Abstract
Haemocyanin (Hc) is a copper-containing respiratory protein, floating freely dissolved in the hemolymph of many arthropod species. A typical haemocyanin is a hexamer or oligohexamer of six identical or similar subunits, with a molecular mass around 75 kDa each. In the crustaceans, the haemocyanins appear to be restricted to the remipedes and the malacostracans. We have investigated the haemocyanins of two freshwater shrimps, the Amano shrimp Caridina multidentata and the bamboo shrimp Atyopsis moluccensis. We obtained three full-length and one partial cDNA sequences of haemocyanin subunits from the Amano shrimp, which were assigned to the α- and γ-types of decapod haemocyanin subunits. Three complete and two partial haemocyanin cDNA sequences were obtained from the bamboo shrimp, which represent subunit types α, β and γ. This is the first time that sequences of all three subunit types of the decapod haemocyanins were obtained from a single species. However, mass spectrometry analyses identified only α- and γ-type subunits, suggesting that a β-subunit is not a major component of the native haemocyanin of the bamboo shrimp. Phylogenetic and molecular clock analyses showed that malacostracan haemocyanins commenced to diversify into distinct subunit types already ~515 million years ago. β-subunits diverged first, followed by α- and γ-type subunits ~396 million years ago. The haemocyanins of phyllocarids and peracarids form distinct clades within the α/γ-cluster. Within the Caridea, an early divergence of distinct α-type subunits occurred ~200 MYA. The tree of the γ-subunits suggests a common clade of the Caridea (shrimps) and Penaeidae (prawns).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oxygen supply in many arthropod species is mediated by haemocyanin, which is floating freely dissolved in the hemolymph (van Holde and Miller 1995; Terwilliger 1998; Burmester 2002). Arthropod haemocyanins are composed of six identical or similar subunits of about 620–660 amino acids, with molecular masses in the range of 75 kDa each (van Holde and Miller 1995; Burmester 2002). The homo- or heterohexamers of about 450 kDa may associate with large multihexameric structures of up to 8 × 6 subunits (Markl et al. 1986; Markl and Decker 1992; van Holde and Miller 1995). Each subunit contains two Cu2+ ions, which are stabilized by six histidine residues, forming the O2-binding site, and every pair of Cu2+ ions will bind one O2 molecule (Gaykema et al. 1984; Linzen et al. 1985). Subunit composition and arrangement are typically taxon-specific and may have been conserved in some arthropod taxa for several 100 million years (Burmester 2002; Markl and Decker 1992; Rehm et al. 2012).
Haemocyanins have been identified in all arthropod subphyla (Burmester 2001, 2002; Kusche et al. 2002; Pick et al. 2009). Within the Crustacea, haemocyanins appear to be restricted to the Malacostraca and Remipedia (Mangum 1985; Markl and Decker 1992; Ertas et al. 2009), while other classes either employ haemoglobin as oxygen transport protein or are devoid of any respiratory protein (Mangum 1985). A putative haemocyanin has been reported in the cirripede Sacculina carcini (Herberts and de Frescheville 1981), but it is unclear whether this was actually an endogenous protein or derived from the decapod host, Carcinus maenas.
Haemocyanins have been identified in all malacostracan subclasses, including Phyllocarida (Leptostraca), Hoplocarida (mantis shrimps), Pericarida (Isopoda, Amphipoda, Mysidacea, Cumacea), Syncarida (Anaspidacea, Bathynellacea) and Eucarida (Decapoda, Euphausiacea) (Mangum 1983; Markl 1986; Burmester 2002). Most malacostracan haemocyanins form either mono- or di-hexamers, with the exceptions of the thalassinid shrimps Callianassa californiensis (Miller et al. 1976; Markl et al. 1986; Markl and Decker 1992) and Upogebia pusilla (Paoli et al. 2007; Micetic et al. 2010), which possess 4 × 6 haemocyanins. While structure and subunit composition of chelicerate and myriapod haemocyanins are strikingly conserved and may have been retained for more than 500 million years (Markl 1986; Markl et al. 1986; Burmester 2002; Rehm et al. 2012), malacostracan haemocyanins show much higher variability, e.g., subunits may be present only in some developmental stages or under certain physiological conditions (Durstewitz and Terwilliger 1997b; Terwilliger 1998; Decker et al. 2007). Moreover, subunit assemblies may differ between closely related species or even within populations (Markl 1986; Markl et al. 1986; Stöcker et al. 1988; Mangum and Joy 1997).
Employing immunological methods, Markl and co-workers classified the decapod haemocyanin subunits into three distinct types referred to as α, β and γ (Markl 1986; Markl et al. 1986). This classification is essentially supported by more recent molecular phylogenetic analyses (Burmester 2002; Hagner-Holler et al. 2005; Scherbaum et al. 2010). The α- and γ-subunits are more closely related to each other than to the β-type (Markl 1986; Markl et al. 1986; Kusche et al. 2003; Hagner-Holler et al. 2005; Scherbaum et al. 2010). α-and γ-type haemocyanin subunits separated close to 400 million years ago (MYA), while β-types diverged from the α/γ-clade already in the early Cambrian period ~520 MYA. The peracarid haemocyanins split from the β-type before the emergence of the α- and γ-subunits about 420 MYA (Scherbaum et al. 2010), whereas the non-respiratory pseudo-haemocyanins (cryptocyanins) (Burmester 1999; Terwilliger et al. 1999) diverged from γ-type haemocyanins.
