elevated in the gills of IPW exposed salt-fed fish versus IPW-exposed control fish, while cldn-8b, -10c, -10d, -29a, and -31 exhibited significantly lower mRNA levels in salt-fed fish gills versus control fish gills.
Discussion
Overview
This study provides evidence that chronic dietary salt loading in a freshwater fish can change the molecular physiology of the gill epithelium paracellular pathway by modifying the abundance of gill epithelium TJ protein mRNA. As a result, the original hypothesis that alterations in the molecular physiology of the TJ complex will occur in salt-fed fishes can be accepted. These observations are in line with the important role that TJ proteins, TJs, and paracellular ion movement play in the maintenance of ionoregulatory homeostasis in fishes (for review see Chasiotis et al. 2012a; Kolosov et al. 2013). Therefore in this regard, salt-feeding does appear to be a useful tool that can help to advance our understanding of how the molecular physiology of the TJ complex contributes to salt and water balance in fishes. In addition to this it was found that salt-loading appeared to be neither deleterious nor ameliorative in terms of the overall response of fish to IPW 24 h after abrupt exposure (i.e. no significant differences were found in serum Na+, Cl− or muscle moisture content between salt-fed and control fish following IPW exposure). This was a surprising result considering that salt feeding significantly altered a number of endpoints related to gill transcellular and paracellular ion transport in FW fish. However, in salt-fed animals several measured parameters were found to be responding to 24 h IPW exposure in a manner different from control-fed animals. This would suggest that despite the fact that serum Na+ and Cl− levels as well as muscle moisture content were not significantly different in salt-fed and control fish exposed to IPW, salt-fed fish deal with the challenge of an ion-poor environment differently from the control group. Indeed, it was interesting to note when looking at TJ protein mRNA, a number of cldns were found to be responsive to IPW only, irrespective of dietary salt content, while only one mRNA encoding a cldn (i.e. cldn-10d) responded to salt feeding irrespective of water ion content. In addition to this, a small number of cldns altered in response to the combined treatment of dietary salt and IPW exposure or to both experimental parameters. Therefore the experimental manipulations of dietary salt loading combined with IPW exposure allows greater insight into the role of specific TJ protein mRNA in the ionoregulatory physiology of fishes than salt feeding or IPW exposure alone.
Dietary salt loading and systemic endpoints of ionoregulatory homeostasis
Following 4 weeks of dietary salt loading, muscle moisture content as well as serum [Na+] and [Cl−] in salt-fed fish were not significantly different from control fish (Fig. 1). Previous studies report conflicting results with regard to the effect of the dietary salt loading on plasma/serum ion levels. For example, salt feeding is reported not to alter plasma Na+ and Cl− levels in some studies (e.g. Pelletier and Besner 1992; Staurnes and Finstad 2000; Perry and Rivero-Lopez 2012; Alam et al. 2015), while other accounts report elevated levels of circulating Na+ and Cl− (e.g. Basulto 1976; Smith et al. 1995; Pyle et al. 2003). However, information missing from most of the aforementioned studies is when fish were last fed prior to sampling. One exception is the work of Smith et al. (1995), where fish were sampled 7 h post-feeding. Smith et al. (1995) reported that diffusive Na+ loss (Na+ efflux) increased approximately 6.5-fold 1 h after salt feeding and that this rate of ion loss was maintained for up to 7 h, at which point Na+ levels in the stomach contents and serum of salt-fed fish were still significantly elevated. Therefore it seems likely that time of sampling is of particular importance with regard to whether an elevation in plasma or serum ion levels will be seen in salt-fed fish. In the current work, an examination of samples taken 24 h following feeding may have been too late in the peri-prandial time frame to observe elevated serum [Na+] and [Cl−]. This could potentially be resolved by conducting peri-prandial time course studies on salt-fed rainbow trout.