To better understand the subunit evolution of crustacean haemocyanins, we investigated the haemocyanin from two freshwater shrimps, the Amano shrimp Caridina multidentata and the bamboo shrimp Atyopsis moluccensis. Both animals belong to the infraorder Caridea (Decapoda), a taxon that includes a large variety of shrimp-like crustaceans with a “primitive” body plan. The Caridea are assumed to be the sister group to all other Pleocyemata, which represent the large majority of the decapod crustaceans.
Materials and methods
Protein biochemistry
Adult specimens of C. multidentata and A. moluccensis were purchased from “Aquarium Tonndorf”, Hamburg, Germany. Hemolymph of one adult A. moluccensis and two adult C. multidentata specimens was withdrawn with a syringe and pooled where necessary. To avoid degradation, the hemolymph was diluted with an equal volume of stabilizing buffer consisting of 100 mM Tris–HCl, pH 7.8, 10 mM DTT, 10 mM protease inhibitor (Pefabloc, Roth), 10 mM MgCl2, and 10 mM CaCl2. Hemocytes and other particles were removed by a centrifugation for 20 min at 10,000×g (4 °C). The protein content of the hemolymph samples was quantified using the Bradford microassay (Roti-Quant) with BSA as a standard protein (Bradford 1976) and the OD280 method with BSA as a reference. The hemolymph proteins were separated by SDS-PAGE on 10 % gels under standard conditions (Laemmli 1970), using 10 μg protein per lane. The gels were stained with 0.1 % Coomassie Brilliant Blue in 10 % acetic acid/25 % isopropanol. For Western Blotting, the proteins were electrotransferred on nitrocellulose membranes and dried. Non-specific binding sites were blocked with 5 % non-fat dry milk in TBS (20 mM Tris–HCl, pH 7.5, 150 mM NaCl) for 2 h. We employed polyclonal antisera against the haemocyanins from the decapods Astacus astacus, Cancer pagurus, Galathea sp., Homarus americanus, Matuta sp., Ocypode sp., Pagurus bernhardus and Panulirus interruptus (Markl 1986), as well as antisera against haemocyanin subunits 1 and 2 from the cockroach Blaptica dubia (Pick et al. 2010). The antisera were diluted 1:2,500 in blocking solution and incubated at 4 °C overnight. After three successive washing steps with 0.1 % Tween 20 in TBS, the second antibody, a goat-anti-rabbit alkaline phosphatase conjugated IgG (L+H) (Dianova, Hamburg), was applied in a dilution of 1:10,000 in TBS for 1.5 h at room temperature. After washing as described above, the enzyme was detected with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate in 100 mM Tris–HCl, pH 9.5 and 100 mM NaCl in the dark.
The two-dimensional (2D) gel electrophoresis was performed with an immobilized pH gradient for the first dimension, ranging from pH 4.1 to 6.5 over a distance of 14 cm (Immobiline Dry Strip pH 4–7, GE Healthcare). The dry strips were rehydrated at room temperature over night in a fresh prepared solution, containing 8 M urea, 2 M thiourea, 1 % CHAPS, 19.4 M DTT, 0.5 % Pharmalyte 3–10, Bromophenol blue, plus 20 μg hemolymph protein from Atya moluccensis. The first dimension was prefocused at 300 V for 2 h, followed by a rise to 3,500 V in a linear gradient over 3 h and finished at constant 3,500 V for 2 h. Thereafter, the IEF strip was equilibrated in a buffer consisting of 50 mM Tris–HCl, pH 8.8, 6 M urea, 30 % glycerol and 4 % SDS. For the second dimension a 10 % PAGE was used and stained with Coomassie Brilliant Blue as described above.
Protein identification by nanoLC–ESI-ion trap analysis
Tryptic digestion of the bands from SDS-PAGE and selected spots from the 2D gel electrophoresis was done according to Shevchenko et al. (2006). Identification was performed on an Agilent 1100 LC/MSD Trap XCT Ultra equipped with a Chip Cube system using a Large Capacity Chip (II) (Agilent Technologies, Santa Clara, USA). Sample loading onto the enrichment column was performed at a flow rate of 4 μl/min (98 % mobile phase A: 0.2 % formic acid in H2O. 2 % mobile phase B: 100 % acetonitrile). Tryptic peptides were eluted from the reversed-phase column with a flow rate of 200 nl/min using a linear gradient elution of 2–40 % B in 23 min. For MS experiments, the following mode and tuning parameters were used: Scan range: 300–1,500 m/z, polarity: positive, capillary voltage 1,730 V. Flow and temperature of the drying gas were 3 l/min and 350 °C. The MS/MS experiments were carried out in data-dependent acquisition mode using the following parameters: scan range: 100–1,800 m/z, window for precursor ion selection: 4 Da, fragmentation amplitude: 1.25 V. After 3 MS/MS spectra the precursor ions were excluded from fragmentation for at least 0.2 min. The mgf files for database searching were generated by Data Analysis software 6.2 (Agilent Technologies, Santa Clara, USA). Protein identification was performed with the Mascot software (Perkins et al. 1999) using the NCBI nr database. The searches were performed with the following parameters: only tryptic peptides with up to one miss cleavage were allowed, 1.2 Da mass tolerances for precursor ions and 0.6 Da for fragment ions and carbamidomethyl cysteine and oxidized methionine were permitted as variable modifications.
Cloning of haemocyanin cDNA
Adult individuals of C. multidentata and A. moluccensis were killed with fluid N2. The cephalothorax region with a partially removed cuticula was thoroughly crushed with a mortar and pestle. The total RNA from single specimens was extracted using the RNeasy Kit (Qiagen, Hilden), following the manufacturer’s instructions. cDNA was generated by reverse transcription. For the first-strand cDNA synthesis, Super Script III reverse transcriptase (Invitrogen) was used. The subsequent PCR reactions were carried out with Platinum Supermix (Invitrogen) according to the manufacturer’s instructions. Two degenerated oligonucleotide primer pairs were designed after conserved regions in the centre of crustacean haemocyanins with the sequences 5′-GTNGCGGTYTCRAARTGYTCCAT-3′ and 5′-ATGGAYTTYCCNTTYTGGTGGAA-3′ or the second primer pair 5′-CAYCAYGTNCANTGGCA-3′ and 5′-DATRTTRTCCATRTAYTTRTG-3′, respectively. The PCR products of about 530 base pairs with primer pair one or 556 nucleotides with primer pair two were cloned using the pGEM-T Easy vector (Promega) into E. coli JM109 (Promega) and sequenced by a commercial service with the help of vector-specific primers (GATC, Germany). The reconstruction of the complete haemocyanin sequences was carried out using the GeneRacer Kit for full-length, RNA ligase-mediated rapid amplification of 5′ and 3′ cDNA ends (Invitrogen), following the manufacturer’s instructions. The gene-specific primers were designed to obtain the highest possible specificity for each subunit with the program Primerlyze (Janus Borner, unpublished) and are shown in Supplemental Table 1. PCR products of the expected length were cloned and sequenced as described above.
Sequence and phylogenetic analyses
The program provided at the ExPASy server (Swiss Institute of Bioinformatics; http://www.expasy.org/translate) was used for the prediction of the open reading frames. The sequences were assembled by hand and translated into amino acids with GeneDoc 2.7 (Nicholas et al. 1997). Putative signal peptides were identified with the program SignalP 4.0 (Petersen et al. 2011) using the standard options (“input sequences may include TM regions” without D-cutoff values).
A multiple sequence alignment was built by MAFFT (Katoh et al. 2005), employing the L-INS-i method. We included 107 haemocyanin sequences from 43 arthropod species. A complete list of haemocyanin subunits is given in the Supplemental Table 2. Minor corrections of the alignment were done according to the known 3D-structure of P. interruptus haemocyanin (Gaykema et al. 1984). For phylogenetic analyses, signal peptides and other short N-terminal extensions were removed. A phylogenetic tree was reconstructed with MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001). The tree was visualized by Treeview 1.6.6 (Page 1996).
Molecular clock estimates
PhyloBayes 3.3 was applied for molecular clock estimates (Lartillot et al. 2009). MrBayes consensus tree was used as input. We assumed the log-normal auto-correlated model (Thorne et al. 1998). Rates across sites were modelled assuming a discrete gamma distribution with four categories. Divergence time priors were either uniform or modelled with a birth death process. Rates across sites were modelled assuming a discrete gamma distribution with four categories. The calculations were run for 50,000 cycles.
The calibration of the tree was done essentially as described before (Scherbaum et al. 2010). Stratigraphic information was obtained from http://www.fossilrecord.net (Benton and Donoghue 2007; Briggs et al. 1993). Numerical ages derived from the “International Stratigraphic Chart 2009” (http://www.stratigraphy.org). Specifically, we assumed that the Ordovician phyllocarid fossils define the minimum age of Malacostraca 488 MYA, whereas the first crustacean fossils from the lower Cambrian period 542 MYA represent the maximum age (Benton and Donoghue 2007; Briggs et al. 1993; Wills 1997; Wills et al. 2009). The first appearance of the Hoplocarida defines the minimum of the split of Hoplocarida, Phyllocarida and Eumalacostraca about 375 MYA in the upper Devonian. Penaeid shrimps emerged in the fossil record more than 240 MYA. The branches leading to the Astacura on the one hand and the clade of Palinura (Achelata) plus Brachyura split at least 251 MYA, and Brachyura emerged more than 190 MYA.
Results
Haemocyanin in the hemolymph of C. multidentata and A. moluccensis
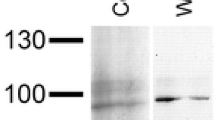
The total protein concentration in the hemolymph of both C. multidentata and A. moluccensis was 105 ± 20 mg/ml. The hemolymph proteins were separated by SDS-PAGE, applying 10 μg of protein per lane. Haemocyanin subunits were detected by Western blotting, employing various antisera against different crustacean and insect haemocyanins, as described in the “Materials and methods” section. A typical result is displayed in Fig. 1. In the Coomassie-stained polyacrylamide gel, two strong bands were visible in the range of 77 and 87 kDa, which agrees with the predicted mass of a typical haemocyanin subunit. In all cases, the anti-haemocyanin antisera detected these two bands. Thus haemocyanin is the most predominant protein in the hemolymph of both freshwater shrimps.