Dietary salt loading and endpoints of gill transcellular ion transport
Alterations in gill morphology as well as active ion transport machinery of the trout gill were noted in response to dietary salt loading (Figs. 2, 3, 4). With regard to gill morphology, a reduction in the number of gill epithelium NKA-ir cells was observed and this was driven by an apparent reduction in the presence of NKA-ir cells in the primary gill filament of salt-fed fish as well as an almost complete disappearance of NKA-ir cells from the secondary lamella. Consistent with the former observation, a decrease in the number of MRCs exposed at the surface of the primary gill filament epithelium was also observed by morphometric analysis of SEM images and this, combined with a reduction in individual MRC surface area, led to an overall decrease in the fractional surface area of MRC exposure at the gill epithelium. In a FW environment, a reduction in the surface area of MRC exposure has been correlated with reduced ion uptake across the gill of fishes (Perry 1997; Laurent and Perry 1990; Perry and Laurent 1989; Perry et al. 1992) and previous studies have also noted decreased Na+ and Cl− uptake across the gills of salt-fed fish (Perry and Rivero-Lopez 2012; Smith et al. 1995). Therefore the morphological changes observed in the current study seem consistent with the expected physiological consequences of longer term salt feeding (i.e. a reduction in active Na+ and Cl− uptake). In addition, the rare presence of a NKA-ir cell in the lamellar epithelium of salt-fed fish in the current study is also consistent with an almost complete disappearance of lamellar epithelium NKA-ir cells following acclimation of salmonids from FW to SW (Pisam et al. 1988; Uchida et al. 1996). However, Salman and Eddy (1987) reported an increase in rainbow trout gill ‘chloride cell’ (=mitochondrion-rich cell) numbers (described as a fraction of total gill cell numbers) in response to salt feeding and similarly, Perry et al. (2006) reported an increase in rainbow trout gill NKA-ir cell numbers and NKA-ir cell surface area following salt feeding. In addition, Perry et al. (2006) also reported that NKA-ir cells were more numerous in the lamellar epithelium following salt feeding. In both cases, these authors saw an increase in gill NKA activity in association with the changes observed in gill NKA-ir/chloride cells, whereas in this study no change in gill NKA activity was observed. The reason for these differences are not entirely clear as the dietary salt content and duration of feeding used in this study are consistent with both the work of Salman and Eddy (1987) and Perry et al. (2006). However, Pyle et al. (2003) did not find significant differences in gill NKA activity after 6 days of dietary Na+ loading and in the current study it was found that transcript abundance of FW-associated NKA catalytic subunit α1a was reduced, coupled with no change in SW-associated α1b subunit. Downregulation of rainbow trout gill tissue FW associated NKA catalytic subunit has been associated with a switchover to hypo-osmoregulation from hyperosmoregulation following acclimation to SW (Richards et al. 2003). In contrast, an increase in mRNA abundance of the SW-associated NKA catalytic subunit was suggested to be the determining factor in the ability of fish to acclimate to SW (Bystriansky et al. 2007). Therefore, it seems that by reducing the transcript abundance of the FW-associated NKA subunit, salt-fed fish in this study may be reconfiguring NKA so as to reduce its participation in ion uptake, but in addition, not yet switching over to SW active ion extrusion.
Additional alterations in the biochemistry and molecular physiology of the gill of salt-fed fish in this study support the idea that the gill epithelium is exhibiting a reduced capacity for active ion acquisition. For example, active Na+ and Cl− uptake across the gill epithelium of FW salmonids are proposed to be electrogenically coupled with VA activity in the gill (Evans and Claiborne 2009), therefore observations of a salt-feeding induced reduction in VA activity seems likely to contribute to a reduced capacity for Na+ and Cl− uptake. Moreover, nbc and cftrII transcript abundance decreased in the gills of salt-fed rainbow trout. NBC has been correlated with active Na+ uptake in the gill of freshwater rainbow trout (Parks et al. 2007). Therefore, a reduction in its transcript abundance may be indicative of reduced capacity for active Na+ uptake in salt-fed rainbow trout. CFTR has been classically associated with the apical membrane of the SW fish gill MRCs where it facilitates Cl− secretion (Marshall and Singer 2002), but it has also been detected in the FW fish gill where it is thought to be associated with the acquisition of Cl− (Marshall and Singer 2002; Madsen et al. 2007; Tang and Lee 2011). Therefore, it seems plausible that a decrease in cftrII transcript abundance in the gill of FW salt-fed rainbow trout may reflect a reduced capacity for Cl− uptake via the latter mechanism.
Dietary salt loading and endpoints of gill paracellular ion transport
Transcript abundance of cldn-6, -8d, -10d and -10e increased in response to dietary salt-loading, while that of tric decreased. Based on studies conducted using primary cultured rainbow trout gill epithelia, increased cldn-6 and -8d mRNA abundance and decreased tric mRNA abundance (coupled with increased Tric protein abundance) have been associated with reduced paracellular permeability (Kelly and Chasiotis 2011; Kolosov and Kelly 2013; Kolosov et al. 2014). Therefore, these proteins are proposed to be barrier forming in the gill epithelium of rainbow trout. Furthermore, in the puffer fish (Tetraodon nigroviridis) gill epithelium, Cldn-6 is found to be expressed only in MRCs where its abundance is greater in FW versus a SW environment (Bui et al. 2010; Bui and Kelly 2014, 2015). In addition, the subcellular localization of Cldn-6 is apicolateral in MRCs of FW fish and cytoplasmic in the MRCs of SW fish (Bui and Kelly 2014). In this regard, it has been suggested that in FW-acclimated puffer fish, Cldn-6 plays a role in reducing paracellular permeability associated with gill MRCs but in SW, it disperses from the junction to the cytoplasm and allows other Cldns to establish a ‘leaky’ pathway for the paracellular movement of Na+ (Bui and Kelly 2014). These other Cldn proteins are Cldn-10d and -10e, which are also MRC-specific in the gill epithelium of T. nigroviridis (Bui et al. 2010; Bui and Kelly 2014, 2015). Therefore it is noteworthy that in this study, transcript abundance of cldn-10d and -10e are also elevated in salt-fed rainbow trout and that previous studies have reported that cldn-10d is MRC-specific in the rainbow trout gill epithelium and proposed that cldn-10e is highly expressed in MRCs (although not exclusive to them) (Kolosov et al. 2014). Therefore, keeping in mind that salt-fed rainbow trout were sampled 24 h after last feeding, when serum ion levels were no different from control animals and when excess dietary salt load had presumably been eliminated (see “Dietary salt loading and systemic endpoints of ionoregulatory homeostasis”), it seems reasonable to suggest that salt-fed animals may be enhancing the barrier properties of the gill epithelium as the animal once again becomes faced with the problem of obligatory ion loss to hypoosmotic surroundings. In contrast, in the hours immediately following a salt load when the fish increase ion efflux rates to eliminate excess salt, Cldn-10d and -10e may play a role in facilitating this, perhaps in conjunction with a decrease in the abundance or shift in the subcellular localization of barrier proteins such as Cldn-6 and -8d. In future studies, it will be very interesting to examine this possibility in more detail.
IPW exposure, dietary salt loading and systemic endpoints of ionoregulatory homeostasis
IPW exposure reduced serum [Na+] and [Cl−] in trout and this was coupled with tissue hydration as muscle moisture content was found to have increased. Exposure to an ion-poor environment typically results in a reduction in circulating Na+ and Cl− levels in the plasma or serum of fishes (Laurent et al. 1994; Greco et al. 1996; Chasiotis et al. 2009, 2012a, b; Flores and Shrimpton 2012; Duffy et al. 2011) and following an abrupt exposure of trout to an ion-poor environment, this is most likely due to reduced Na+ and Cl− influx coupled with increased efflux rates (Perry and Laurent 1989). In fish fed a salt-rich diet, serum Na+ and Cl− levels as well as muscle moisture content were not significantly different from control fish 24 h after abrupt exposure to IPW, suggesting that salt feeding did not impact systemic salt and water balance in rainbow trout abruptly exposed to IPW (at least at a 24 h time point). In this regard, we are unaware of any previous study that has examined serum ion levels or muscle moisture content of salt-fed rainbow trout (or any other freshwater fish) following abrupt exposure to IPW.
IPW exposure, dietary salt loading and endpoints of gill transcellular ion transport
A number of alterations in gill morphology were observed in trout exposed to IPW for 24 h. This did not include a significant change in the number of NKA-ir cells, but a significant (albeit modest) decrease in the number of MRCs exposed at the surface of the gill epithelium was seen. More importantly, a significant increase in individual MRC surface area occurred, resulting in an increase in MRC fractional surface area. These changes are in accord with observations of Perry and Laurent (1989) and were likely aimed at facilitating ion uptake by maximizing the surface area of MRCs interfacing with IPW. In salt-fed fish exposed to IPW, NKA-ir cells were also not observed to significantly alter in number and as such remained low relative to numbers found in FW fish. The number of MRCs exposed at the surface of the gill also did not alter in salt-fed fish, and although individual MRC surface area increased, this response was not as robust as seen in control fish. The net result was that salt-fed fish exhibited a MRC fractional surface area that was not significantly altered 24 h following IPW exposure. This would be in line with a reduced capacity for active ion uptake across the gill epithelium of salt-fed IPW-exposed fish, but at this stage in the acclimation process these modest changes in MRC exposure do not appear to adversely affect the ability of the fish to maintain salt and water balance. This seems likely to be related to the availability of elevated levels of dietary ions early in the exposure period.
No changes in NKA and VA activity were observed in response to IPW. For NKA, this response is consistent with the observations of Flores and Shrimpton (2012). However, the response of these enzymes to IPW seems to be species specific as both NKA and VA activity have been reported to increase in the gill of IPW-exposed moonlight gourami Trichogaster microlepis (Huang et al. 2010), whereas Duffy et al. (2011) reported unaltered activity in puffer fish (Tetraodon biocellatus) and Chasiotis et al. (2009) reported lowered levels in goldfish (Carassius auratus). In this study, transcript abundance of molecular components of NKA remained largely unaltered in response to the IPW exposure. However, an increase in α1c mRNA abundance was observed, and this increase was absent in the gill of salt-fed trout. Although a substantial amount of research has been done regarding nka α1a and α1b subunits and their switching during the FW-SW transition in rainbow trout (e.g. Richards et al. 2003; McCormick et al. 2009; Flores and Shrimpton 2012), no function has been assigned to the α1c isoform in the gill of rainbow trout. Indeed, this isoform has not been found to vary in response to salinity change in Atlantic salmon (Nilsen et al. 2007). Therefore an IPW-induced increase in α1c transcript abundance in the current study may indicate a role for this subunit in ion acquisition under ion-poor conditions. The fact that α1c mRNA abundance was not observed to alter in the gills of salt-fed fish exposed to IPW supports this view as quantitative changes in gill epithelium morphology of salt-fed fish exposed to IPW are not typical of fish in ion-poor surroundings.
A significant IPW-induced decrease in the mRNA abundance of nbc was also observed in control fish. This was surprising as NBC has been associated with active Na+ uptake in the gill of freshwater rainbow trout, where it has been localized in a subpopulation of MRCs (Parks et al. 2007). Therefore, a reduction in transcript abundance may be indicative of reduced capacity for active Na+ uptake in salt-fed rainbow trout or a reduction in the number of NBC-containing MRCs. The functional significance of this change in gill of IPW-challenged fish will require further investigation.