SDS-PAGE and western blot of the C. multidentata and the A. moluccensis hemolymph. Two very prominent bands are visible in the electrophoresis at 77 and 87 kDa. Both bands were detected with antibodies against the haemocyanin subunits of an insect, the cockroach Blaptica dubia (Bdu) and of a crustacea, the European crayfish Astacus astacus (Aas)
cDNA sequences of C. multidentata and A. moluccensis haemocyanin subunits
Total RNA was prepared from each of the shrimps and a reverse transcription reaction was carried out to obtain cDNA of C. multidentata and A. moluccensis, respectively. Partial cDNA sequences were obtained by different pairs of degenerated primers, which had been constructed against the conserved middle region of crustacean haemocyanins. 22 clones that derived from C. multidentata cDNA and that showed an insert of the expected length were sequenced. A BLAST search of their sequences confirmed their identity as haemocyanin. The sequences could be assigned to four distinct groups of three to seven sequences each. The sequences were assigned to one of the crustacean subunit types by a BLAST search and named CmuHcA1, CmuHcA2, CmuHcA3 and CmuHcC1, respectively. We used the abbreviations A, B and C to represent the three subunit types α, β and γ. In the case of A. moluccensis, 21 clones with inserts of the expected lengths were sequenced. The cDNA fragments were assembled to five groups of three to six sequences each and assigned to distinct subunit types as described, resulting in three α, one β and one γ subunit (AmoHcA1, AmoHcA2, AmoHcA3, AmoHcB1, and AmoHcC1).
To obtain the full-length sequences of the subunits, 5′ and 3′ RACE were performed. With the exception of CmuHcA3, AmoHcA2 and AmoHcA3, the full-length sequences were recovered. While for CmuHcA3 and AmoHcA3 neither RACE was successful, for AmoHcA2 only the 3′ end was found. The assembled haemocyanin subunits gave rise to full-length sequences of 2037-2325 base pairs (Table 1). The open reading frames (ORF) covered between 1,992 and 2,010 bp and code for 681 amino acids (CmuHcA1, CmuHcA2, AmoHcA1), 664 amino acids (AmoHcB1), or 670 amino acids (CmuHcC1, AmoHcC1). All sequences (including the partial ones) have been submitted to the EMBL Nucleotide Sequence Database with the accession numbers HE650707 to HE650715. After translation into amino acids, an N-terminal signal peptide required for export into the hemolymph, stretching 15–21 amino acids, was identified in all complete subunits with high probability (Table 1). Putative glycosylation sites are only found in CmuHcA2 at position 391 (NMT) and in CmuHcC1 at position 350 (NYS).
Analyses of the full-length sequences by BLAST showed that the α-type haemocyanins CmuHcA1, CmuHcA2, AmoHcA1 and the incomplete sequence AmoHcA2 display the highest similarity with the oriental river prawn Macrobrachium nipponense haemocyanin (MniHc, JF683437) with identities between 75 and 78 %. The β-type subunit AmoHcB1 shared 68 % identity with the freshwater crayfish Pacifastacus leniusculus haemocyanin (PleHc, AF522504), and the γ-type subunits CmuHcC1 and AmoHcC1 were to 70 % identical with the haemocyanin from the Chinese white shrimp Fenneropenaeus chinensis (FchHc1, FJ594414).
Comparison with selected haemocyanin subunits of other crustaceans showed that the histidines required for copper-binding are present in all identified sequences (Fig. 2). A multiple sequence alignment from 107 haemocyanin sequences of 43 arthropod species is shown in Supplemental Fig. 1. The pairwise identities and similarities between the three complete subunits from C. multidentata and A. moluccensis as well as one representative of each subunit type [Panulirus interruptus HcA (α-type), Cancer magister Hc1 (β-type) and P. interruptus HcC (γ-type subunit)] are given in Table 2. As expected, the putative α-type subunits of the fresh water shrimps show the highest identity to the P. interruptus α-subunit (63–65 %), the putative β-type subunit to the β-type subunit of C. magister (64 %) and the putative γ-type subunits to the P. interruptus γ-subunit (65–67 %) (Table 2).
Multiple sequence alignment of selected decapod haemocyanin sequences. The three complete haemocyanin subunits from C. multidentata (CmuHcA1, HE650712; CmuHcA2, HE650713; CmuHcC1, HE650715) and the three complete haemocyanin subunits from A. moluccensis (AmoHcA1, HE650707; AmoHcB1, HE650710; AmoHcC1, HE650711) are compared with one representative each of the α-type subunits (Panulirus interruptus, PinHcA, P04254) the β-type (Cancer magister, CmaHc1, AY861676), and the γ-type subunits (PinHcC, P80096). Strictly conserved regions are shaded grey and the conserved copper-binding histidines are shaded black. Potential signal regions and glycosylation sites are underlined and the secondary structure elements of PinHcA (Volbeda and Hol 1989) are given at the bottom
Identification of haemocyanin subunits of A. moluccensis by mass-spectrometry
To assign the two protein bands of the bamboo shrimp A. moluccensis found in SDS-PAGE (Fig. 1) to certain subunit types, nanoLC-ESI-ion trap analyses were performed. The upper band was identified as γ-subunit(s) and the lower band as α-subunit(s). In 2D gel electrophoreses of A. moluccensis hemolymph about 30 distinct spots in the mass range of the haemocyanin subunits were identified (Supplemental Fig. 2). Ten representative spots were selected and further analysed by nanoLC-ESI-ion trap mass-spectrometry. In all cases, the Mascot search results gave the highest scores (p < 0.05) with different haemocyanin subunits of A. moluccensis. They were either identified as α- or γ-subunits, corresponding to AmoHcA1-3 or AmoHcC1, respectively. No signature of any β-type subunit was found.