IPW exposure, dietary salt loading and endpoints of gill paracellular ion transport
Tight junction depth as well as mRNA abundance of select TJ proteins have both been reported to increase in the gill of IPW-acclimated fish (Chasiotis et al. 2012b; Duffy et al. 2011), and this is proposed to contribute to reduced passive ion loss to the surrounding water. Indeed, if serum acquired from fish acclimated to IPW is used as a media supplement for promoting the growth and development of a primary cultured model gill epithelium, the result is a model tissue that exhibits greater transepithelial resistance and lower paracellular permeability than preparations supplemented with either fetal bovine serum or serum acquired from FW fish (Chasiotis et al. 2012b). Therefore in the current study it comes as no surprise that select genes encoding TJ proteins are elevated 24 h following IPW exposure. In addition to this, Perry and Laurent (1989) reported that rainbow trout abruptly exposed to IPW water exhibited a reduction in ion efflux rates by 24 h which can be presumed to be (or primarily be) a reduction in diffusive loss through the paracellular pathway.
Of particular interest in the current study is that (1) mRNA abundance of several genes encoding TJ proteins (i.e. cldn-7, -23a, -27b, -28b and -32a) increased in response to the IPW challenge regardless of the amount of NaCl present in the diet and (2) transcript abundance of ZO-1, cldn-8b, -29a and -31 were also increased in response to IPW exposure, but only in control fish and not in salt-loaded fish. In addition to this, cldn-30 mRNA abundance increased in control fish 24 h following IPW exposure, but in salt-fed fish exposed to IPW cldn-30 transcript abundance increased even more. Therefore 24 h following IPW exposure, cldn-30 mRNA abundance was statistically elevated in salt-fed fish versus control, which contrasted with circumstances in FW where cldn-30 mRNA abundance did not statistically differ between control and salt-fed fish. In gill tissue, mRNA encoding cldn-7 and -30 are amongst the most abundant of all cldns transcript. In a primary cultured trout gill epithelium model, cldn-7 and -30 have been associated with restructuring of the gill epithelium during development but have not been directly associated with development of the electroresistive barrier properties of the tissue (Kolosov et al. 2014). Therefore keeping in mind that fish held in IPW for 24 h, irrespective of prior salt-feeding, seem likely to have returned to circumstances where they are now facing the problems of obligatory ion loss to hypoosmotic surroundings, cldn-7 and -30 may be involved in restructuring of the gill epithelium in IPW. That this process is more emphasized in fish that have been salt-fed (i.e. elevated cldn-30 mRNA levels in salt-fed IPW exposed fish) seems reasonable as presumably the gill tissue of these fish would have more changes to make in the face of an extreme hyposmotic challenge.
Of the other transcripts that increase in response to IPW (i.e. ZO-1, cldn-8b, -23a, -27b, -28b, -29a, -31 and -32a), all but one have been demonstrated to increase in association with the development of electroresistive barrier properties of a cultured rainbow trout gill epithelium (Kolosov et al. 2014). Therefore an elevation in the mRNA abundance of these TJ proteins in the gills of fish exposed to IPW may indicate enhancement gill barrier properties. The exception to this is cldn-32a. In studies conducted in vitro, cldn-32a transcript abundance remains stable for around 72 h following the seeding of primary cultured rainbow trout gill cells in cell culture flasks (Kolosov et al. 2014). At 72 h, cultured gill cells are confluent and at this point cldn-32a mRNA levels decrease. Levels also decrease after the formation of a confluent epithelium in insert cultured gill epithelia, and this occurs in association with an increase in transepithelial resistance (Kolosov et al. 2014). Therefore it would appear that Cldn-32a may also be involved in gill restructuring under conditions of environmental change, but this possibility will require further examination. In addition, further studies will also be required to address another important question. Why do some TJ proteins exhibit a transcriptional response to IPW irrespective of salt feeding, while others respond to IPW in control animals, but not in animals that have been salt-fed? It is possible that a lack of response in salt-fed animals exposed to IPW for 24 h may reflect how TJ proteins in the gills of these fish have responded to salt loading in the early stages of meal processing when blood salt levels are (based on observations of others) presumed to increase and salt efflux rates are elevated. Therefore what is being observed in the current study may be a delayed response to IPW by cldn-8b, -29a and -31. Future time-course studies will help to resolve this possibility. It is also interesting to note that changes in cldn-31 mRNA abundance mirror changes in MRC fractional surface area (Fig. 6e). This may be of importance because transcript abundance of cldn-31 has been reported to be ~60 times more abundant in MRC-containing cultured trout gill epithelia versus those that are composed only of PVCs (Kolosov et al. 2014). This would suggest that changes in the transcript abundance of cldn-31 may reflect changes that are taking place in the MRC population of the primary gill filament and/or suggest that differences in cldn-31 mRNA abundance between IPW-exposed control and IPW-exposed salt-fed fish may reflect differences in MRC-PVC TJ area at the gill surface.