Phylogeny of crustacean haemocyanins
A phylogenetic tree was reconstructed from 107 haemocyanin sequences of 43 arthropod species (Supplemental Fig. 3). 48 haemocyanins derive from malacostracan crustaceans. The general arrangement of the clades agreed with recent studies on arthropod haemocyanin evolution (Ertas et al. 2009; Pick et al. 2009; Rehm et al. 2012). The malacostracan haemocyanins form a well-supported monophylum (1.0 Bayesian posterior probability), which is displayed in Fig. 3. Within this subtree, all deeper nodes received high support, as reflected by support values of 1.00. Only in some terminal branches were the support values lower. In agreement with previous studies, the malacostracan haemocyanin subunits can be classified into three distinct types α, β and γ. In addition, the haemocyanin subunits of the Peracarida (here, woodlice and amphipods) and the haemocyanin of the Nebalia kensleyi (Phyllocarida) each form separate clades. Among these five major clades, the β-type subunits branched first and are the sister group to all others. The next branch is formed by the haemocyanin of Nebalia kensleyi, followed by the peracarid haemocyanins, the α-type subunits and the γ-type subunits.
Simplified Bayesian phylogenetic tree from the amino acid sequences of 48 haemocyanin subunits of 22 crustacean taxa. The tree was rooted with haemocyanin sequences of insects, a remipede, myriapods and chelicerates (data not shown). Partial sequences were not included. The sequences from C. multidentata (Cmu) and A. moluccensis (Amo) are shaded grey. The Bayesian posterior probabilities all were 1.0, unless given otherwise. See Table 3 for the abbreviations of the species. Scale bar 0.1 PAM distance
In agreement with the BLAST results, the haemocyanin subunits of C. multidentata and A. moluccensis were assigned to the α-, the β- and the γ-type subunits, respectively. One partial and two complete subunits of C. multidentata belong to the α-type subunits; another one groups with the γ-types. Of the five subunits from A. moluccensis, one complete and two incomplete subunits are α-types, one is a γ-type and another one a β-type. Within the clades of subunits, the topology essentially follows the expected phylogeny of the Decapoda. However, we found an unexpected grouping within the γ-type subunits of the haemocyanins from the penaeid shrimps (Dendrobranchiata) and the true shrimps (Caridea), rendering the Pleocyemata paraphyletic. Because neither α- nor β-type haemocyanin subunits are known from the Penaeoidea, this topology could not be verified in the other clades.
A molecular clock of malacostracan haemocyanins
To estimate the timing of malacostracan haemocyanin subunits, we employed a Bayesian relaxed clock model, which was calibrated by known fossil dates (Fig. 4). We calculated that the malacostracan haemocyanins commenced to diversify into distinct subunit types ~515 MYA (494–539 MYA). The β-type haemocyanins of Hoplocarida and Eucarida (Decapoda) diverged 407 MYA (377–451 MYA). Leptostracan and eumalacostracan haemocyanin subunits (α and γ) separated 500 MYA (488–524 MYA), and peracarid and eucarid haemocyanins (subunits α and γ) split ~434 MYA (405–463 MYA). The eucarid subunit types α- and γ-type subunits diverged ~396 MYA (366–424 MYA). The divergence of the haemocyanins of penaeid and caridid shrimps, as estimated from the γ-subunits, was 270 MYA (217–310 MYA). In case that AmoHcA2 and CmuHcA1 are orthologs, the divergence of the genera Caridina and Atyopsis took place ~86 MYA.
Discussion
Both the Amano shrimp C. multidentata and the bamboo shrimp A. moluccensis are members of the decapod crustaceans of the family Atyidae, infraorder Caridea, and live in the Indo-Pacific region. These amphidromous animals migrate from the freshwater to the sea, or vice versa, not for the purpose of breeding, but as a regular event in life (Hamano and Hayashi 1992; Iwata et al. 2003).
The haemocyanin repertoire of the caridean shrimps
We have investigated for the first time the haemocyanins of the caridean shrimp on the molecular level and found mRNAs for at least four distinct haemocyanin subunits in C. multidentata and five in A. moluccensis (Table 1). Although not all cDNA sequences could be completed at their 5′ and 3′ ends, each of them was unequivocally allocated to one of the subunit types (α, β, or γ) known from the malacostracan crustaceans (Table 2; Fig. 3) (Markl 1986; Markl et al. 1986). Because of our PCR-based approach, we cannot state with confidence that this represents the full haemocyanin repertoire of these species. Thus the subunit composition 3/0/1 and 3/1/1 (α/β/γ) should be considered as the minimal inventory of C. multidentata and A. moluccensis, respectively.
Three different α-type and one γ-type subunits were found in both species. The high sequence divergence suggests that the different α-subunits do not represent alleles but distinct genes, which code for polypeptides that are different components of the native haemocyanin hexamer. In the hemolymph of A. moluccensis, we identified by 2D electrophoresis at least ~30 polypeptides in the range of a typical haemocyanin subunit, of which ten were assigned either to an α- or to a γ-subunit (Supplementary Fig. 2). This high diversity of subunit proteins was unexpected and—to the best of our knowledge—has not been found in any other haemocyanin. While we could rather exclude a high contribution of allelic variations (we used only hemolymph from a single specimen per analysis), it is unknown whether the diversity is due to a large number of unidentified gene copies or, which we consider more likely, results from posttranslational modifications (acylation, deamidation, phosphorylation, glycosylation, etc.). Because the patterns in 2D gels were the same with freshly prepared and stored hemolymph (data not shown), we consider protein degradation as an unlikely explanation for the high diversity.
The evolution of distinct α-type subunits does not follow the evolution of the caridean taxa. Rather distinct α-subunits commenced to diversify early in the evolution of the Caridea: subunit AmoHcA2 is more closely related to CmuHcA1 than to the AmoHcA1 subunit of A. moluccensis (Fig. 3). Likewise, the α-subunits of C. multidentata are paraphyletic in terms of α-subunits of A. moluccensis. Moreover, the molecular clock estimates suggest that distinct α-subunits within the Caridea emerged more than 200 MYA. Together, these results provide for a divergent role of these subunits, e.g. in the structure of the native haemocyanin.
The loss of one of the subunit types was frequently observed in decapod taxa (Markl et al. 1986). In fact, β-type haemocyanin subunits had been unknown in the suborder Caridea. Employing immunological methods, it has been demonstrated that the glass prawn Palaemon elegans (Palaemonoidea) harbours a 1 × 6 haemocyanin with only α- and γ-type subunits (Stöcker 1984; Markl et al. 1986). In addition, none of the expressed sequence tags (ESTs) of Caridea (Neocaridina denticulata and Macrobrachium nipponense), which are present in the databases, represent s a β-subunit, while multiple α- and γ-sequences were found. A similar pattern was derived here for C. multidentata. Therefore, the identification of an mRNA encoding a β-subunit in the bamboo shrimp A. moluccensis was surprising. This demonstrates that the corresponding gene is actually present in Caridea.
Notably, the β-subunit could not be identified in the haemocyanin protein of A. moluccensis using mass spectrometry. This is in accordance with the two major bands in SDS-PAGE (Fig. 1). We cannot rule out that a β-subunit is hidden in one of the spots of the 2D gel that we did not analyse by mass spectrometry. In any case, our results show that the β-subunit is not a major component of the native haemocyanin. This is in contrast to various other decapod haemocyanins, in which the β-subunit represents an essential component (Markl 1986; Markl et al. 1986; Stöcker et al. 1988; Durstewitz and Terwilliger 1997a, b; Scherbaum et al. 2010). It is unknown whether the expression levels of the β-subunit in the bamboo shrimp relate to certain environmental conditions or developmental stages.
It is unknown which environmental pressure led to the formation of the three different haemocyanin subunit types in the Decapoda. So far, it has not been possible to correlate the presence or absence of a particular type to certain conditions or demands of a habitat (Giomi and Beltramini 2007). However, functional adaptations of crustacean haemocyanins to different environmental conditions are well established (Terwilliger 1998), e.g., environmental salinity has an effect on the concentration of inorganic ions in the hemolymph (Mantel and Farmer 1983), which in turn affects the oxygen-binding characteristics of haemocyanin (Brown and Terwilliger 1998). Among others, in the Dungeness crab Cancer magister two haemocyanin subunits change during development (Durstewitz and Terwilliger 1997a, b). The juvenile crab haemocyanin differs from adult haemocyanin in its structure as well as its function (Brown and Terwilliger 1998, 1999; Terwilliger 1998). These environmental challenges might have led to the emergence of the large diversity in subunit composition of malacostracan haemocyanins, which is in sharp contrast to the low variation of spider haemocyanin (Burmester 2002; Rehm et al. 2012).
Implications for haemocyanin and decapod evolution
The phylogenetic tree derived from the malacostracan haemocyanins (Fig. 3; Supplemental Fig. 3) is in general accordance with previous studies (Scherbaum et al. 2010; Burmester 2002; Kusche et al. 2003; Hagner-Holler et al. 2005). The occurrence of three haemocyanin subunit types α, β and γ was confirmed, as well the separate branch of the peracarid haemocyanins. We also identified a common branch of haemocyanin subunits γ of the Penaeidae (prawns) and Caridea (true shrimps), thereby supporting the taxon “Natantia” (swimming decapods), as opposed to the Reptantia (crawling decapods). This classification has been originally proposed by Boas (1880), but more recent taxonomic work agrees that the Natantia are a paraphyletic assemblage (Burkenroad 1963). This has led to the erection of the superorder Dendrobranchiata (prawns) and Pleocyemata (all remaining decapods, including the Caridea). While the monophyly of the Pleocyemata received wide support by morphological studies, molecular approaches did not gave conclusive results, supporting either topology (Porter et al. 2005; Crandall et al. 2009; Liu and Cui 2011). We must also note that a single haemocyanin subunit type is not sufficient for re-erecting the taxon Natantia. Unfortunately, the haemocyanins of the Penaeidae apparently only consist of γ-subunits and the topology cannot be confirmed with the α- and β-subunits.
Although we applied a different method than that used for molecular clock estimates, the calculated divergence of the crustacean haemocyanin subunits led to similar ages as before (Scherbaum et al. 2010). These dates largely agree with the fossil record (Benton and Donoghue 2007; Briggs et al. 1993) and previous molecular clock calculations (Crandall et al. 2009). However, the phylogenetic tree of the malacostracan haemocyanins is complex and far from being complete. Hidden paralogy may mask the true divergence times and may also lead to wrong calibrations. Additional sequences, which may derive from genome and EST (expressed sequence tags) projects, may provide better knowledge on the true diversity of crustacean haemocyanins. This will allow obtaining a detailed view on the evolution of the subunit diversity in this taxon, as it has been derived for the Chelicerata (Rehm et al. 2012).