Statistical analysis indicated that transcript encoding Cldn-8c and -10c was not affected by salt-loading in FW. In control fish gill, cldn-8c and -10c mRNA were also unaffected by IPW water. However, in salt-fed fish exposed to IPW for 24 h, cldn-8c and -10c mRNA significantly decreased. In our view, this response was modest (arguably marginal) for cldn-8c but particularly pronounced for cldn-10c. Cldn-10c exhibits a very restricted expression pattern in discrete tissues of rainbow trout, and is expressed predominantly in the gill (with low levels being found in the kidney and skin). But this Cldn is not found in the brain, eye or any organ within the gastrointestinal tract (Kolosov et al. 2014). In the gill epithelium of rainbow trout, cldn-10c is also not found in PVCs, but does appear in primary cultured preparations when MRCs are introduced (Kolosov et al. 2014). Therefore this TJ protein is presumed to be associated with MRCs in the gill epithelium of rainbow trout. In this study, a reduction in gill NKA-ir cells (i.e. MRCs) is observed in salt-fed fish whether they are in FW or IPW. Therefore if cldn-10c is generically associated with gill MRCs, it might be expected that a reduction would be seen in salt-fed fish irrespective of the ion content of surrounding water. Therefore, it seems possible that cldn-10c may be associated with a specific type of gill MRC that reduces in abundance when salt-fed fish are exposed to IPW. Alternately, although cldn-10c is not found in gill PVCs, it may be present in MRCs as well as other types, or another type of non-PVC gill cell, and these cells may reduce in number or require a reduction in cldn-10c transcript abundance in this group of animals.
Perspectives
The contribution of the TJ complex to ionoregulatory homeostasis in fishes is broadly accepted to be an important one (for reviews see Evans et al. 2005; Chasiotis et al. 2012b; Kolosov et al. 2013; Günzel and Yu 2013). The TJ complex is well characterized in the gill epithelium because the gross electroresistive properties of the gill are reversed in FW (volume-loaded, ion-deprived) versus SW (volume-deprived, ion-loaded) animals and this is generally accepted to be driven by changes in TJ properties. Therefore a fundamental role for TJ proteins in aquatic vertebrates can be considered by examining FW and SW residing vertebrates. In contrast, using the diet to salt-load FW fish produces a peculiar physiological condition that is different from that found in FW and SW animals. Salt-loaded fish are both volume- and ion-loaded (Chen et al. 2007; Johnson and Olson 2009a, b) and exposing salt-loaded fish to IPW accentuates aspects of this physiological condition. In this study, the combination of these experimental variables unquestionably altered the molecular physiology of the gill TJ complex in several distinct ways. Therefore experimental manipulations such as these seem likely to be useful in terms of understanding mechanistic aspects of paracellular as well as transcellular solute movement. In future studies it would be useful to couple observations on the molecular physiology of the gill epithelium TJ with measurements of paracellular permeability. Furthermore, and in a broader sense, this general approach can be extended to other tissues, organs and systems involved in the regulation of salt and water balance. For example, salt-loading causes high blood pressure in fish (Olson and Hoagland 2008; Perry et al. 2011). Therefore, mechanisms and endocrine axes involved in the regulation of blood pressure and volume may influence the molecular physiology of the TJ complex in salt loaded animals and manipulations such as those used in this study may help to provide insight into such possibilities.
References
Alam MS, Watanabe WO, Myers AR, Rezek TC, Carroll PM, Skrabal SA (2015) Effects of dietary salt supplementation on growth, body composition, tissue electrolytes, and gill and intestinal Na+/K+ ATPase activities of black sea bass reared at low salinity. Aquaculture 250:250–258
Al-Amoudi MM (1987) The effects of high salt diet on the direct transfer of Oreochromis mossambicus, O. spilurus and O. aureus/O. niloticus hybrids to seawater. Aquaculture 64:333–338
Basulto S (1976) Induced saltwater tolerance in connection with inorganic salts in the feeding of Atlantic salmon (Salmo salar L.). Aquaculture 8:45–55
Bui P, Kelly SP (2014) Claudin-6, -10d, and -10e contribute to seawater acclimation in the euryhaline puffer fish Tetraodon nigroviridis. J Exp Biol 217:1758–1767
Bui P, Kelly SP (2015) Claudins in a primary cultured puffer fish (Tetraodon nigroviridis) gill epithelium model alter in response to acute seawater exposure. Comp Biochem Physiol A Mol Integr Physiol 189:91–101