Abbreviations
- Hc:
-
Haemocyanin
- MYA:
-
Million years ago
- PCR:
-
Polymerase chain reaction
- SDS-PAGE:
-
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- Cmu:
-
Caridina multidentata
- Amo:
-
Atyopsis moluccensis
References
Benton MJ, Donoghue PC (2007) Paleontological evidence to date the tree of life. Mol Biol Evol 24:26–53
Boas JEV (1880) Studier over Decapodernes Slægtskabsforhold. Kongelige Danske Videnskabernes Selskabs Skrifter, 6 Række. Naturvidenskabelig og Mathematisk Afdeling 1(2):25–210
Bradford M (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye-binding. Anal Biochem 72:248–254
Briggs DEG, Weedon MJ, White MA (1993) Arthropoda (Crustacea excluding Ostracoda). In: Benton MJ (ed) The fossil record 2. Chapman and Hall, London, pp 321–342
Brown A, Terwilliger N (1998) Ontogeny of hemocyanin function in the Dungeness crab Cancer magister: hemolymph modulation of hemocyanin oxygen-binding. J Exp Biol 201:819–826
Brown AC, Terwilliger N (1999) Developmental changes in oxygen uptake in Cancer magister (Dana) in response to changes in salinity and temperature. J Exp Mar Biol Ecol 241:179–192
Burkenroad MD (1963) The evolution of the Eucarida (Crustacea, Eumalacostraca), in relation to the fossil record. Tulane Stud Geol 2:2–17
Burmester T (1999) Identification, molecular cloning, and phylogenetic analysis of a non-respiratory pseudo-hemocyanin of Homarus americanus. J Biol Chem 274:13217–13222
Burmester T (2001) Molecular evolution of the arthropod hemocyanin superfamily. Mol Biol Evol 18:184–195
Burmester T (2002) Origin and evolution of arthropod hemocyanins and related proteins. J Comp Physiol B 172:95–107
Crandall KA, Porter ML, Pérez-Losada M (2009) Crabs, shrimps, and lobsters (Decapoda). In: Hedges SB, Kumar S (eds) The timetree of life. Oxford University Press, Oxford, pp 293–297
De Haan W (1833–1850) Crustacea. In: von Siebold PF (ed) Fauna Japonica sive Descriptio Animalium, quae in Itinere per Japoniam, Jussu et Auspiciis Superiorum, qui Summum in India Batava Imperium Tenent, Suspecto, Annis 1823-1830 Collegit, Notis, Observationibus et Adumbrationibus Illustravit: i-xxxi, ix-xvi, 1–243, plates A–J, L–Q, 1–55. Lugduni-Batavorum
Decker H, Hellmann N, Jaenicke E, Lieb B, Meissner U, Markl J (2007) Minireview: recent progress in hemocyanin research. Integr Comp Biol 47:631–644
Durstewitz G, Terwilliger NB (1997a) Developmental changes in hemocyanin expression in the Dungeness crab, Cancer magister. J Biol Chem 272:4347–4350
Durstewitz G, Terwilliger NB (1997b) cDNA cloning of a developmentally regulated hemocyanin subunit in the crustacean Cancer magister and phylogenetic analysis of the hemocyanin gene family. Mol Biol Evol 14:266–276
Ertas B, von Reumont BJ, Wägele JW, Misof B, Burmester T (2009) Hemocyanin suggests a close relationship of Remipedia and Hexapoda. Mol Biol Evol 26:2711–2718
Gaykema WPJ, Hol WGJ, Vereifken JM, Soeter NM, Bak HJ, Beintema JJ (1984) 3.2 Å structure of the copper-containing oxygen-carrying protein Panulirus interruptus hemocyanin. Nature 309:23–29
Giomi F, Beltramini M (2007) The molecular heterogeneity of hemocyanin: its role in the adaptive plasticity of Crustacea. Gene 398:192–201
Hagner-Holler S, Kusche K, Hembach A, Burmester T (2005) Biochemical and molecular characterisation of hemocyanin from the amphipod Gammarus roeseli: complex pattern of hemocyanin subunit evolution in Crustacea. J Comp Physiol B 175:445–452
Hamano T, Hayashi KI (1992) Ecology of an atyid shrimp C. multidentata (de Man, 1982). Crustac Carcinol Soc Japan 21:1–13
Herberts C, de Frescheville J (1981) Occurrence of hemocyanin in the rhizicephalan crustacea Sacculina carcini Thompson. Comp Biochem Physiol B 70:657–659
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Iwata T, Inoue M, Nakano S, Miyasaka H, Doi A, Covich AP (2003) Shrimp abundance and habitat relationships in tropical rain-forest streams, Sarawak, Borneo. J Tropic Ecol 19:387–395
Katoh K, Kumal K, Tohl H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518
Kusche K, Ruhberg H, Burmester T (2002) A hemocyanin from the Onychophora and the emergence of respiratory proteins. Proc Natl Acad Sci USA 99:10545–10548
Kusche K, Hembach A, Milke C, Burmester T (2003) Molecular characterisation and evolution of the hemocyanin from the European spiny lobster, Palinurus elephas. J Comp Physiol B 173:319–325
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lartillot N, Lepage T, Blanquart S (2009) PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288
Linzen B, Soeter NM, Riggs AF, Schneider HJ, Schartau W, Moore MD, Yokata E, Behrens PQ, Nakashima H, Takagi T, Nemoto T, Vereijken JM, Bak HJ, Beintema JJ, Volbeda A, Gaykema WPJ, Hol WGJ (1985) The structure of arthropod hemocyanins. Science 229:519–524
Liu Y, Cui Z (2011) Complete mitochondrial genome of the Chinese spiny lobster Panulirus stimpsoni (Crustacea: Decapoda): genome characterization and phylogenetic considerations. Mol Biol Rep 38:403–410
Mangum CP (1983) Oxygen transport in blood. In: Mantel LH (ed) The biology of the Crustacea. Academic Press, New York, pp 373–429
Mangum CP (1985) Oxygen transport in invertebrates. Am J Physiol 248:505–514
Mangum CP, Joy PJ (1997) Hemocyanin subunit composition in the American lobster Homarus americanus. J Crust Biol 17:1–5
Mantel LH, Farmer LL (1983) Osmotic and ionic regulation. In: Mantel LH (ed) The biology of Crustacea, vol 5., Internal Anatomy and Physiological Regulation Academic Press, New York, pp 53–161
Markl J (1986) Evolution and function of structurally diverse subunits in the respiratory protein hemocyanin from arthropods. Biol Bull 171:90–115
Markl J, Decker H (1992) Molecular structure of the arthropod hemocyanins. In: Mangum CP (ed) Advances in comparative and environmental physiology, vol 13., Blood and Tissue Oxygen Carriers Springer, Heidelberg, pp 325–376
Markl J, Stocker W, Runzler R, Precht E (1986) Immunological correspondences between the hemocyanin subunits of 86 arthropods: evolution of a multigene protein family. In: Linzen B (ed) Invertebrate oxygen carriers. Springer, Heidelberg, pp 281–292
Mičetić I, Losasso C, Di Muro P, Tognon G, Benedetti P, Beltramini M (2010) Solution structures of 2 x 6-meric and 4 x 6-meric hemocyanins of crustaceans Carcinus aestuarii, Squilla mantis and Upogebia pusilla. J Struct Biol 171:1–10
Miller KI, Neal WE, Fumio A, Van Holde KE (1976) Structure and function of hemocyanin from thalassinid Shrimp. J Comp Physiol 115:171–184
Nicholas KB, Nicholas HBJ, Deerfield DWI (1997) GeneDoc: analysis and visualization of genetic variation. EMBnet NEWS 4:14
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Paoli M, Giomi F, Hellmann N, Jaenicke E, Decker H, Di Muro P, Beltramini M (2007) The molecular heterogeneity of hemocyanin: structural and functional properties of the 4 × 6-meric protein of Upogebia pusilla (Crustacea). Gene 398:177–182
Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 18:3551–3567
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Pick C, Schneuer M, Burmester T (2009) The occurrence of hemocyanin in Hexapoda. FEBS J 276:1930–1941
Pick C, Schneuer M, Burmester T (2010) Ontogeny of hemocyanin in the ovoviviparus cockroach Blaptica dubia suggests an embryo-specific role in oxygen supply. J Insect Physiol 56:455–460
Porter ML, Pérez-Losada M, Crandall KA (2005) Model-based multi-locus estimation of decapod phylogeny and divergence times. Mol Phylogenet Evol 37:355–369
Rehm P, Pick C, Borner J, Markl J, Burmester T (2012) The diversity and evolution of chelicerate hemocyanins. BMC Evol Biol 12:19
Scherbaum S, Beyhan E, Gebauer W, Burmester T (2010) Characterization of hemocyanin from the peacock mantis shrimp Odontodactylus scyllarus (Malacostraca: Hoplocarida). J Comp Physiol B 180:1235–1245
Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860
Stimpson W (1860) Prodromus descriptionis animalium evertebratorum, quae in Expeditione ad Oceanum Pacificum Septemtrionalem, a Republica Federata missa, C. Ringgold et J. Rodgers, observavit et descriptist. Proc Acad Nat Sci Phila 1860 (January):22–47
Stöcker W (1984) Immunologische Verwandtschaft und Quartärstruktur von Crustaceen-Hämocyaninen. University of München, Dissertation
Stöcker W, Raeder U, Bijlholt MCM, Wichertjes T, Bruggen EFJ, Markl J (1988) The quaternary structure of four crustacean two-hexameric hemocyanins: immunocorrelation, stoichiometry, reassembly and topology of individual subunits. J Comp Physiol B 158:271–289
Terwilliger NB (1998) Functional adaptations of oxygen-transport proteins. J Exp Biol 201:1085–1098
Terwilliger NB, Lawrence D, Ryan M (1999) Cryptocyanin, a crustacean molting protein: evolutionary link with arthropod hemocyanins and insect hexamerins. Proc Natl Acad Sci USA 96:2013–2018
Thorne JL, Kishino H, Painter IS (1998) Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol 15:1647–1657
Van Holde KE, Miller KI (1995) Hemocyanins. Adv Prot Chem 47:1–81
Volbeda A, Hol WGJ (1989) Crystal structure of hexameric hemocyanin from Panulirus interruptus refined at 3.2 Å resolution. J Mol Biol 209:249–279
Wills MA (1997) A phylogeny of recent and fossil Crustacea derived from morphological characters. In: Fortey RA, Thomas RH (eds) Arthropod relationships. Chapman and Hall, London, pp 189–209
Wills MA, Jenner RA, Ni Dhubhghaill C (2009) Eumalacostracan evolution: Conflict between three sources of data. Arthropod Syst Phylog 67:71–90
Acknowledgments
We thank Janus Borner for providing access to the program Primerlyze, which helps designing highly specific primers. We thank Peter Rehm for his help with PhyloBayes. This work was supported in part by the Deutsche Forschungsgemeinschaft (Bu 956/9).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Heldmaier.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Marxen, J.C., Pick, C., Kwiatkowski, M. et al. Molecular characterization and evolution of haemocyanin from the two freshwater shrimps Caridina multidentata (Stimpson, 1860) and Atyopsis moluccensis (De Haan, 1849). J Comp Physiol B 183, 613–624 (2013). https://doi.org/10.1007/s00360-013-0740-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00360-013-0740-9