Bui P, Bagherie-Lachidan M, Kelly SP (2010) Cortisol differentially alters claudin isoform mRNA abundance in a cultured gill epithelium from puffer fish (Tetraodon nigroviridis). Mol Cell Endocrinol 317:120–126
Bystriansky JS, Frick NT, Richards JG, Schulte PM, Ballantyne JS (2007) Failure to up-regulate gill Na+, K+-ATPase α-subunit isoform α1b may limit seawater tolerance of land-locked Arctic char (Salvelinus alpinus). Comp Biochem Physiol A Mol Integr Physiol 148:332–338
Chasiotis H, Kelly SP (2008) Occludin immunolocalization and protein expression in goldfish. J Exp Biol 211:1524–1594
Chasiotis H, Effendi JC, Kelly SP (2009) Occludin expression in goldfish held in ion-poor water. J Comp Physiol B 179:145–154
Chasiotis H, Wood CM, Kelly SP (2010) Cortisol reduces paracellular permeability and increases occludin abundance in cultured trout gill epithelia. Mol Cel Endocrinol 323:232–238
Chasiotis H, Kolosov D, Bui P, Kelly SP (2012a) Tight junctions, tight junction proteins and paracellular permeability across the gill epithelium of fishes: a review. Resp Physiol Neurobiol 184:269–281
Chasiotis H, Kolosov D, Kelly SP (2012b) Permeability properties of the teleost gill epithelium under ion-poor conditions. Am J Physiol Regul Integr Comp Physiol 302:R727–R739
Chen X, Moon TW, Olson KR, Dombkowski RA, Perry SF (2007) The effects of salt-induced hypertension on α1-adrenoreceptor expression and cardiovascular physiology in the rainbow trout (Oncorhynchus mykiss). Am J Physiol Regul Integr Comp Physiol 293:R1384–R1392
Chen CC, Kolosov D, Kelly SP (2015) Effect of the liquorice root derivatives on salt and water balance in a teleost fish, rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol A Mol Integr Physiol 180:86–97
Cnaani A, Barki A, Slosman T, Scharcanski A, Milstein A, Harpaz S (2010) Dietary salt supplement increases the growth rate in freshwater cultured tilapia hybrids. Aquacult Res 41:1545–1548
D’Cruz LM, Wood CM (1998) The influence of dietary salt and energy on the response to low pH in juvenile rainbow trout. Physiol Zool 71:642–657
Donini A, O’Donnell MJ (2005) Analysis of Na+, Cl−, K+, H+ and NH4 + concentration gradients adjacent to the surface of anal papillae of the mosquito Aedes aegypti: application of self-referencing ion-selective microelectrodes. J Exp Biol 208:603–610
Duffy NM, Bui P, Bagherie-Lachidan M, Kelly SP (2011) Epithelial remodeling and claudin mRNA abundance in the gill and kidney of puffer fish (Tetraodon biocellatus) acclimated to altered environmental ion levels. J Comp Physiol B 181:219–238
Duston J (1993) Effects of dietary betaine and sodium chloride on seawater adaptation in Atlantic salmon parr (Salmo salar L.). Comp Biochem Physiol 105A:673–677
Eroldoğan OT, Kumlu M, Kir M, Kiris GA (2005) Enhancment of growth and feed utilization of the European sea bass (Dicentrarchus labrax) fed supplementary dietary salt in freshwater. Aquacult Res 36:361–369
Evans DH, Claiborne JB (2009) Osmotic and Ionic Regulation in Fishes. In: Evans DH (ed) Osmotic and ionic regulation: cells and animals. CRC Press, Boca Raton, pp 295–367
Evans DH, Piermarini PM, Choe KP (2005) The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev 85:97–177
Farquhar MG, Palade GE (1963) Junctional complexes in various epithelia. J Cell Biol 17:375–412
Flores AM, Shrimpton MJ (2012) Differential physiological and endocrine responses of rainbow trout, Oncorhynchus mykiss, transferred from fresh water to ion-poor or salt water. Gen Comp Endocrinol 175:244–250
Fontainhas-Fernandes A, Russell-Pinto F, Gomes E, Reis-Henriques MA, Coimbra J (2001) The effect of dietary sodium chloride on some osmoregulatory parameters of the teleost, Oreochromis niloticus, after transfer from freshwater to seawater. Fish Physiol Biochem 23:307–316
Garcia LDO, Becker AG, Copatti CE, Baldisserotto B, Neto JR (2007) Salt in the food and water as a supportive therapy for Ichthyophthirius multifiliis infestation on silver catfish, Rhamdia quelen, fingerlings. J World Aqua Soc 38:1–11
Gatlin DM III, MacKenzie DS, Craig SR, Neill WH (1992) Effects of dietary sodium chloride on red drum juveniles in waters of various salinities. Prog Fish Cult 54:220–227
Glover CN, Bucking C, Wood CM (2013) The skin of fish as a transport epithelium: a review. J Comp Physiol B 183:877–891
Greco AM, Fenwick JC, Perry SF (1996) The effects of soft-water acclimation on gill structure in the rainbow trout Oncorhynchus mykiss. Cell Tissue Res 285:75–82
Günzel D, Fromm M (2012) Claudins and other tight junction proteins. Compr Physiol 2:1819–1852
Günzel D, Yu ASL (2013) Claudins and the modulation of tight junction permeability. Physiol Rev 93:525–569
Harpaz S, Hakim Y, Slosman T, Eroldogan OT (2005) Effects of adding salt to the diet of Asian sea bass Lates calcarifer reared in fresh or salt water recirculating tanks, on growth and brush border enzyme activity. Aquaculture 248:315–324
He FJ, MacGregor GA (2009) A comprehensive review on salt and health and current experience of worldwide salt reduction programmes. J Hum Hypertens 23:363–384
Huang CY, Chao PL, Lin HC (2010) Na+/K+-ATPase and vacuolar-type H+-ATPase in the gills of the aquatic air-breathing fish Trichogaster microlepis in response to salinity variation. Comp Biochem Physiol A 155:309–318
Johnson KR, Olson KR (2009a) Responses of the trout cardiac natriuretic peptide system to manipulation of salt and water balance. Am J Physiol Regul Integr Comp Physiol 296:R1170–R1179
Johnson KR, Olson KR (2009b) The response of non-traditional natriuretic peptide production sites to salt and water manipulations in the rainbow trout. J Exp Biol 212:2991–2997
Kelly SP, Chasiotis H (2011) Glucocorticoid and mineralocorticoid receptors regulate paracellular permeability in a primary cultured gill epithelium. J Exp Biol 214:2308–2318
Kelly SP, Peter RE (2006) Prolactin-releasing peptide, food intake and hydromineral balance in goldfish. Am J Physiol Regul Integr Comp Physiol 291:R1474–R1481
Keshavanath P, Oishi CA, Leao da Fonesca FA, Affonso EG, Filho MP (2012) Growth response of tambaqui (Colossoma macropomum) fingerlings to salt (sodium chloride) supplemented diets. J Fish Aqua Sci 7:439–446
Kolosov D, Kelly SP (2013) A role for tricellulin in the regulation of gill epithelium permeability. Am J Physiol Regul Integr Comp Physiol 304:R1139–R1148
Kolosov D, Bui P, Chasiotis H, Kelly SP (2013) Claudins in teleost fishes. Tissue Barrier 1:e25391
Kolosov D, Chasiotis H, Kelly SP (2014) Tight junction protein gene expression patterns and changes in transcript abundance during development of model fish gill epithelia. J Exp Biol 217:1667–1681
Laurent P, Perry SF (1990) Effects of cortisol on gill chloride cell morphology and ionic uptake in the freshwater trout, Salmo gairdneri. Cell Tissue Res 259:429–442
Laurent P, Dunel-Erb S, Chevalier C (1994) Gill epithelial cells kinetics in a freshwater teleost, Oncorhynchus mykiss during adaptation to ion-poor water and hormonal treatments. Fish Physiol Biochem 13:353–370
Loh YH, Christoffels A, Brenner S, Hunziker W, Venkatesh B (2004) Extensive expansion of the claudin gene family in the teleost fish, Fugu rubripes. Genom Res 14:1248–1257
Madsen SS, Jensen LN, Tipsmark CK, Kiilerich P, Borski RJ (2007) Differential regulation of cystic fibrosis transmembrane conductance regulator and Na+, K+-ATPase in gills of striped bass, Morone saxatilis: effect of salinity and hormones. J Endocrinol 192:249–260
Marshall WS, Grosell M (2006) Ion transport, osmoregulation, and acid-base balance. In: Evans DH, Claiborne JB (eds) The physiology of fishes, 3rd edn. Taylor and Francis Group, Boca Raton, pp 177–210
Marshall WS, Singer TD (2002) Cystic fibrosis transmembrane conductance regulator in teleost fish. Biochim Biophys Acta 1566:16–27
McCormick SD (1993) Methods for nonlethal gill biopsy and measurement of Na+, K+-ATPase activity. Can J Fish Aquat Sci 50:656–658
McCormick SD, Regish AM, Christensen AK (2009) Distinct freshwater and seawater isoforms of Na+/K+-ATPase in gill chloride cells of Atlantic salmon. J Exp Biol 212:3994–4001
Meneton P, Jeunemaitre X, De Wardener HE, MacGregor GA (2005) Links between dietary salt intake, renal salt handling, blood pressure, and cardiovascular diseases. Physiol Rev 85:679–715
Nilsen TO, Ebbesson LOE, Madsen SS, McCormick SD, Andersson E, Bjornsson BT, Prunet P, Stefansson SO (2007) Differential expression of gill Na+, K+-ATPase α and β subunits, Na+, K+, 2Cl− cotransporter and CFTR anion channel in juvenile anadromous and landlocked Atlantic salmon Salmo salar. J Exp Biol 210:2885–2896
Olson KR, Hoagland TM (2008) Effects of freshwater and saltwater adaptation and dietary salt on fluid compartments, blood pressure, and venous capacitance in trout. Am J Physiol Regul Integr Comp Physiol 294:R1061–R1067
Parks SK, Tresguerres M, Goss GG (2007) Interactions between Na+ channels and Na+-HCO3 − cotransporters in the freshwater fish gill MR cell: a model for transepithelial Na+ uptake. Am J Physiol Cell Physiol 292:C935–C944
Pelletier D, Besner M (1992) The effect of salty diets and gradual transfer to sea water on osmotic adaptation, gill Na+, K+-ATPase activation, and survival of brook charr, Salvelinus fontinalis, Mitchill. J Fish Biol 41:791–803
Perry SF (1997) The chloride cell: structure and function in the gills of freshwater fishes. Annu Rev Physiol 59:325–347
Perry SF, Laurent P (1989) Adaptational responses of rainbow trout to lowered external NaCl concentration: contribution of the branchial chloride cell. J Exp Biol 147:147–168
Perry SF, Rivero-Lopez L (2012) Does the presence of a seawater gill morphology induced by dietary salt loading affect Cl− uptake and acid-base regulation in freshwater rainbow trout Oncorhynchus mykiss. J Fish Biol 80:301–311
Perry SF, Goss GG, Fenwick JC (1992) Interrelationships between gill chloride cell morphology and calcium uptake in freshwater teleosts. Fish Physiol Biochem 10:327–337
Perry SF, Rivero-Lopez L, McNeill B, Wilson J (2006) Fooling a freshwater fish: how dietary salt transforms the rainbow trout gill into a seawater gill phenotype. J Exp Biol 209:4591–4596
Perry SF, Ellis K, Russell J, Bernier NJ, Montpetit C (2011) Effects of chronic dietary salt loading on the rennin angiotensin and adrenergic systems of rainbow trout (Oncorhynchus mykiss). Am J Physiol Regul Integr Comp Physiol 301:R811–R821
Pisam M, Prunet P, Boeuf G, Rambourg A (1988) U1trastructural features of chloride cells in the gill epithelium of the Atlantic salmon, Salmo salar, and their modifications during smoltification. Am J Anat 183:235–244
Pyle GG, Kamunde CN, McDonald DG, Wood CM (2003) Dietary sodium inhibits aqueous copper uptake in rainbow trout (Oncorhynchus mykiss). J Exp Biol 206:609–618
Richards JG, Semple JW, Bystriansky JS, Schulte PM (2003) Na+/K+-ATPase-isoform switching in gills of rainbow trout (Oncorhynchus mykiss) during salinity transfer. J Exp Biol 206:4475–4486
Salman NA, Eddy FB (1987) Response of chloride cell numbers and gill Na+ K+ ATPase activity of freshwater rainbow trout (Salmo gairdneri Richardson) to salt feeding. Aquaculture 61:41–48
Salman NA, Eddy FB (1988) Effect of dietary sodium chloride on growth, food intake and conversion efficiency in rainbow trout (Salmo gairdneri Richardson) to salt feeding. Aquaculture 70:131–144
Salman NA, Eddy FB (1990) Increased sea-water adaptability of non-smolting rainbow trout by salt feeding. Aquaculture 86:259–270
Sardet C, Pisam M, Maetz J (1979) The surface epithelium of teleostean fish gills. Cellular and junctional adaptations of the chloride cell in relation to salt adaptation. J Cell Biol 80:96–117
Smith NF, Eddy FB, Talbot C (1995) Effect of dietary salt load on transepithelial Na+ exchange in freshwater rainbow trout (Oncorhynchus mykiss). J Exp Biol 198:2359–2364
Staurnes M, Finstad B (2000) The effects of dietary NaCl supplement on hypo-osmoregulatory ability and seawater performance of Arctic charr (Salvelinus alpinus L.) smolts. Aqua Res 31:737–743
Tang CH, Lee TH (2011) Ion-deficient environment induces the expression of basolateral chloride channel, ClC-3-like protein, in gill mitochondrion-rich cells for chloride uptake of the tilapia Oreochromis mossambicus. Physiol Biochem Zool 84:54–67
Trombetti F, Ventrella V, Pagliarani A, Ballestrazzi R, Galeotti M, Trigari G, Pirini M, Borgatti AR (1996) Response of rainbow trout gill (Na++K+)-ATPase and chloride cells to T3 and NaCl administration. Fish Physiol Biochem 15:265–274
Uchida K, Kaneko T, Yamauchi K (1996) Morphometrical analysis chloride cell activity in the gill filaments and lamellae and changes in Na+, K+-ATPase activity during seawater adaptation in chum salmon fry. J Exp Zool 276:193–200
Wood CM, Bucking C (2011) The role of feeding in salt and water balance. In: Grosell M, Farrell AP, Brauner CJ (eds) Fish physiology, vol 30. Academic Press, San Diego, pp 166–213
Zall DM, Fisher D, Garner MD (1956) Photometric determination of chlorides in water. Anal Chem 28:1665–1678
Zaugg WS, Roley DD, Prentice EF, Gores KX, Waknitz FW (1983) Increased seawater survival and contribution to the fishery of Chinook salmon (Oncorhynchus tshawytscha) by supplemental dietary salt. Aquaculture 32:183–188
Acknowledgments
This work was supported by a NSERC Discovery Grant to SPK. DK was supported by an Ontario Graduate Scholarship followed by a York University Provost Dissertation Scholarship. The α5 monoclonal antibody was developed by DM Fambrough and obtained from the Developmental Studies Hybridoma Bank (University of Iowa, Department of Biological Sciences, Iowa City, IA, 52242, USA).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Heldmaier.
Rights and permissions
About this article
Cite this article
Kolosov, D., Kelly, S.P. Dietary salt loading and ion-poor water exposure provide insight into the molecular physiology of the rainbow trout gill epithelium tight junction complex. J Comp Physiol B 186, 739–757 (2016). https://doi.org/10.1007/s00360-016-0987-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00360-016-0987-z